枣(Ziziphus jujuba Mill.)是鼠李科枣属植物,源产于中国,是中国传统特色果品之一,已有7000多年的栽培历史[1]。酸枣又名山枣、荆棘、野荆棘等,多处于野生环境条件,作为适应性极强的古生植物,主要分布在我国黄河流域中下游地区[2]。酸枣是中国特有的生态经济型树种,开发潜力大。酸枣仁是重要的药材,酸枣果肉含有多糖、氨基酸、黄酮、维生素等营养元素,具有较高的营养价值[3]。酸枣特有的萌蘖能力和发达的根系,增强了其对环境的适应能力及繁殖能力,对抗旱、水土保持、防风固沙和绿化荒山起到十分重要的作用。此外,酸枣富含抗逆基因,作为枣的优良砧木,与其他枣树嫁接,可发挥自身优良的抗旱耐盐碱、根系深广发达的特性[4]。
5-氨基乙酰丙酸(ALA)是叶绿素、细胞色素、光敏素等重要生物分子的关键合成前体,在植物生长发育中起着重要作用。此外,ALA通过调节植物的生长状态,提高植物的抗逆性和适应能力[5],在植物的抗氧化代谢中扮演着非常重要的角色[6]。ALA处理过的小白菜叶片中抗氧化酶如超氧化物歧化酶(SOD)等的活性升高,同时叶绿素含量和光合速率也明显升高[7]。此外,研究发现低浓度ALA可促进某些关键抗氧化酶活性的升高,如抗坏血酸过氧化物酶(APX)、SOD和过氧化氢酶(CAT)等的活性,表明低浓度的ALA能够有效地提高植物的抗氧化能力[8]。
microRNA(miRNA)是一类源自真核生物的内源性非编码小分子RNA,长度为18~24个核苷酸。miRNA通常在植物转录后发挥负调控功能[9-10],主要通过靶基因切割降解或抑制翻译两种方式[11],在植物的生长发育、非生物胁迫和生物胁迫等生物过程中发挥作用[12]。现有研究表明,在植物对非生物胁迫(如温度[13]、干旱[14]和盐胁迫[15])的响应过程中,miRNA起着重要调控作用[16]。目前关于提高作物抗逆性的研究主要集中在光合作用、叶绿素合成[7]、抗氧化系统[17-18]和离子吸收[19]方面。然而,关于ALA如何通过miRNA缓解酸枣盐胁迫的研究少有报道。因此,本研究以酸枣组培幼苗为试验材料,设置CK、ALA、NaCl及ALA+NaCl等4个处理。通过miRNA测序,构建12个sRNA文库,鉴定响应盐胁迫的miRNA,解析miRNA及其核心靶基因在ALA缓解盐胁迫中的作用。
1 材料和方法
1.1 试验材料与处理
以酸枣(Ziziphus jujuba var. spinosa)组培幼苗为试验材料。组培苗生根后,选择长势一致的幼苗移至水培盒中培养,温度为28 ℃(昼)/20 ℃(夜),湿度为55% (昼)/85%(夜),光照度12 000 lx,每隔3 d更换一次营养液。长至第6枚叶片完全展开、大小均一的酸枣组培幼苗进行试验,共设置4个处理:(1)叶面喷施蒸馏水(CK);(2)叶面喷施100 mg·L-1 ALA(ALA);(3)添加150 mmol·L-1 NaCl溶液和叶面喷施蒸馏水(NaCl);(4)添加150 mmol·L-1 NaCl溶液和叶面喷施100 mg·L-1 ALA(ALA+NaCl)。每个处理3次重复,每个重复15株酸枣苗。为避免盐激效应,以每天50 mmol·L-1 NaCl溶液的梯度逐渐增加至150 mmol·L-1 NaCl溶液,记为处理的第0天。每个处理重复分别标注为CK-1、CK-2、CK-3、ALA-1、ALA-2、ALA-3、NaCl-1、NaCl-2、NaCl-3、ALA+NaCl-1、ALA+NaCl-2、ALA+NaCl-3。于处理第6天进行一次性取样。将每个重复的15株幼苗叶片混合后,用锡纸包好于-80 ℃冰箱保存。酸枣幼苗具体培养过程、NaCl(150 mmol·L-1)和ALA(100 mg·L-1)所施用的浓度及取样时间均为本课题组前期试验筛选确定的结果[20-21]。
1.2 形态指标和生理生化指标测定
使用游标卡尺从基部测量植株生长量。使用根系扫描仪(Winrhizo Pro,Quebec,Canada)进行扫描获得图像,使用扫描仪配套的WinRHIZO Pro根系结构分析系统分析根系相关指标。POD活性使用微量检测试剂盒(科铭,中国苏州)并按照操作说明进行测定。
1.3 small RNA文库的构建及测序分析
酸枣叶片总RNA提取、RNA的质量和浓度检测由百迈克生物科技有限公司(百迈克生物,中国北京)完成。构建12个small RNA(sRNA)文库,使用Illumina平台测序。
1.4 sRNA注释和miRNA鉴定
对测序获得的原始数据(raw data)进行处理,计算出clean tags的长度分布。使用bowtie [22]将长度筛选后的sRNA定位到参考序列上,分析small RNA在参考序列上的分布情况。使用miRNA预测软件mirdeep2[23]对新miRNA进行分析,预测样品中novel miRNA。
1.5 miRNA差异基因筛选
基于MA-plot法[24-25]计算差异表达基因(DEGs)。每个基因的P值再用Q-value进行多重假设检验校正,对于符合差异在2倍以上且Q-value值≤0.001,认为是显著的差异表达基因。
1.6 靶基因功能富集分析
利用TargetFinder软件[26]进行miR408a靶基因预测,通过BLAST软件将预测靶基因序列与GO[27]、KEGG[28]数据库进行比对获得靶基因的注释信息,筛选特定生物学功能的基因。
1.7 差异表达的miRNA及其靶基因的RT-qPCR验证
按照Trizol(Invitrogen,CA,United States)的标准方案,从CK、ALA、NaCl及ALA+NaCl处理后的叶片中提取总RNA。使用PrimeScript™RT reagent Kit(TaKaRa)试剂盒去除gDNