枇杷(Eriobotrya japonica Lindl.)属蔷薇科(Rosaceae)苹果亚科(Maloideae)枇杷属(Eriobotrya)植物,是原产于中国的一种常绿果树,秋萌冬花,春末夏初成熟,也是一种重要的亚热带水果。枇杷果可供食用,有轻微的甜酸味,富含维生素A、维生素B6、钾、镁和膳食纤维;叶可入药,有化痰止咳之效。因其富含营养和药用特性而受到高度赞赏,是食药两用水果[1-2]。枇杷在中国中南部、日本、巴基斯坦、印度和韩国地区种植最为广泛,而中国是主要生产国和出口国,产量占全球的80%以上[3-4]。近年来,随着枇杷栽培面积的不断扩大,枇杷病害的发生也越发严重,尤其是叶部病害,种类繁多,常见的有灰斑病、斑点病、角斑病、炭疽病等10多种[5]。其中,枇杷灰斑病是由拟盘多毛孢属(Pestalotiopsis)引起的真菌性病害[6],在叶部常见,还会感染枇杷的花朵和果实,进而造成经济损失[7]。因此,开展灰斑病抗性机制研究,筛选抗灰斑病的枇杷种质,是枇杷产业发展的重要保障。灰斑病抗性作为枇杷遗传育种中的重要抗性指标之一,一直受到人们的重视。李金萍等[8]调查北京地区主要栽培的南方果树的病害发病情况,在确诊的11种病害中,由枇杷拟盘多毛孢(Pestalotiopsis eriobotrifolia)引起的灰斑病危害最为严重。该病害一年多次侵染,每个时期都会发生,尤其危害叶片,导致叶片僵化变小、易碎,造成早期大量落叶,影响新梢生长;还能危害花和果实,引起花果的腐烂,最终导致枇杷产量严重下滑[9]。有关灰斑病的发病规律以及防治方法,前人开展了大量研究,在苹果[10]、柑橘[11-12]、杧果[13]等研究中发现植株的形态结构特征,如叶片形态、气孔形态、结构和密度、叶片表皮覆盖物、叶片解剖结构等与抗病性相关。陈依丽等[14]在枇杷叶片显微结构、气孔形态学指数与抗病性的相关性方面进行了一定研究,为枇杷抗病种质资源创新利用和抗病品种早期选择提供了参考,但具体的抗病机制尚不清楚。
转录组测序技术(RNA-Seq)因高通量、高准确性、覆盖度广、可独立分析及价格便宜等优点在多种植物中广泛应用,是当前挖掘抗病相关基因的重要技术之一[15]。挖掘优异基因是加快改良品种选育进程的基础[16]。目前,RNA-Seq技术己应用在草莓[17]、柑橘[18]、猕猴桃[19]和葡萄[20]等果树研究中,但在枇杷上的相关研究较少。因此,笔者利用RNA-Seq技术研究枇杷与灰斑病病原菌互作时基因表达水平的变化,进而挖掘抗性相关基因。研究结果将为阐明枇杷抗、感病品种灰斑病抗性差异的分子机制提供新视角,对枇杷产业健康发展和抗病育种具有重要意义。
1 材料和方法
1.1 试验材料
试验材料均采自广东省广州市华南农业大学枇杷种质资源圃,树龄5~10 a(年),生长期内采用常规管理。52份供试枇杷种质包括15个枇杷野生种(变种)和37个普通枇杷资源,种质资源详情见表1和表2。
表1 枇杷野生种(变种)信息表
Table 1 Information sheet for wild varieties of E. japonica
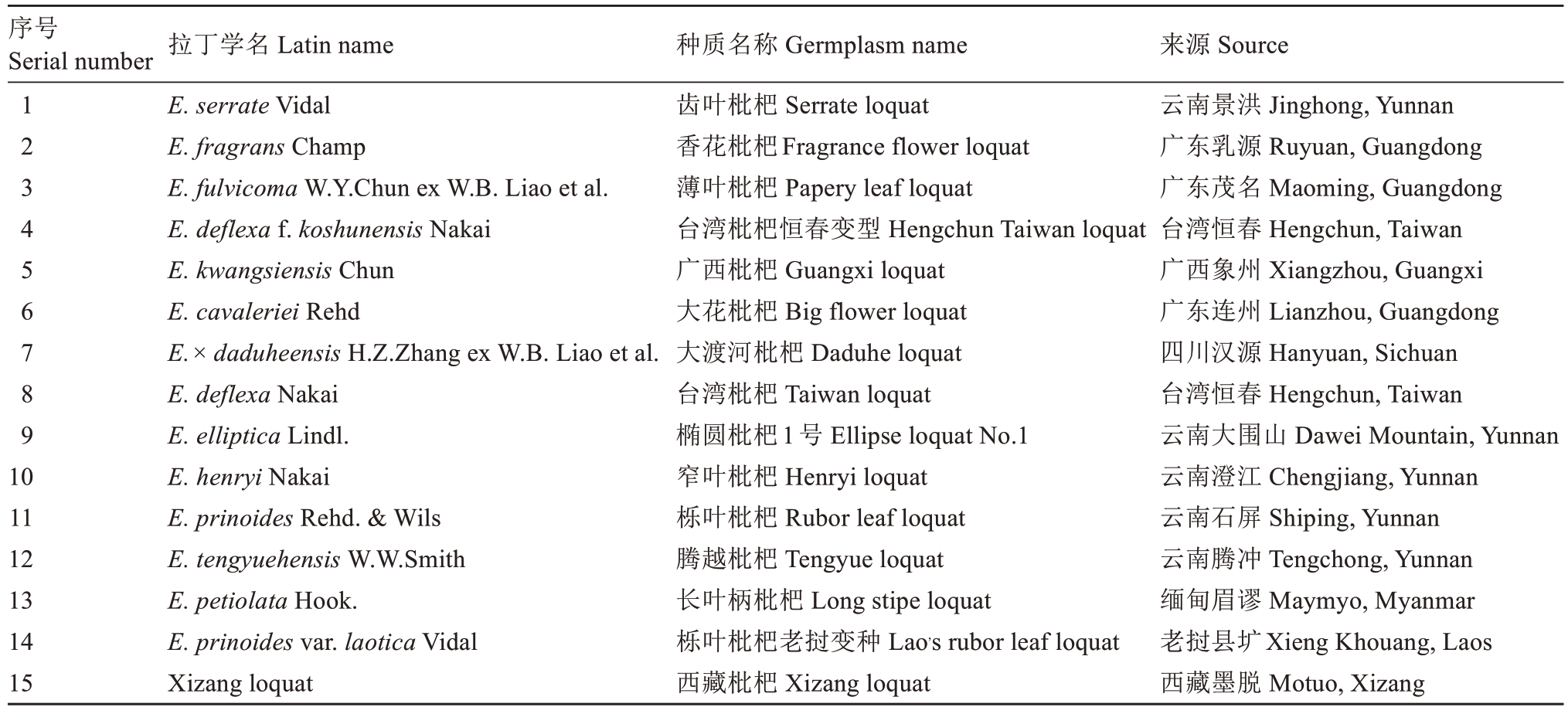
序号Serial number 123456789 10 11 12 13 14 15拉丁学名 Latin name E. serrate Vidal E. fragrans Champ E. fulvicoma W.Y.Chun ex W.B. Liao et al.E. deflexa f. koshunensis Nakai E. kwangsiensis Chun E. cavaleriei Rehd E.× daduheensis H.Z.Zhang ex W.B. Liao et al.E. deflexa Nakai E. elliptica Lindl.E. henryi Nakai E. prinoides Rehd. & Wils E. tengyuehensis W.W.Smith E. petiolata Hook.E. prinoides var. laotica Vidal Xizang loquat种质名称 Germplasm name齿叶枇杷 Serrate loquat香花枇杷Fragrance flower loquat薄叶枇杷 Papery leaf loquat台湾枇杷恒春变型 Hengchun Taiwan loquat广西枇杷 Guangxi loquat大花枇杷 Big flower loquat大渡河枇杷 Daduhe loquat台湾枇杷 Taiwan loquat椭圆枇杷1号 Ellipse loquat No.1窄叶枇杷 Henryi loquat栎叶枇杷 Rubor leaf loquat腾越枇杷 Tengyue loquat长叶柄枇杷 Long stipe loquat栎叶枇杷老挝变种 Lao,s rubor leaf loquat西藏枇杷 Xizang loquat来源 Source云南景洪 Jinghong, Yunnan广东乳源 Ruyuan, Guangdong广东茂名 Maoming, Guangdong台湾恒春 Hengchun, Taiwan广西象州 Xiangzhou, Guangxi广东连州 Lianzhou, Guangdong四川汉源 Hanyuan, Sichuan台湾恒春 Hengchun, Taiwan云南大围山 Dawei Mountain, Yunnan云南澄江 Chengjiang, Yunnan云南石屏 Shiping, Yunnan云南腾冲 Tengchong, Yunnan缅甸眉谬 Maymyo, Myanmar老挝县圹Xieng Khouang, Laos西藏墨脱 Motuo, Xizang
表2 普通枇杷种质信息表
Table 2 Germplasm information sheet of common E. japonica
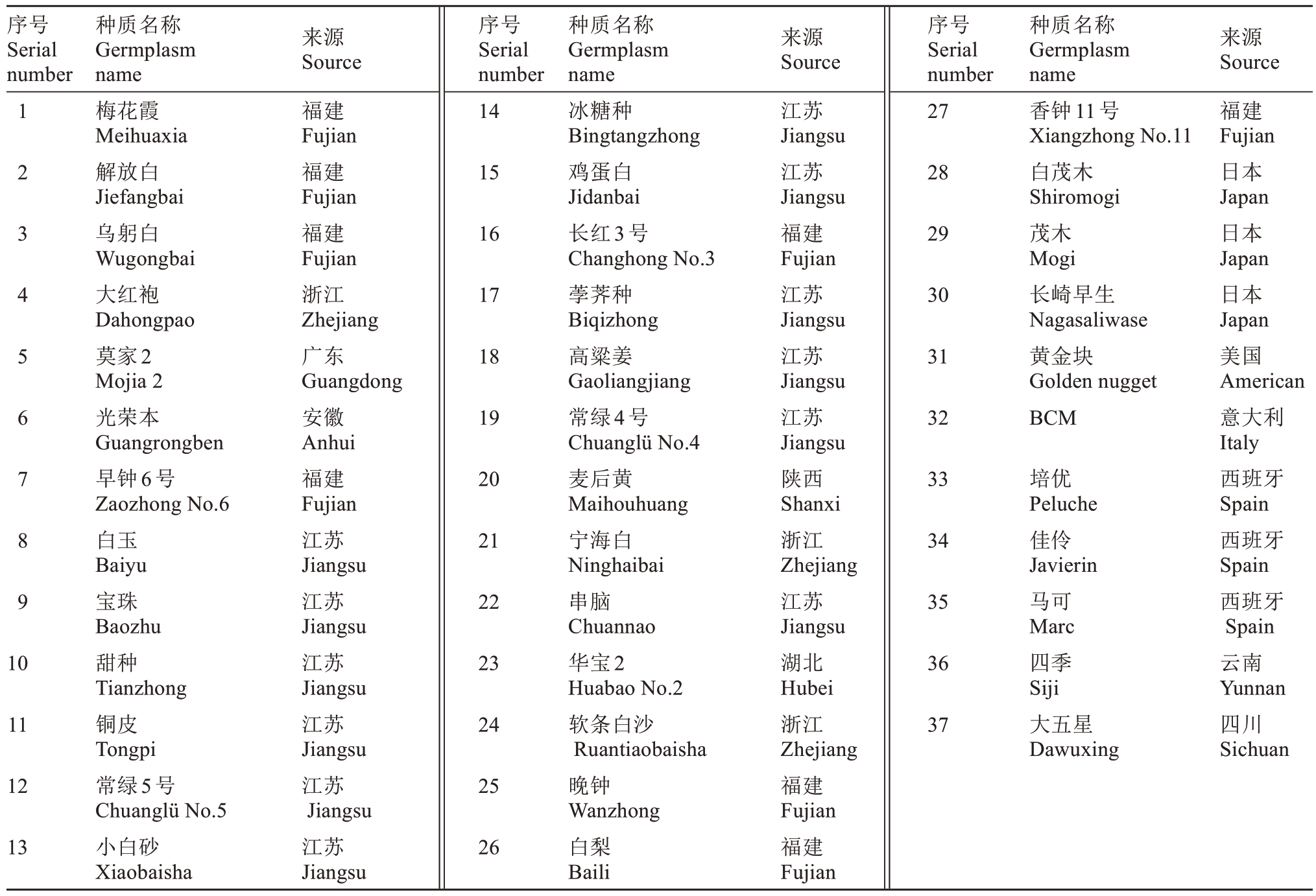
序号Serial number来源Source来源Source来源Source 123456789序号Serial number 14序号Serial number 27种质名称Germplasm name梅花霞Meihuaxia解放白Jiefangbai乌躬白Wugongbai大红袍Dahongpao莫家2 Mojia 2光荣本Guangrongben早钟6号Zaozhong No.6白玉Baiyu宝珠Baozhu甜种Tianzhong铜皮Tongpi常绿5号Chuanglü No.5小白砂Xiaobaisha福建Fujian福建Fujian福建Fujian浙江Zhejiang广东Guangdong安徽Anhui福建Fujian江苏Jiangsu江苏Jiangsu江苏Jiangsu江苏Jiangsu江苏Jiangsu江苏Jiangsu 1528 1629 1730 1831 1932种质名称Germplasm name香钟11号Xiangzhong No.11白茂木Shiromogi茂木Mogi长崎早生Nagasaliwase黄金块Golden nugget BCM 2033 2134 2235 102336 112437培优Peluche佳伶Javierin马可Marc四季Siji大五星Dawuxing福建Fujian日本Japan日本Japan日本Japan美国American意大利Italy西班牙Spain西班牙Spain西班牙Spain云南Yunnan四川Sichuan 1225 1326种质名称Germplasm name冰糖种Bingtangzhong鸡蛋白Jidanbai长红3号Changhong No.3荸荠种Biqizhong高粱姜Gaoliangjiang常绿4号Chuanglü No.4麦后黄Maihouhuang宁海白Ninghaibai串脑Chuannao华宝2 Huabao No.2软条白沙Ruantiaobaisha晚钟Wanzhong白梨Baili江苏Jiangsu江苏Jiangsu福建Fujian江苏Jiangsu江苏Jiangsu江苏Jiangsu陕西Shanxi浙江Zhejiang江苏Jiangsu湖北Hubei浙江Zhejiang福建Fujian福建Fujian
1.2 枇杷灰斑病田间调查评价和室内接种鉴定
本研究于2019年10月至2021年10月,开展连续2年的田间调查评价和室内接种鉴定。其中田间调查评价按照《枇杷种质资源描述规范和数据标准》[21]的分级标准,采用目测法和Photoshop软件法[22]调查上述52份枇杷种质的灰斑病发病情况。枇杷灰斑病的病症识别参照《枇杷病虫害诊治图谱》[23],每株按照东南西北中5个方向取样,每个方向从上至下调查10枚叶片,抗病性通过病情指数评价,病情指数=100×[Σ(各级发病级别×相应发病级别的叶片数)] ÷(5×调查总叶片数)。室内接种鉴定采用针刺法,具体操作方法参考文献[3] 。
根据抗性评价结果选取高抗种质白梨和高感种质光荣本接种灰斑病病原菌,具体接种方法参考周蓓[9]的报道。根据前期接种1、1.5、2、3、6、12、24、36、48、72、96、120 h的试验结果,发现在36 h时,白梨与光荣本的超氧阴离子自由基和过氧化氢含量的变化存在显著差异,因此在接种后33、36、48 h采样,每个处理设置3次重复。用无菌水冲洗接种叶片后,避开叶脉取样,对照样品和试验处理样品经液氮速冻后放入-80 ℃冰箱保存。
1.3 枇杷灰斑病病原菌的分离纯化及分子生物学鉴定
采用组织分离法对部分种质进行病菌分离,取大小约5 mm的小块感病组织于PDA培养基上培养,待产生分生孢子后,采用稀释纯化法进行单孢纯化。采用2×T5 Direct PCR Kit(Plant)试剂盒提取病原菌的DNA。利用Bryan等[24]报道的真菌通用引物ITS1(TCCGTAGGTGAACCTGCGC)和ITS4(TCCTCCGCTTATTGATATGC)进行引物合成,然后参照周蓓[9]的方法进行PCR扩增。PCR反应体系为25 μL,其中包含12.5 μL的2×T5 Direct PCR Mix(Plant)、1 μL引物ITS1(10 μmol·L-1)、1 μL引物ITS4(10 μmol·L-1)、1 μL模板 DNA、9.5 μL ddH2O。
1.4 RNA提取、文库构建及测序
总RNA提取采用快速通用植物RNA提取试剂盒(华越洋生物)。转录组测序由北京百迈客生物科技有限公司完成。测序数据分析及差异表达基因鉴定方法:使用FastQC v0.11.9软件对所有测序样品的原始测序数据进行质量评估,并使用fastp软件[25]过滤原始测序数据中的低质量碱基以及一些接头序列,获得干净数据;使用HISAT2软件[26]将每个文库的干净读数映射到七星枇杷基因组[27]中,使用String Tie软件[28]对这些映射结果文件进行分析,输出每个基因的counts值以及TPM(Transcripts per million)值。
1.5 差异表达基因筛选
在R中使用DESeq2包[29],以每个基因的counts值为输入数据,以|log2FoldChange|≥1和P-value≤0.01为筛选条件,输出差异表达基因。使用R对样本表达量数据进行Spearman相关性分析,使用TBtools软件[30]绘制venn图、upset图和基因表达量热图。
1.6 差异表达基因GO和KEGG富集分析
GO(Gene Ontology)和KEGG(Kyoto Encyclopedia of Genes and Genomes)富集分析:使用Egg-NOG-mapper v2[31]对七星枇杷基因组所有蛋白序列进行GO注释,以此为富集背景,使用TBtools软件中的GO Enrichment程序对差异表达基因集进行GO富集分析。利用KOBAS v3.0[32]的注释功能以苹果为参照物种对七星枇杷基因组所有蛋白序列进行KEGG注释,并利用富集功能对差异表达基因集进行KEGG富集分析,对KEGG富集分析结果使用R中的ggplot2 v3.3.5包进行气泡图的绘制。
1.7 实时荧光定量验证
为验证RNA-seq技术所获得的差异表达基因的可靠性,在抗、感种质的处理组和对照组的差异基因中,选取7个差异基因,利用qRT-PCR试验检测7个差异基因在接种前后的表达变化情况。使用primer 3 plus网站[33]设计qPCR引物,并通过溶解曲线分析和PCR产物测序来确认引物特异性。使用LightCyclerR480罗氏荧光定量PCR仪(Roche,USA)和iTaqTM universal SYBR® Green SuperMix(Bio-Rad)(Bio-Rad,USA)荧光定量预混液,进行qPCR试验。以枇杷β-actin作为内参基因,试验结果采用2-△△CT法[34]计算(引物详见表3)。qPCR反应体系10 μL,具体方法参照iTaqTM universal SYBR® Green SuperMix荧光定量预混液说明书。
表3 qPCR引物列表
Table 3 Primer list of qPCR

引物名称Primers name β-actin EVM0022518 EVM0039675 EVM0014520 EVM0020330 EVM0006863 EVM0022064 EVM0024983正向引物Forward primer GGATTTGCTGGTGATGATGC GTCTCTCCCTCGTCTCAGGT GAACCCGTGTCTCTCATCCC GCTGCCCCGTCAAAAAGAAG AAGAGCATGGTGGTGGGATG TACGCGATCCACACAAAGCT GCACTAACAACCAGACCCCA ACAGGGATGGACTTCTTGCG反向引物Reverse primer CCGTGCTCAATGGGATACTT GGAGCTGCTACCTCATGTGG TAGCCATATACGCGCACCTC ATCCTATGGTCATGCAGCGG CCAAAAGCCGTTGCAAGTGA GAGTTGGTGGTGGTGGTTGA CCTCGCAGTCCCTTGATCTC AGGCAACACATCTACGTCGG
2 结果与分析
2.1 抗性评价
根据2年的平均病情指数,对52份枇杷种质资源的抗病性进行了聚类分析,共划分为5类(表4)。第1类表现为高抗(HR),包含香花枇杷、齿叶枇杷、白梨等8份资源,占调查总数的15.4%;第2类表现为抗病(R),包含薄叶枇杷、台湾枇杷恒春变型、茂木、白茂木等14份资源,占调查总数的26.9%;第3类表现为中抗(MR),包含栎叶枇杷、白玉、窄叶枇杷等17份资源,种质资源最多,占调查总数的32.7%;第4类表现为感病(S),包含广西枇杷、大渡河枇杷、大红袍等10份资源,占调查总数的19.2%;第5类表现为高感(HS),数目最少,仅有大花枇杷、大五星和光荣本3份资源,占调查总数的5.8%。结合室内接种鉴定结果,得到11份抗性评价一致的种质资源,其中包括高抗的齿叶枇杷、香花枇杷、白梨;抗病的薄叶枇杷、台湾枇杷恒春变型、茂木、白茂木;感病的广西枇杷、大红袍;高感的光荣本、大花枇杷。
表4 52份枇杷种质资源对灰斑病的抗病性评价
Table 4 Evaluation of the resistance of 52 loquat germplasm resources to gray spot disease
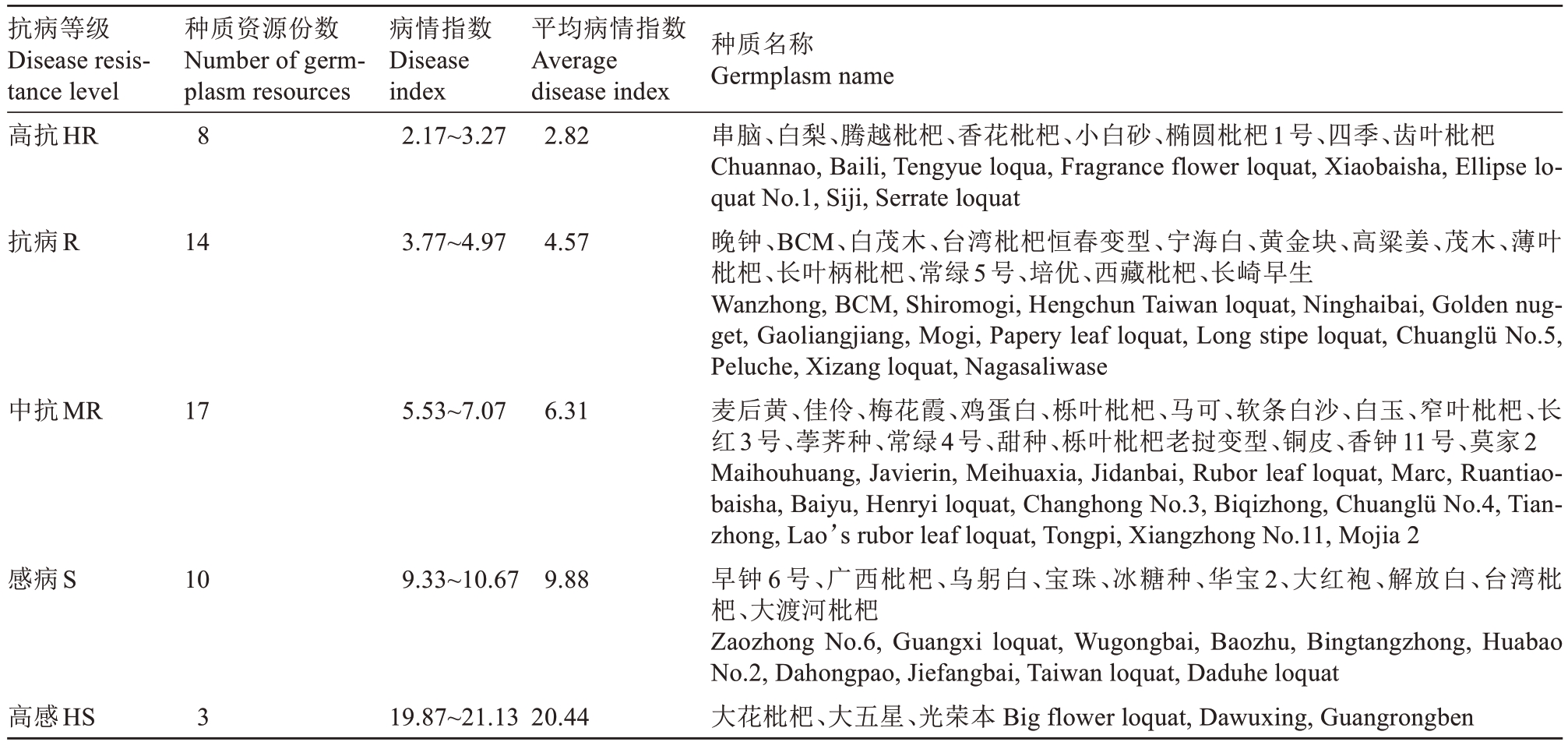
抗病等级Disease resistance level高抗HR种质资源份数Number of germplasm resources 8病情指数Disease index 2.17~3.27平均病情指数Average disease index 2.82抗病R 143.77~4.974.57中抗MR 175.53~7.076.31感病S 109.33~10.679.88高感HS 319.87~21.1320.44种质名称Germplasm name串脑、白梨、腾越枇杷、香花枇杷、小白砂、椭圆枇杷1号、四季、齿叶枇杷Chuannao, Baili, Tengyue loqua, Fragrance flower loquat, Xiaobaisha, Ellipse loquat No.1, Siji, Serrate loquat晚钟、BCM、白茂木、台湾枇杷恒春变型、宁海白、黄金块、高粱姜、茂木、薄叶枇杷、长叶柄枇杷、常绿5号、培优、西藏枇杷、长崎早生Wanzhong, BCM, Shiromogi, Hengchun Taiwan loquat, Ninghaibai, Golden nugget, Gaoliangjiang, Mogi, Papery leaf loquat, Long stipe loquat, Chuanglü No.5,Peluche, Xizang loquat, Nagasaliwase麦后黄、佳伶、梅花霞、鸡蛋白、栎叶枇杷、马可、软条白沙、白玉、窄叶枇杷、长红3号、荸荠种、常绿4号、甜种、栎叶枇杷老挝变型、铜皮、香钟11号、莫家2 Maihouhuang, Javierin, Meihuaxia, Jidanbai, Rubor leaf loquat, Marc, Ruantiaobaisha, Baiyu, Henryi loquat, Changhong No.3, Biqizhong, Chuanglü No.4, Tianzhong, Lao’s rubor leaf loquat, Tongpi, Xiangzhong No.11, Mojia 2早钟6号、广西枇杷、乌躬白、宝珠、冰糖种、华宝2、大红袍、解放白、台湾枇杷、大渡河枇杷Zaozhong No.6, Guangxi loquat, Wugongbai, Baozhu, Bingtangzhong, Huabao No.2, Dahongpao, Jiefangbai, Taiwan loquat, Daduhe loquat大花枇杷、大五星、光荣本 Big flower loquat, Dawuxing, Guangrongben
2.2 病原菌的分离纯化及鉴定
2.2.1 病原菌的分离纯化 在枇杷灰斑病病叶上分离得到4个不同的菌株D-2、D-3、D-4和D-5,但致病性测定试验显示均能引起枇杷叶片全部发病且与田间发病症状相同(图1),说明D-2、D-3、D-4和D-5可能是同一种菌。初期产生褐色圆形小病斑,而后转为灰白色,表皮干枯,容易与下部组织脱离,多数病斑可形成不规则形状的大病斑。从发病部位重新分离出的病原菌与接种的病菌形态一致,表明分离出的菌株确实为枇杷灰斑病致病菌。从中筛选出致病性较强的菌株,并将此病原菌命名为D-4,其形态学特征如图1-A~D所示。培养数天后菌丝表面产生黑色小点即分生孢子盘(图1-A),菌株D-4的分生孢子呈直或稍弯曲的纺锤形,顶端有2~3根附属丝,基细胞具有1根中生式柄(图1-B),在PDA培养基上,菌落正面呈现纯白色、绒毛状(图1-C),背面呈现橘黄色(图1-D)。

图1 田间发病症状及回接症状(左),菌株D-4的形态学特征(右)
Fig. 1 Symptoms of field onset and reinfection (left), morphological characteristics of strain D-4 (right)
2.2.2 病原菌的分子生物学鉴定 电泳结果显示(图2),PCR产物片段长度约500 bp。将测序结果在NCBI数据库上进行BLAST同源性比对,发现D-4菌株与小孢拟盘多毛孢菌Pestalotiopsis microspora(KU720061)的同源性高达97%,其片段长度为520 bp,与电泳结果基本相符。综合形态学特征和分子生物学鉴定,可推断分离的病原菌D-4为拟盘多毛孢菌。
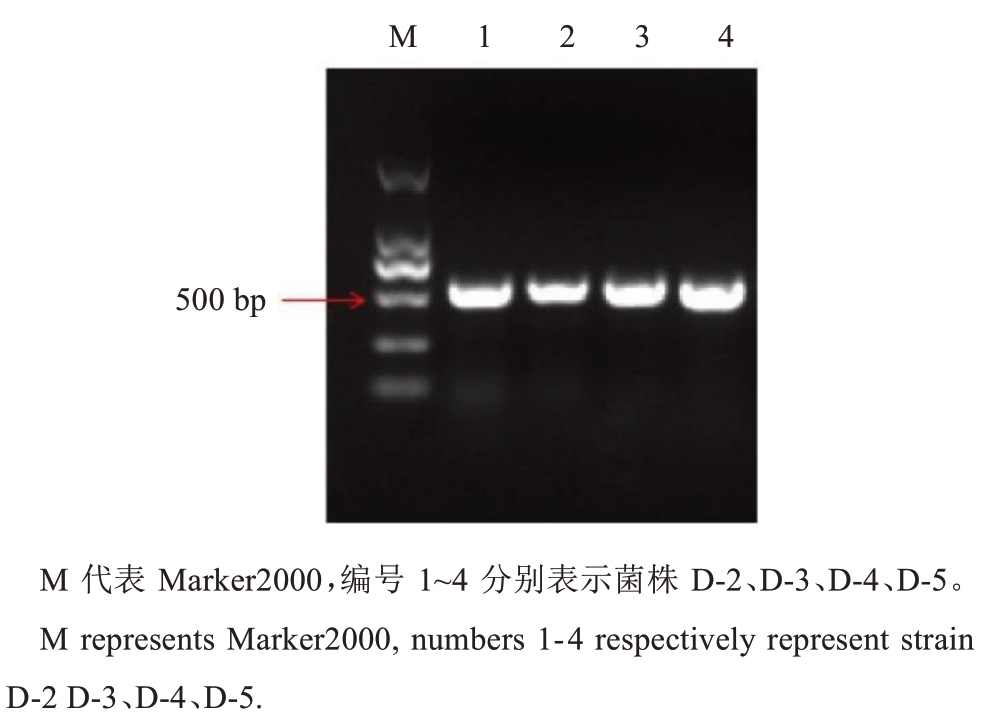
图2 引物扩增结果
Fig. 2 Primer amplification results
2.3 枇杷叶响应灰斑病菌侵染的RNA-Seq分析
2.3.1 转录组测序数据统计 将白梨和光荣本接种灰斑病菌后33、36、48 h的幼嫩叶片样本送往公司进行测序,共36份样本(表5)。将测序数据进行数据过滤后,每个样本产生4000~5360万条干净读数,所有样本都至少有95.62%条干净读数被定位到七星枇杷基因组。
表5 表达谱文库测序质量和产量
Table 5 Sequencing quality and yield of expression profile library
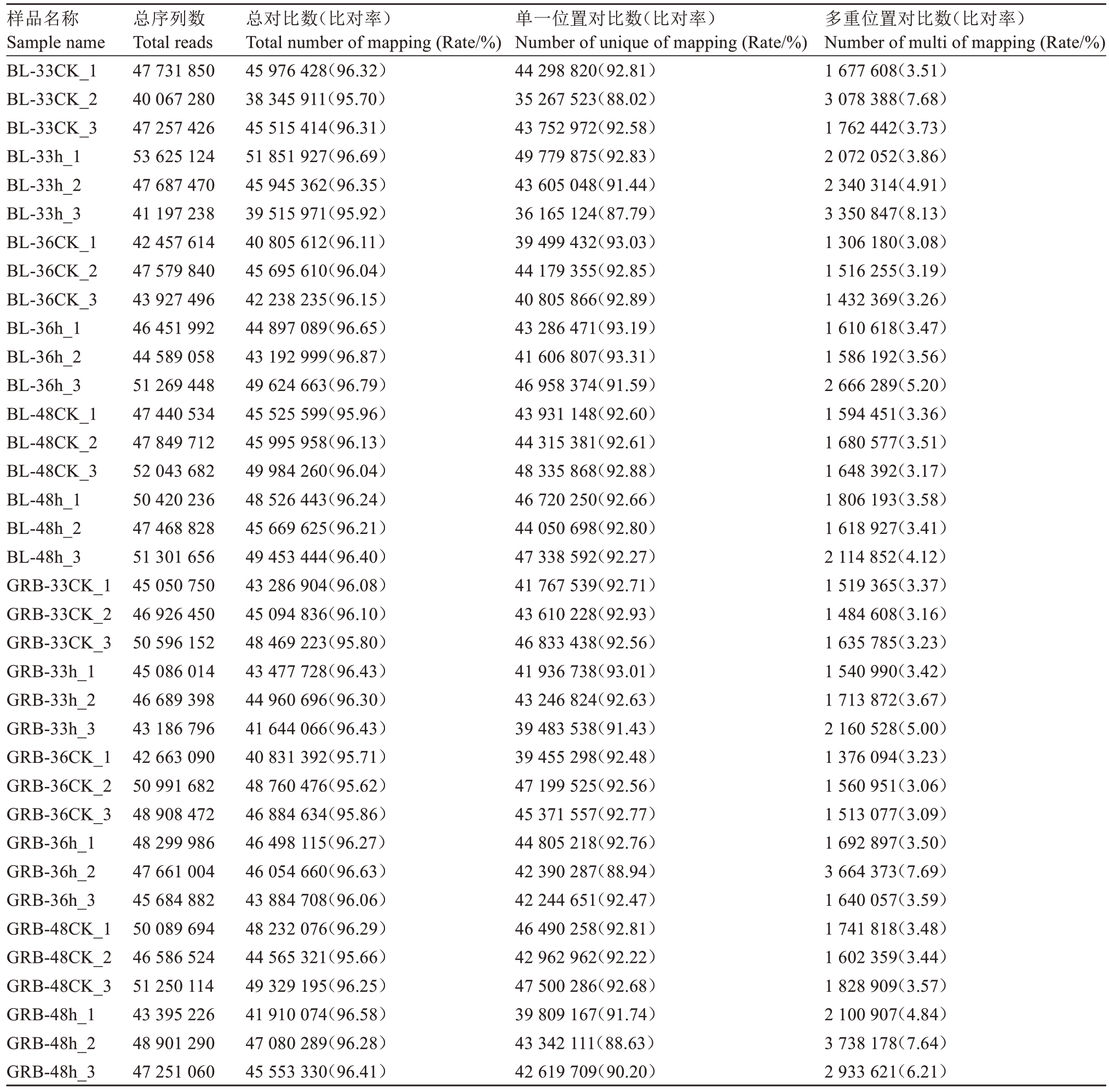
注:送测样品名称由枇杷品种名+接种后时间+序号组成。
Note:The name of the sample to be tested consists of loquat variety name+time after inoculation+serial number.
样品名称Sample name BL-33CK_1 BL-33CK_2 BL-33CK_3 BL-33h_1 BL-33h_2 BL-33h_3 BL-36CK_1 BL-36CK_2 BL-36CK_3 BL-36h_1 BL-36h_2 BL-36h_3 BL-48CK_1 BL-48CK_2 BL-48CK_3 BL-48h_1 BL-48h_2 BL-48h_3 GRB-33CK_1 GRB-33CK_2 GRB-33CK_3 GRB-33h_1 GRB-33h_2 GRB-33h_3 GRB-36CK_1 GRB-36CK_2 GRB-36CK_3 GRB-36h_1 GRB-36h_2 GRB-36h_3 GRB-48CK_1 GRB-48CK_2 GRB-48CK_3 GRB-48h_1 GRB-48h_2 GRB-48h_3多重位置对比数(比对率)Number of multi of mapping (Rate/%)1 677 608(3.51)3 078 388(7.68)1 762 442(3.73)2 072 052(3.86)2 340 314(4.91)3 350 847(8.13)1 306 180(3.08)1 516 255(3.19)1 432 369(3.26)1 610 618(3.47)1 586 192(3.56)2 666 289(5.20)1 594 451(3.36)1 680 577(3.51)1 648 392(3.17)1 806 193(3.58)1 618 927(3.41)2 114 852(4.12)1 519 365(3.37)1 484 608(3.16)1 635 785(3.23)1 540 990(3.42)1 713 872(3.67)2 160 528(5.00)1 376 094(3.23)1 560 951(3.06)1 513 077(3.09)1 692 897(3.50)3 664 373(7.69)1 640 057(3.59)1 741 818(3.48)1 602 359(3.44)1 828 909(3.57)2 100 907(4.84)3 738 178(7.64)2 933 621(6.21)总序列数Total reads 47 731 850 40 067 280 47 257 426 53 625 124 47 687 470 41 197 238 42 457 614 47 579 840 43 927 496 46 451 992 44 589 058 51 269 448 47 440 534 47 849 712 52 043 682 50 420 236 47 468 828 51 301 656 45 050 750 46 926 450 50 596 152 45 086 014 46 689 398 43 186 796 42 663 090 50 991 682 48 908 472 48 299 986 47 661 004 45 684 882 50 089 694 46 586 524 51 250 114 43 395 226 48 901 290 47 251 060总对比数(比对率)Total number of mapping (Rate/%)45 976 428(96.32)38 345 911(95.70)45 515 414(96.31)51 851 927(96.69)45 945 362(96.35)39 515 971(95.92)40 805 612(96.11)45 695 610(96.04)42 238 235(96.15)44 897 089(96.65)43 192 999(96.87)49 624 663(96.79)45 525 599(95.96)45 995 958(96.13)49 984 260(96.04)48 526 443(96.24)45 669 625(96.21)49 453 444(96.40)43 286 904(96.08)45 094 836(96.10)48 469 223(95.80)43 477 728(96.43)44 960 696(96.30)41 644 066(96.43)40 831 392(95.71)48 760 476(95.62)46 884 634(95.86)46 498 115(96.27)46 054 660(96.63)43 884 708(96.06)48 232 076(96.29)44 565 321(95.66)49 329 195(96.25)41 910 074(96.58)47 080 289(96.28)45 553 330(96.41)单一位置对比数(比对率)Number of unique of mapping (Rate/%)44 298 820(92.81)35 267 523(88.02)43 752 972(92.58)49 779 875(92.83)43 605 048(91.44)36 165 124(87.79)39 499 432(93.03)44 179 355(92.85)40 805 866(92.89)43 286 471(93.19)41 606 807(93.31)46 958 374(91.59)43 931 148(92.60)44 315 381(92.61)48 335 868(92.88)46 720 250(92.66)44 050 698(92.80)47 338 592(92.27)41 767 539(92.71)43 610 228(92.93)46 833 438(92.56)41 936 738(93.01)43 246 824(92.63)39 483 538(91.43)39 455 298(92.48)47 199 525(92.56)45 371 557(92.77)44 805 218(92.76)42 390 287(88.94)42 244 651(92.47)46 490 258(92.81)42 962 962(92.22)47 500 286(92.68)39 809 167(91.74)43 342 111(88.63)42 619 709(90.20)
对36个样本进行Spearman相关性分析表明,样本生物学重复间的基因表达水平有着高度相关性。样本相关性聚类分析结果(图3)表明了36个样本之间的全局相对关系,所有样本之间的生物学重复被聚类在一起,同时除BL-48 h的3个生物学重复外,本试验中的所有对照组BL-33 CK、BL-36 CK、BL-48 CK、GRB-33 CK、GRB-36 CK、GRB-48 CK和所有的处理组BL-33 h、BL-36 h、GRB-33 h、GRB-36 h、GRB-48 h被聚类在一起,说明经过菌株接种处理后,对照组和处理组在基因表达上存在明显差异,同时也可以发现2个品种间无论是否接种菌株,其基因表达均存在一定差异。

图3 样本相关性热图
Fig. 3 Sample correlation heat map
2.3.2 差异表达基因鉴定与分析 将相同时间点处理组与对照组叶片进行比较,共获得12 924个非冗余的差异表达基因(DEGs)。如图4所示,在BL-33 h与BL-33 CK的比较中,共有8664个DEGs(4049个上调和4615个下调),随后在36 h和48 h,DEGs数目都呈现下降趋势,而光荣本品种刚好相反。在GRB-48 h与GRB-48 CK的比较中,共有7483个DEGs(3098个上调和4385个下调),推测2个品种在不同时间点DEGs数量差异可能与两者对灰斑病的抗性不同有关。
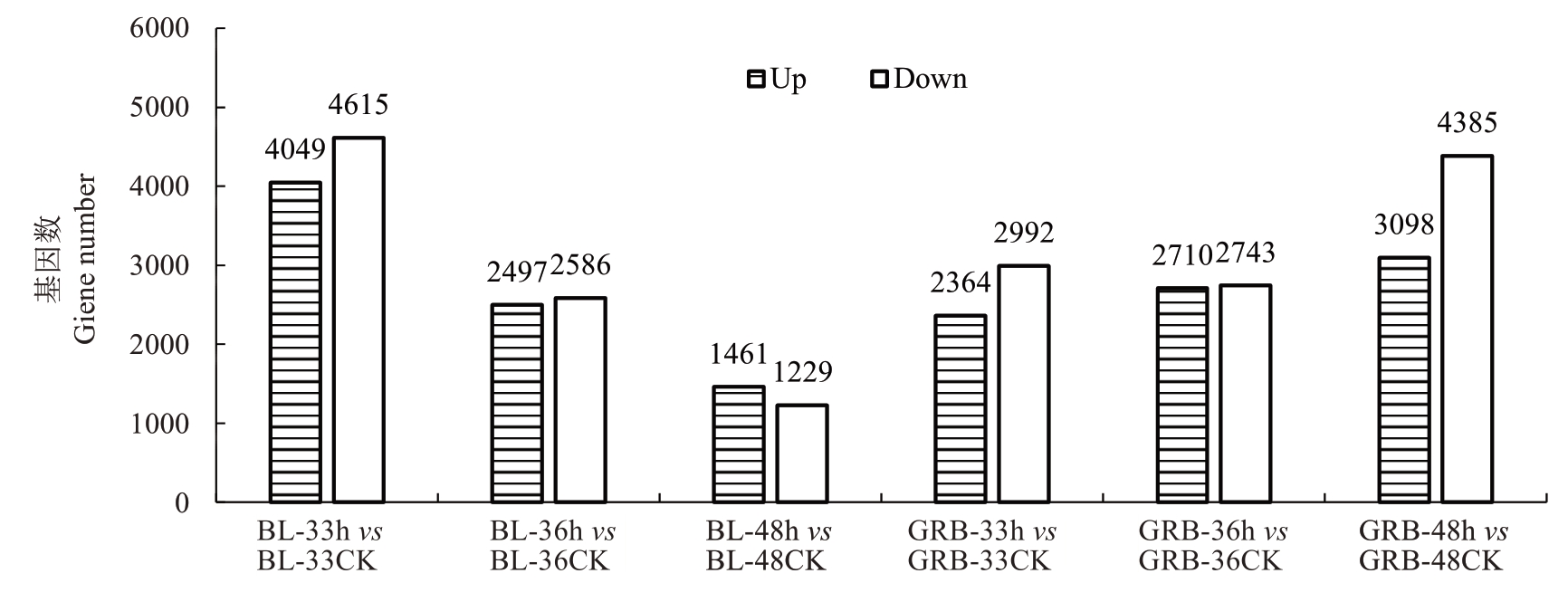
图4 差异比较组合差异基因数目统计柱状图
Fig. 4 Statistical histogram of the number of differential genes in differential comparison combinations
由图5可知,白梨中共有1184个基因在3个时间点中均差异表达,在33 h时特异性差异表达基因最多,共4117个;而光荣本中共有2776个基因在3个时间点中均差异表达,在48 h时特异性差异表达基因最多,共2904个。从这些特异性差异表达基因数量中可知,白梨接种后33 h时,在基因表达上有较大反应;光荣本接种后48 h时,在基因表达上有较大反应。
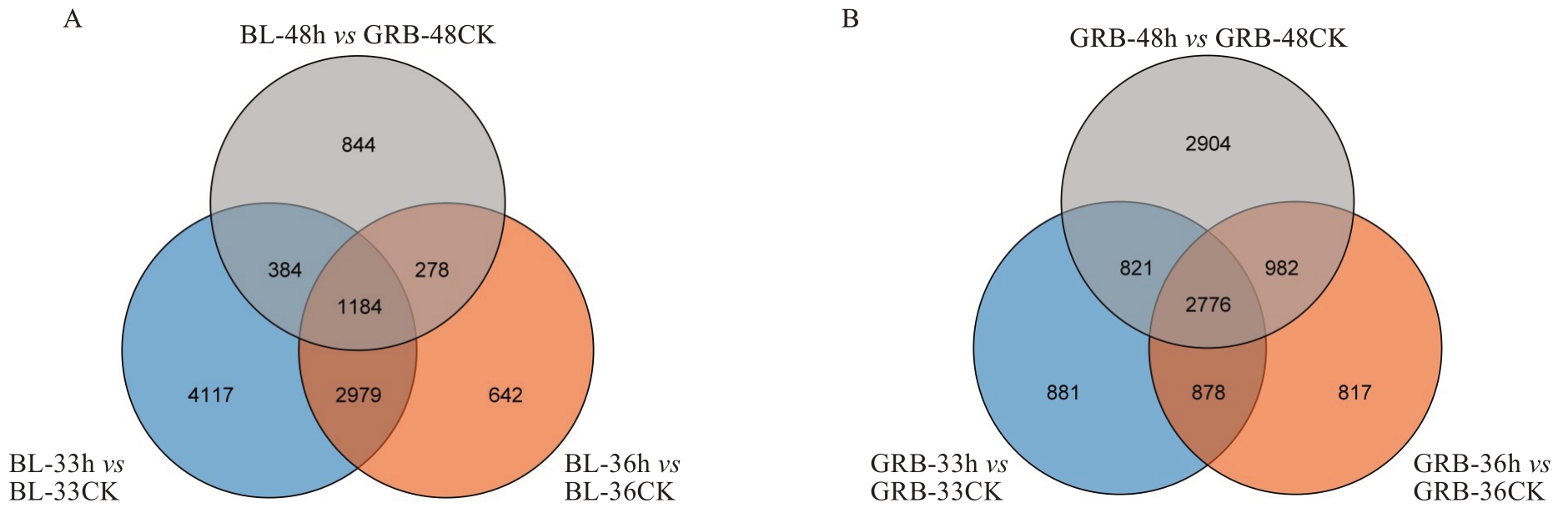
图5 各样品差异表达基因韦恩图
Fig. 5 Wayne diagram of differentially expressed genes in each sample
经分析发现,一共有684个(585个上调和99下调)DEGs均在灰斑病抗性品种白梨和灰斑病易感品种光荣本的3个处理时期中差异表达(图6~图7)。此外,一共有25个(12上调和13个下调)DEGs只在灰斑病抗性品种白梨的3个处理时期中存在差异表达。
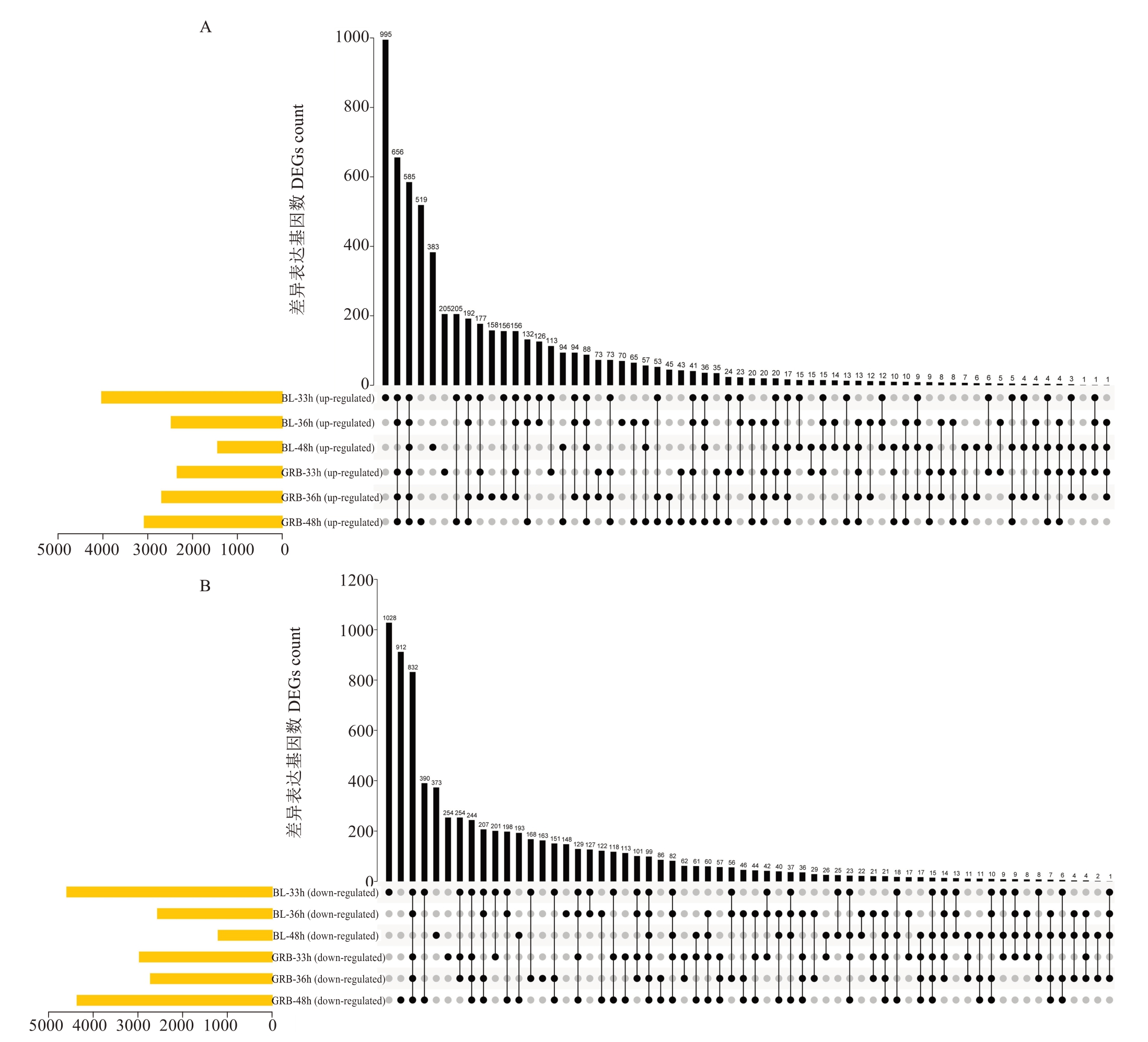
图6 上调(A)和下调(B)差异基因Upset图
Fig. 6 Upset plot of upregulated (A) and downregulated (B) differentially expressed genes
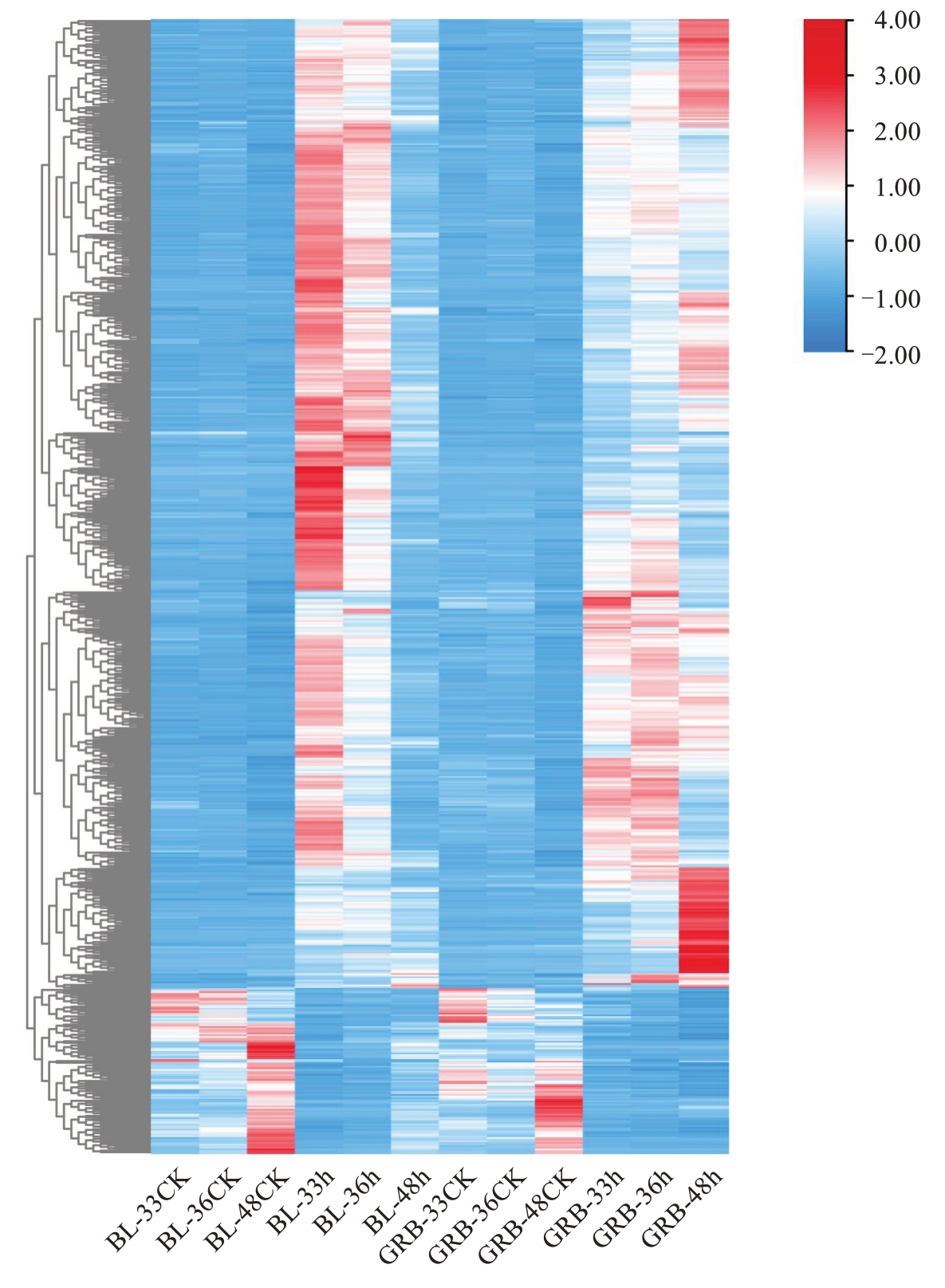
图7 各样品差异表达基因聚类热图
Fig. 7 Clustering heat map of differentially expressed genes in each sample
2.3.3 差异表达基因GO功能富集分析和KEGG通路分析 将上述得到的差异基因进行GO富集分析,所有DEGs被归类为三种类别,即分子功能、细胞成分和生物学过程。从GO富集分析的结果中,选取最显著的50个terms绘制气泡图。由图8可知,除了BL-48 h与BL-48 CK,其他差异基因都显著富集于对外源性刺激的反应、对非生物刺激的反应、对刺激的反应、对外部刺激的反应、对外界生物刺激的反应,表明在灰斑病菌处理后,枇杷叶片受到了显著外来刺激;同时,也显著富集一些生物过程如光合作用光反应、光合作用、退化的氧化还原过程、前体代谢产物的产生和能量、光系统Ⅰ中的光合作用光收获等,说明感染灰斑病后枇杷叶片光合作用等过程会被显著影响。比较特殊的是,BL-48 h与BL-48 CK比较得到的差异基因,多富集到与细胞壁等相关的生物学过程,如植物型次生细胞壁生物发生、植物型细胞壁组织或生物发生、细胞壁生物发生、细胞壁组织或生物发生、植物型细胞壁生物发生等。

图8 灰斑病侵染对枇杷抗、感种质差异表达基因的GO功能富集分析Fig. 8 GO functional enrichment analysis of differentially expressed genes in loquat resistant and susceptible germplasm infected by gray spot disease
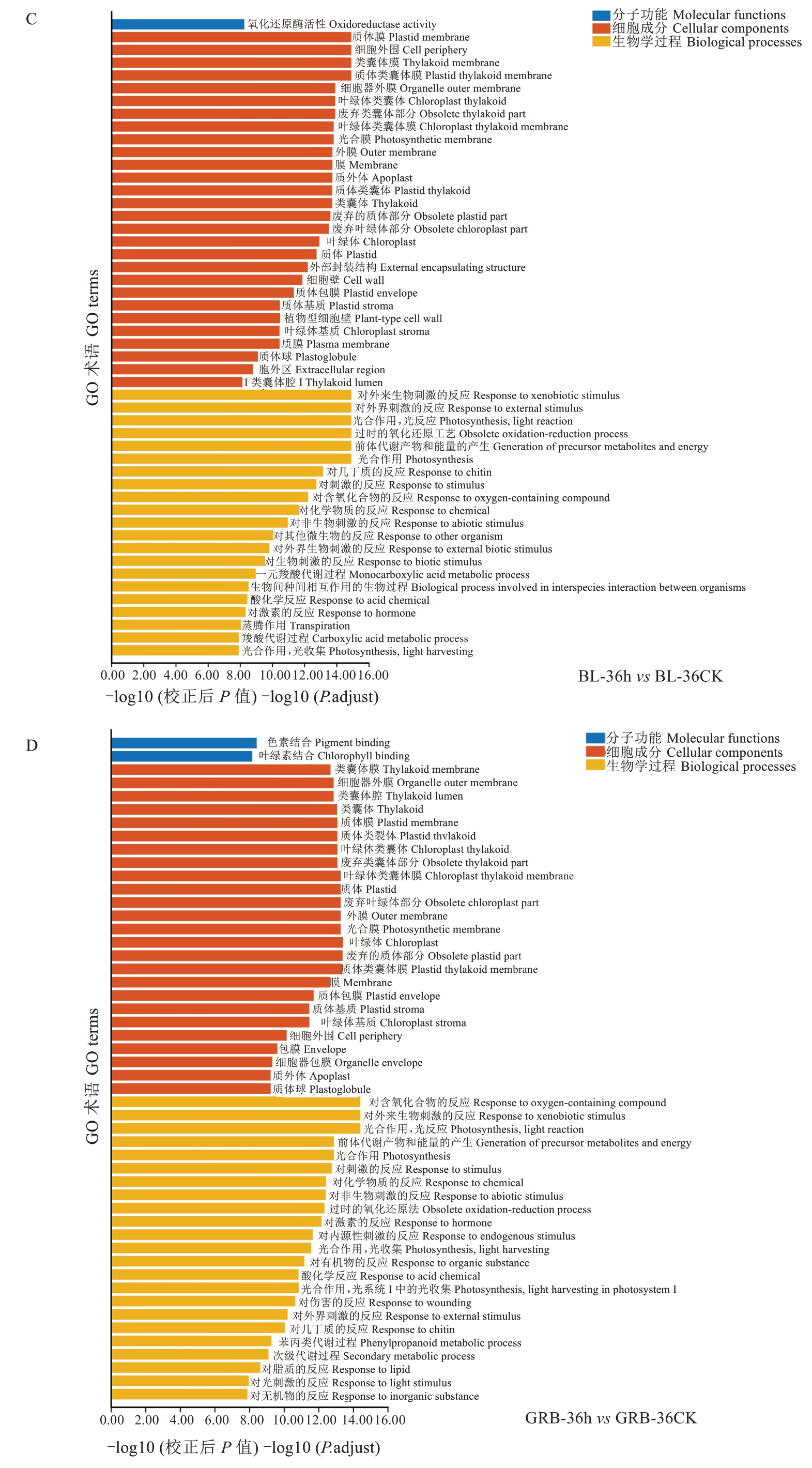
图8 (续) Fig. 8 (Continued)
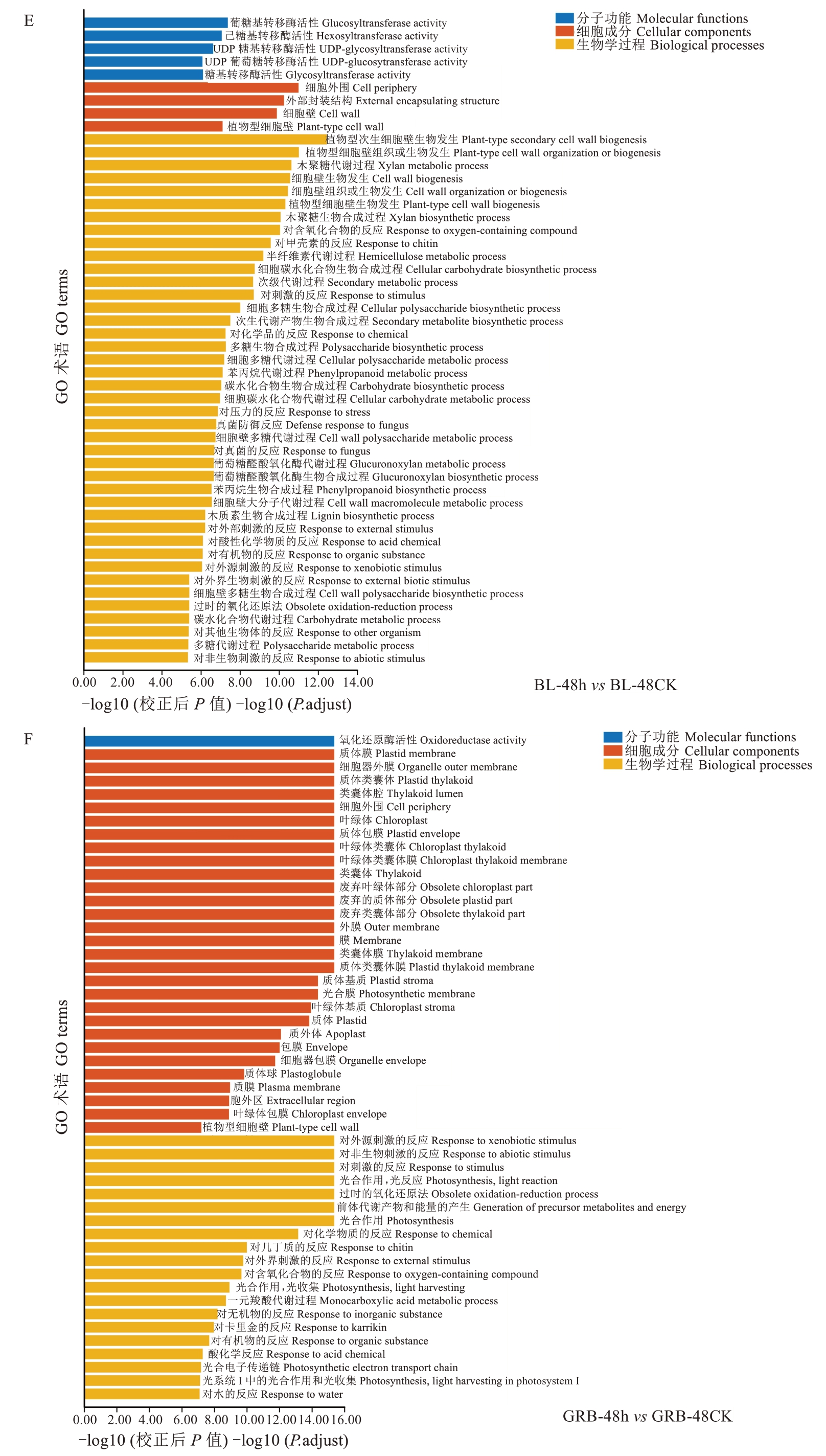
图8 (续) Fig. 8 (Continued)
从KEGG富集分析结果中,选取最显著的30个KEGG通路绘制气泡图。如图9所示,在白梨和光荣本中,最显著富集的KEGG通路为代谢途径、次生代谢产物的生物合成、碳代谢、氨基酸的生物合成、植物激素信号转导、淀粉和蔗糖代谢、植物-病原体相互作用、苯丙烷类生物合成、谷胱甘肽代谢和MAPK信号通路-植物。KEGG富集分析表明,病原菌接种后,白梨和光荣本的差异表达基因多数富集于各种代谢途径或各种合成途径以及植物-病原体相互作用等通路中。

图9 灰斑病侵染对枇杷抗、感种质差异表达基因的KEGG通路富集分析Fig. 9 Enrichment analysis of KEGG pathway of differentially expressed genes in resistant and susceptible loquat germplasm infected by gray spot disease

图9 (续) Fig. 9 (Continued)
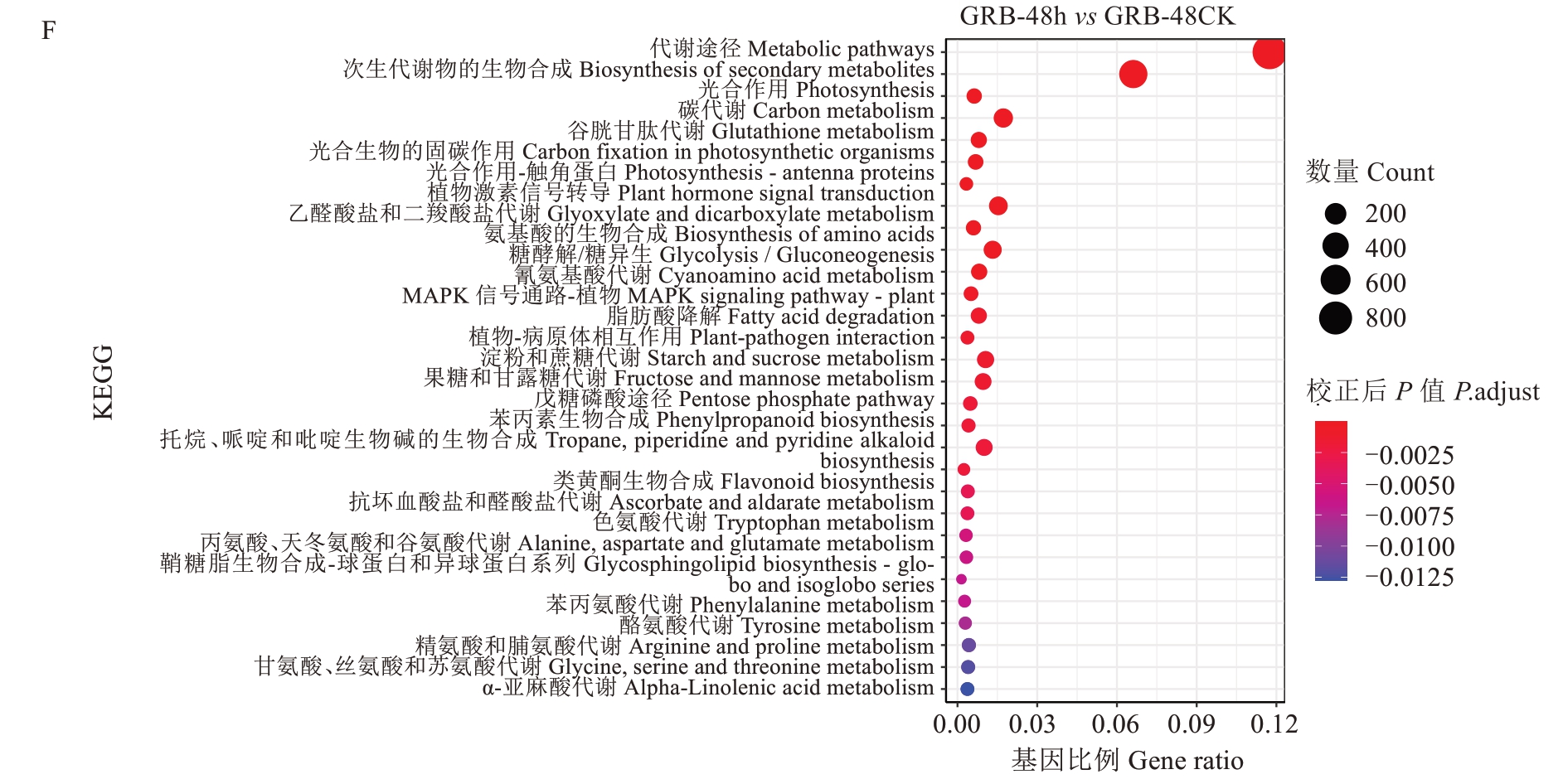
图9 (续) Fig. 9 (Continued)
对白梨和光荣本的684个共响应的DEGs进行转录因子和抗病基因的预测发现,其中共存在64个转录因子,数量较多的是AP2/ERF(17)、WRKY(9)、NAC(6)、MYB(6)。除EVM0001615(AP2/ERF)、EVM0042513(AP2/ERF)、EVM0003490(AUX/IAA)、EVM0024608(AUX/IAA)、EVM0026620(B3-ARF)、EVM0001978(C3H)、EVM0017008(LIM)这些转录因子在接种后33 h、36 h和48 h中均表达下调外,其余转录因子均表达上调。同时,684个DEGs中共存在59个抗病基因,除了EVM0007455、EVM0008538、EVM0032212表达下调外,其他抗病基因均表达上调。
为了进一步阐明枇杷应答灰斑病的基因功能,使用GO和KEGG对684个在枇杷叶灰斑病高抗材料与高感材料共响应的DEGs进行分析(图10)。通过GO富集分析发现,在生物学过程类别中,DEGs主要参与对压力、刺激、激素、防御响应,表明这些DEGs在枇杷对灰斑病的应答中起到重要作用;在细胞组分类别中,这些DEGs大多是胞外区、质外体、细胞壁、膜的相关基因;在分子功能类别中,DEGs主要行使槲皮素3-O-糖基转移酶活性、槲皮素7-O-糖基转移酶活性、谷胱甘肽转移酶活性、葡糖基转移酶活性。通过KEGG分析发现,DEGs最显著富集的通路是代谢途径、谷胱甘肽代谢、次生代谢产物的生物合成、光合作用-天线蛋白、倍半萜和三萜生物合成、MAPK信号通路-植物,表明枇杷叶片在响应灰斑病时,这些DEGs主要参与代谢与生物合成途径。
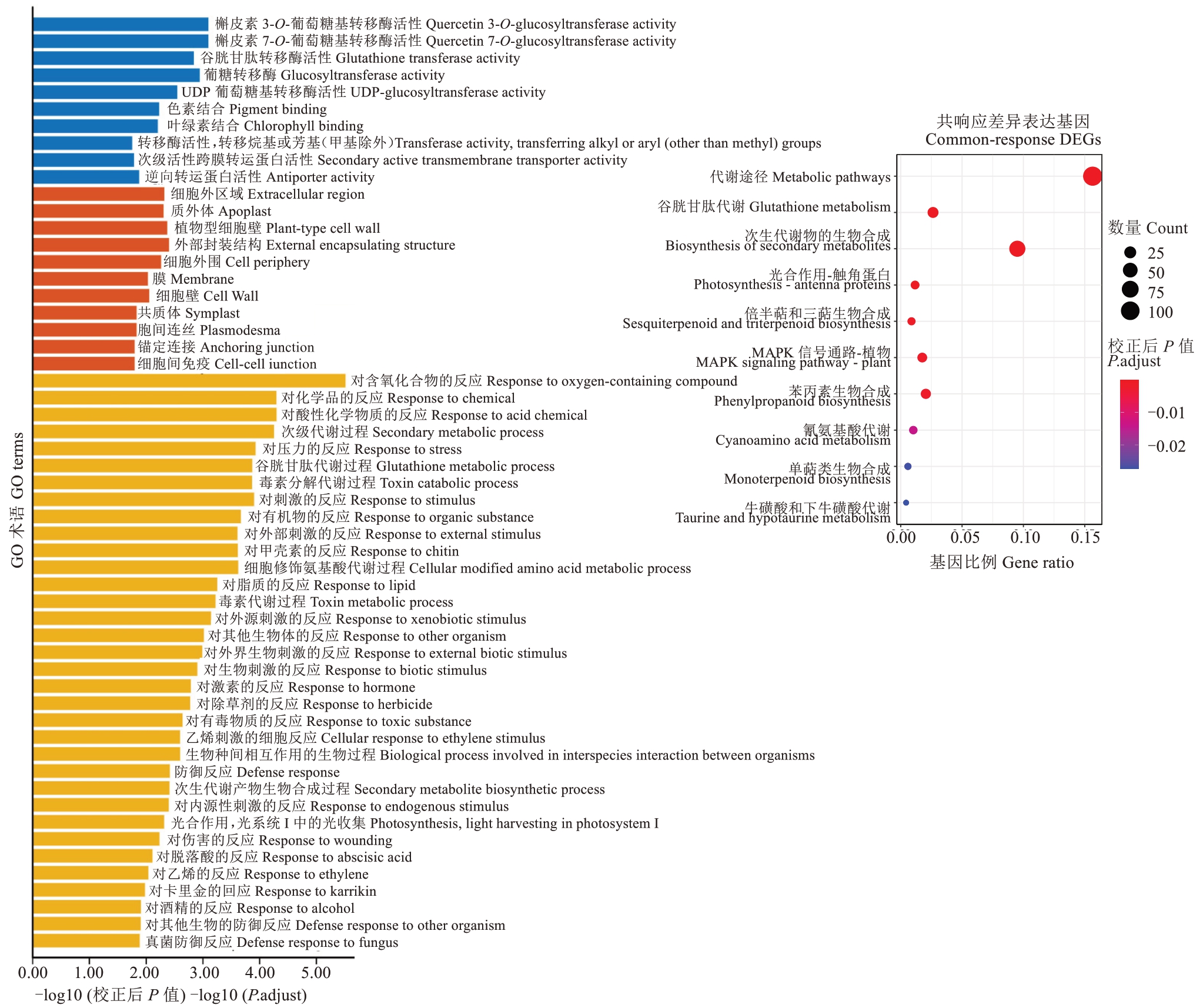
图10 灰斑病抗、感种质共响应的差异表达基因的GO及KEGG富集分析
Fig. 10 Enrichment analysis of KEGG pathway of differentially expressed genes in resistant and susceptible loquat germplasm infected by gray spot disease
根据GO和KEGG富集结果,进一步对关键生物过程和代谢途径中的基因进行了分析。GO富集结果显示一共有21个基因全部被富集到对真菌的反应、真菌的防御反应、其他生物的防御反应、防御反应这4个途径;KEGG富集结果显示,一共有3个基因被富集在植物-病原菌互作途径。
除了灰斑病高抗种质与高感种质共响应的DEGs,在灰斑病抗性种质白梨中还存在25个(12上调和13个下调)特异表达的DEGs。25个基因中存在1个转录因子EVM0037921(AP2/ERF)以及3个抗病基因EVM0015411、EVM0019301、EVM0024983,这4个基因可能在枇杷叶片对灰斑病抗性的提升上行使重要功能(表6)。
表6 4个特异表达差异基因的注释及表达趋势
Table 6 Annotation and expression trend of four differentially expressed genes
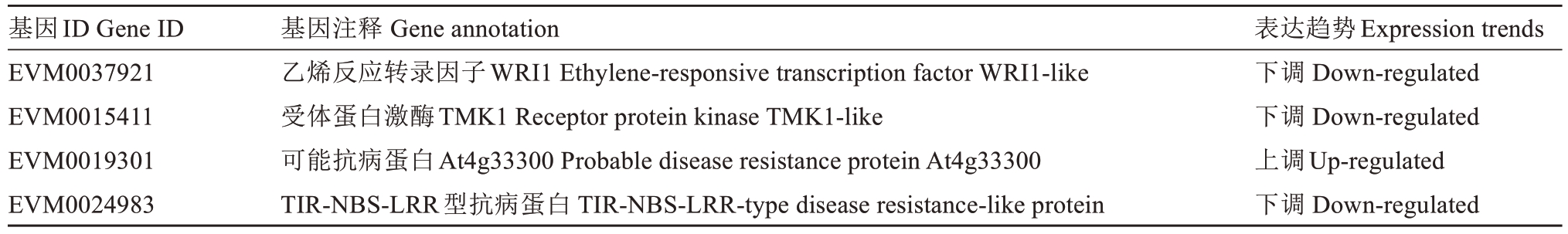
基因ID Gene ID EVM0037921 EVM0015411 EVM0019301 EVM0024983基因注释 Gene annotation乙烯反应转录因子WRI1 Ethylene-responsive transcription factor WRI1-like受体蛋白激酶TMK1 Receptor protein kinase TMK1-like可能抗病蛋白At4g33300 Probable disease resistance protein At4g33300 TIR-NBS-LRR型抗病蛋白 TIR-NBS-LRR-type disease resistance-like protein表达趋势Expression trends下调 Down-regulated下调 Down-regulated上调Up-regulated下调 Down-regulated
2.3.4 差异表达基因的qRT-PCR验证 采用qRTPCR对参与枇杷灰斑病防御反应的7个差异基因进行相对定量表达分析,其中包括转录因子(如WRKY、ERF、MYB)与抗病基因,并与RNA-seq数据进行比较,以验证RNA-seq测序数据的可靠性。如图11所示,7个差异基因在接种病原菌后表现出了与RNA-seq相似的趋势,其中WRKY转录因子在植物抗病以及应对刺激等方面起到重要的调节作用。EVM0014520(WRKY40-like)和EVM0020330(WRKY71-like)在白梨与光荣本中接菌处理后表达量均上调。此外,植物MYB转录因子在激素应答以及胁迫响应等生理生化过程中起着广泛的调控作用,EVM0022518(MYB44-like)同样在白梨或是光荣本中接菌处理后表达量均上调。在抗病基因方面,EVM0039675(PTI5-like)同样在2个品种中均上调,并且在白梨中有着更快的响应。总体来看,荧光定量PCR验证结果与转录组测序中差异基因的表达趋势基本一致,二者之间存在高度相关性,表明了转录组数据的可靠性。
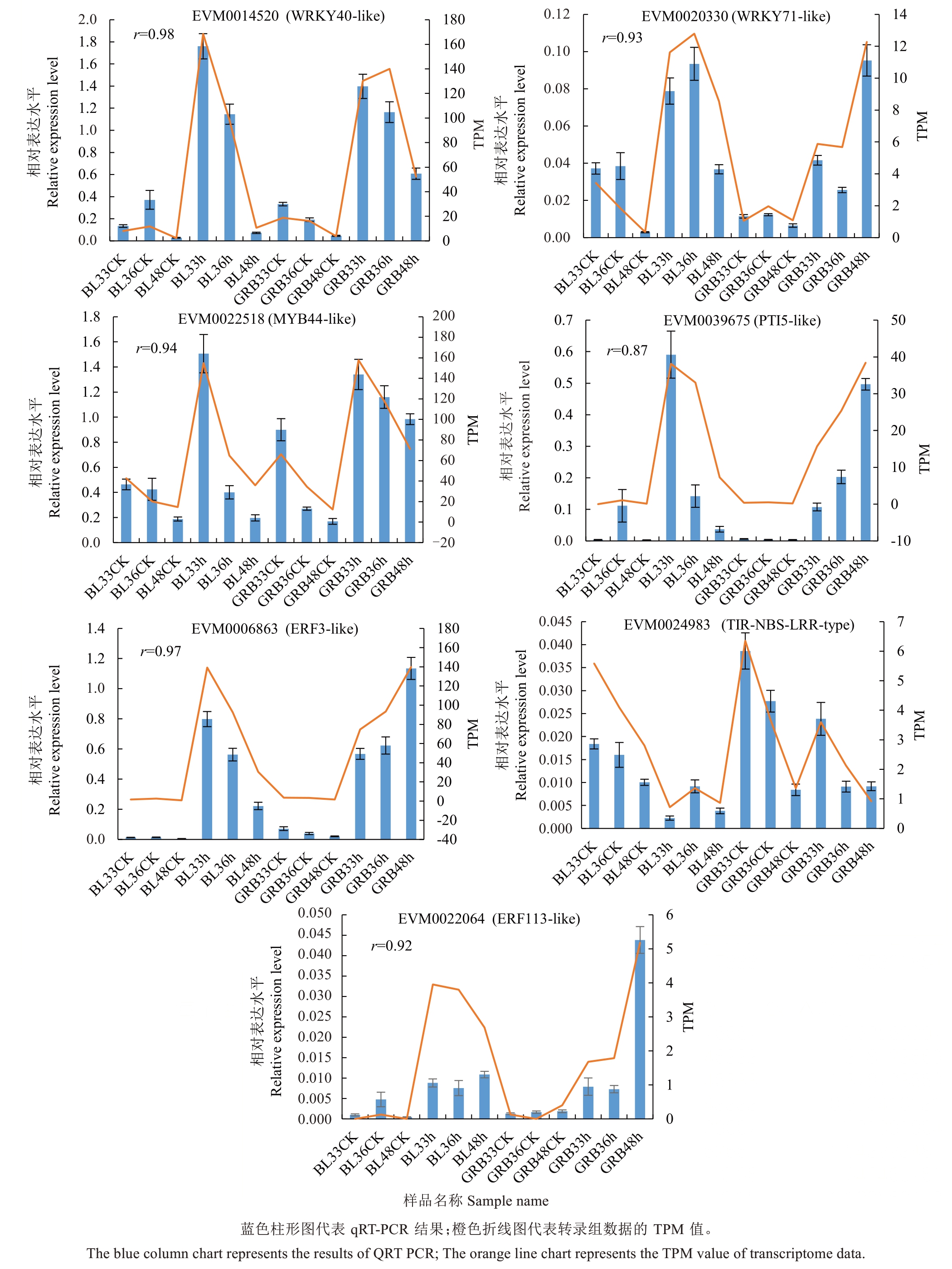
图11 差异基因qRT-PCR验证
Fig. 11 qRT-PCR verification of differential genes
3 讨 论
在进行抗病评价的过程中,计算相对病斑面积是关键一步,其反映了病原真菌或细菌侵染叶片的程度。前人已经提出了很多方法,包括公式计算法、工具法和软件法等[35],采用较多的是图片识别技术[36]。周蓓[9]采用目测估算法对枇杷叶斑病进行调查,即通过目测估算的方式对调查材料的相对病斑面积进行估算,进而做出抗性分级。这种方法简单易行、效率较高,但受主观因素的影响较大。因此,本次试验结合目测和软件法进行枇杷灰斑病抗性调查,可以更加高效、客观地对发病叶片进行相应的病级分类,进而对枇杷材料进行抗性评价,两者相辅相成。笔者将人工接种评价结果与田间调查结果进行比较,发现与大多数抗病性评价筛选方法的结果类似[37-38],在田间自然条件和室内人工接种条件下的评价结果相似但又并不完全一致。尽管离体叶片人工接种评价无法完全复制田间的复杂情况,但离体叶片人工接种作为一种高效的初筛工具是完全可靠的,能够准确筛选有潜力的抗病材料。一般来说,有些种质在田间因环境条件不适于发病而表现出较高水平的抗性,但在人工接种条件下可能出现容易感病的情况。这种差异的原因尚不明确,可能是人工接种对植物产生不利的因素,从而增强病原体的致病力,也可能是人工接种培养为病原体提供了有利的侵染环境[39-40]。在本试验中,没有完全免疫或几乎免疫的种质,这与大多数枇杷抗病性研究的结果一致[41]。研究表明,表型为高抗的种质通常表达丰富的抗病基因,其抗性通常容易被其他病原菌菌株克服,从而导致抗病性丧失[42]。齿叶枇杷、香花枇杷和白梨表现出比其他种质更强的抗病性。因此,一些被病原菌侵染但仍表现出较强抗性的种质在育种中被认为可能具备持久和广泛的抗性,是进行进一步抗病评价的优良材料[43]。
利用转录组技术能够全面地分析作物与病害相互作用过程中的mRNA表达水平的变化,如Deng等[18]利用转录组技术分析发现,在外源水杨酸的诱导下CsWRKY70参与柑橘果实对柑橘绿霉病菌的抗性。笔者利用转录组技术分析了高抗材料白梨与高感材料光荣本,发现两者经过菌株接种处理后,对照组和处理组在基因的表达上出现明显的差异,进一步分析发现两个品种在同一时间点上DEGs的数量也存在差异。对两个品种存在的684个共响应基因分析发现,存在着大量响应枇杷灰斑病转录因子和抗病基因,这些基因大多被富集在响应压力、各种刺激、防御等相关通路、各种代谢途径和植物-病原菌互作途径中,其中WRKY类和AP2/ERF类转录因子起到了重要的调节作用,这与其他植物中的抗病研究结论相一致[44-45]。在灰斑病抗性种质白梨中存在25个特异表达的DEGs,其中包括1个AP2/ERF类转录因子和3个抗病基因,推测这4个基因可能在枇杷叶片对灰斑病抗性的提升上行使重要功能。
4 结 论
通过田间调查和离体接种评价,获得11份抗性结果一致的枇杷种质,其中高抗种质3份、抗病种质4份、感病种质2份、高感种质2份。通过对高抗种质白梨和高感种质光荣本接种组和对照组叶片的转录组进行分析,发现一共有684个(585个上调和99下调)DEGs在白梨和光荣本中共同响应,一共有25个(12个上调和13下调)DEGs只在白梨中响应。在共响应基因中鉴定出64个转录因子以及59个抗病基因,在特异响应基因中鉴定出1个转录因子以及3个抗病基因,这为枇杷种质资源抗性的提高提供了参考基因,并为枇杷抗病品种的选育提供借鉴。
[1] 姜琬. 重庆枇杷灰斑病菌鉴定、抗性评价及防控研究[D] . 重庆:西南大学,2014.JIANG Wan. Study on identification,resistance evaluation and control of Pestalotiopsis eriobotrifolia in Chongqing[D] . Chongqing:Southwest University,2014.
[2] WEI Y Y,XU F,SHAO X F. Changes in soluble sugar metabolism in loquat fruit during different cold storage[J] . Journal of Food Science and Technology,2017,54(5):1043-1051.
[3] ALI M M,ALAM S M,ANWAR R,ALI S,SHI M,LIANG D D,LIN Z M,CHEN F X. Genome-wide identification,characterization and expression profiling of aluminum-activated malate transporters in Eriobotrya japonica Lindl.[J] . Horticulturae,2021,7(11):441.
[4] SHAN Y X,DENG C J,HU W S,CHEN J W,CHEN X P,ZHENG S Q,QIN Q P. First insight into diversity of leaf color of loquat (Eribotrya) and its potential value on taxonomy[J] . Genetic Resources and Crop Evolution,2019,66(1):143-163.
[5] 江旭升,杨勇胜,李庆宏,魏椿,陈树红,罗国伟. 贵阳地区枇杷种质叶斑病的抗性调查[J] . 贵州农业科学,2016,44(8):39-44.JIANG Xusheng,YANG Yongsheng,LI Qinghong,WEI Chun,CHEN Shuhong,LUO Guowei. Resistance investigation of loquat leaf spot disease in Guiyang area[J] . Guizhou Agricultural Sciences,2016,44(8):39-44.
[6] REN Y F,YAN T Y,HU C M,LIU D,HE J Y. Exogenous nitric oxide-induced postharvest gray spot disease resistance in loquat fruit and its possible mechanism of action[J] . International Journal of Molecular Sciences,2023,24(5):4369.
[7] 陈福如,杨秀娟. 福建省枇杷真菌性病害调查与鉴定[J] . 福建农业学报,2002,17(3):151-154.CHEN Furu,YANG Xiujuan. Investigation and identification of the fungal diseases of Eriobotrya japonica in Fujian[J] . Fujian Journal of Agricultural Sciences,2002,17(3):151-154.
[8] 李金萍,郭喜红,侯峥嵘,王璐,解晓军,尹哲. 北京地区南方果树病害调查初报[J] . 中国植保导刊,2015,35(8):21-25.LI Jinping,GUO Xihong,HOU Zhengrong,WANG Lu,XIE Xiaojun,YIN Zhe. Preliminary investigation on southern fruit trees diseases in Beijing[J] . China Plant Protection,2015,35(8):21-25.
[9] 周蓓. 若干枇杷属植物种及种间杂种叶斑病抗性鉴定、天牛危害与根系调查[D] . 广州:华南农业大学,2017.ZHOU Bei. Investigation on leaf spot disease with its identification of resistance,beetles damage and root systemin several Eri‐obotrya species and interspecific hybrids[D] . Guangzhou:South China Agricultural University,2017.
[10] 郑伟,吴亚维,王彬,宋莎,罗昌国. 苹果叶片结构与白粉病抗性的相关性初步研究[J] . 西南农业学报,2017,30(9):2108-2112.ZHENG Wei,WU Yawei,WANG Bin,SONG Sha,LUO Changguo. Preliminary study on correlation between leaf structure and powdery mildew resistance in apple[J] . Southwest China Journal of Agricultural Sciences,2017,30(9):2108-2112.
[11] 李敏,段硕,李中安,周彦,周常勇,谭锦,彭耀武. 叶片微形态结构特征与柑橘溃疡病抗性的关系[J] . 中国南方果树,2013,42(2):1-5.LI Min,DUAN Shuo,LI Zhong’an,ZHOU Yan,ZHOU Changyong,TAN Jin,PENG Yaowu. Analysis of relationship between citrus canker resistance and leaf micro-morphological characteristics[J] . South China Fruits,2013,42(2):1-5.
[12] 潘贞珍,黄运鹏,黄桂香,杨翠红,何新华. 3个柑橘品种叶片结构和生化物质与柑橘溃疡病抗性的相关性研究[J] . 中国果树,2020(4):31-36.PAN Zhenzhen,HUANG Yunpeng,HUANG Guixiang,YANG Cuihong,HE Xinhua. Correlation research of leaf structure and biochemical substances of three varieties of citrus with the resistance to citrus canker[J] . China Fruits,2020(4):31-36.
[13] EBRAHIM S,USHA K,SINGH B. Plant architectural traits and their role in defense mechanism against malformation in mango(Mangifera Indica L.)[J] . Scientia Horticulturae,2012,139:25-31.
[14] 陈依丽,陈雨琼,邓颖,李春雨,彭泽,杨向晖. 枇杷属植物叶片结构与叶斑病抗性的相关性研究[J] . 果树学报,2022,39(11):2133-2140.CHEN Yili,CHEN Yuqiong,DENG Ying,LI Chunyu,PENG Ze,YANG Xianghui. Analysis of correlation between leaf structure and resistance to leaf spot in Eriobotrya[J] . Journal of Fruit Science,2022,39(11):2133-2140.
[15] 张健,唐露,张雅洁,冉启凡. 转录组测序技术在植物水淹胁迫研究中的应用[J] . 分子植物育种,2019,17(4):1191-1202.ZHANG Jian,TANG Lu,ZHANG Yajie,RAN Qifan. Application of transcriptome sequencing technique in the study of waterlogging stress in plants[J] . Molecular Plant Breeding,2019,17(4):1191-1202.
[16] 贾昌路,张瑶,朱玲,张锐. 转录组测序技术在生物测序中的应用研究进展[J] . 分子植物育种,2015,13(10):2388-2394.JIA Changlu,ZHANG Yao,ZHU Ling,ZHANG Rui. Application progress of transcriptome sequencing technology in biological sequencing[J] . Molecular Plant Breeding,2015,13(10):2388-2394.
[17] 陈哲,黄静,赵佳,梁宏. 草莓应答炭疽菌侵染的转录组分析[J] .植物保护,2020,46(3):138-146.CHEN Zhe,HUANG Jing,ZHAO Jia,LIANG Hong. Transcriptomics analysis of strawberry response to Colletotrichum theo‐bromicola infection[J] . Plant Protection,2020,46(3):138-146.
[18] DENG B,WANG W J,RUAN C Q,DENG L L,YAO S X,ZENG K F. Involvement of CsWRKY70 in salicylic acid-induced citrus fruit resistance against Penicillium digitatum[J] .Horticulture Research,2020,7(1):157.
[19] 井赵斌. 猕猴桃抗溃疡病转录组分析和基因功能注释[J] . 分子植物育种,2021,19(6):1830-1838.JING Zhaobin. Transcriptome analysis and gene function annotation of kiwifruit Psa resistance[J] . Molecular Plant Breeding,2021,19(6):1830-1838.
[20] 张玮. 葡萄溃疡病菌侵染葡萄的组学基础与分子机制的初步研究[D] . 北京:中国农业大学,2019.ZHANG Wei. Preliminary study on molecular mechanisms and omics of Lasiodiplodia theobromae infecting grapevine[D] . Beijing:China Agricultural University,2019.
[21] 郑少泉. 枇杷种质资源描述规范和数据标准[M] . 北京:中国农业出版社,2006.ZHENG Shaoquan. Descriptors and data standard for loquat[M] .Beijing:China Agriculture Press,2006.
[22] 崔华威,杨艳丽,黎敬涛,罗文富,苗爱敏,胡振兴,韩小女. 一种基于Photoshop的叶片相对病斑面积快速测定方法[J] . 安徽农业科学,2009,37(22):10760-10762.CUI Huawei,YANG Yanli,LI Jingtao,LUO Wenfu,MIAO Aimin,HU Zhenxing,HAN Xiaonü. A faster method for measuring relative lesion area on leaves based on software photoshop[J] .Journal of Anhui Agricultural Sciences,2009,37(22):10760-10762.
[23] 陈福如,陈元洪,翁启勇. 枇杷病虫害诊治图谱[M] . 福州:福建科学技术出版社,2003.CHEN Furu,CHEN Yuanhong,WENG Qiyong. Loquat pest diagnosis[M] . Fuzhou:Fujian Science & Technology Publishing House,2003.
[24] BRYAN G T,DANIELS M J,OSBOURN A E. Comparison of fungi within the Gaeumannomyces-Phialophora complex by analysis of ribosomal DNA sequences[J] . Applied and Environmental Microbiology,1995,61(2):681-689.
[25] CHEN S F,ZHOU Y Q,CHEN Y R,GU J. Fastp:An ultra-fast all-in-one FASTQ preprocessor[J] . Bioinformatics,2018,34(17):i884-i890.
[26] KIM D,PAGGI J M,PARK C,BENNETT C,SALZBERG S L.Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype[J] . Nature Biotechnology,2019,37(8):907-915.
[27] JIANG S,AN H S,XU F J,ZHANG X Y. Chromosome-level genome assembly and annotation of the loquat (Eriobotrya ja‐ponica) genome[J] . GigaScience,2020,9(3):giaa015.
[28] PERTEA M,PERTEA G M,ANTONESCU C M,CHANG T C,MENDELL J T,SALZBERG S L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads[J] . Nature Biotechnology,2015,33(3):290-295.
[29] LOVE M I,HUBER W,ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J] .Genome Biology,2014,15(12):550.
[30] CHEN C J,CHEN H,ZHANG Y,THOMAS H R,FRANK M H,HE Y H,XIA R. TBtools:An integrative toolkit developed for interactive analyses of big biological data[J] . Molecular Plant,2020,13(8):1194-1202.
[31] CANTALAPIEDRA C P,HERNÁNDEZ-PLAZA A,LETUNIC I,BORK P,HUERTA-CEPAS J. eggNOG-mapper v2:Functional annotation,orthology assignments,and domain prediction at the metagenomic scale[J] . Molecular Biology and Evolution,2021,38(12):5825-5829.
[32] BU D C,LUO H T,HUO P P,WANG Z H,ZHANG S,HE Z H,WU Y,ZHAO L H,LIU J J,GUO J C,FANG S S,CAO W C,YI L,ZHAO Y,KONG L. KOBAS-i:Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis[J] . Nucleic Acids Research,2021,49(W1):W317-W325.
[33] UNTERGASSER A,NIJVEEN H,RAO X Y,BISSELING T,GEURTS R,LEUNISSEN J A M. Primer 3 plus,an enhanced web interface to primer 3[J] . Nucleic Acids Research,2007,35(Suppl. 2):W71-W74.
[34] LIVAK K J,SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△CT Method[J] . Methods,2001,25(4):402-408.
[35] 陈冬梅,袁琳,颜鹏,范姗慧,周贤锋,张竞成,吴开华. 基于自适应模糊阈值的茶炭疽病斑面积计算[J] . 茶叶通讯,2019,46(2):185-191.CHEN Dongmei,YUAN Lin,YAN Peng,FAN Shanhui,ZHOU Xianfeng,ZHANG Jingcheng,WU Kaihua. Calculation of spot area of tea anthracnose based on adaptive fuzzy threshold[J] .Journal of Tea Communication,2019,46(2):185-191.
[36] 任行海,刘博,乔曦,王福宽,钱万强,万方浩,刘怀. 利用图像识别技术计算薇甘菊锈病的相对病斑面积[J] . 生物安全学报,2021,30(1):72-77.REN Xinghai,LIU Bo,QIAO Xi,WANG Fukuan,QIAN Wanqiang,WAN Fanghao,LIU Huai. Calculation of spot area of Mi‐kania micrantha rust based on image processing technology[J] .Journal of Biosafety,2021,30(1):72-77.
[37] FOOLAD M R,SULLENBERGER M T,ASHRAFI H. Detached-leaflet evaluation of tomato germplasm for late blight resistance and its correspondence to field and greenhouse screenings[J] . Plant Disease,2015,99(5):718-722.
[38] CHANDELIER A,HUSSON C,DRUART P,MARÇAIS B. Assessment of inoculation methods for screening black alder resistance to Phytophthora × alni[J] . Plant Pathology,2016,65(3):441-450.
[39] DORRANCE A E,INGLIS D A. Assessment of greenhouse and laboratory screening methods for evaluating potato foliage for resistance to late blight[J] . Plant Disease,1997,81(10):1206-1213.
[40] MICHALSKA A M,ZIMNOCH-GUZOWSKA E,SOBKOWIAK S,PLICH J. Resistance of potato to stem infection by Phy‐tophthora infestans and a comparison to detached leaflet and field resistance assessments[J] . American Journal of Potato Research,2011,88(4):367-373.
[41] MCDONALD B A,LINDE C. Pathogen population genetics,evolutionary potential,and durable resistance[J] .Annual Review of Phytopathology,2002,40(1):349-379.
[42] SHARMA B P,FORBES G A,MANANDHAR H K,SHRESTHA S M,THAPA R B. Determination of resistance to Phytoph‐thora infestans on potato plants in field,laboratory and greenhouse conditions[J] . Journal of Agricultural Science,2013,5(5):148-157.
[43] FORBES G A. Using host resistance to manage potato late blight with particular reference to developing countries[J] . Potato Research,2012,55(3):205-216.
[44] EULGEM T,SOMSSICH I E. Networks of WRKY transcription factors in defense signaling[J] . Current Opinion in Plant Biology,2007,10(4):366-371.
[45] SHAN Y F,LI M Y,WANG R Z,LI X G,LIN J,LI J M,ZHAO K J,WU J. Evaluation of the early defoliation trait and identification of resistance genes through a comprehensive transcriptome analysis in pears[J] . Journal of Integrative Agriculture,2023,22(1):120-138.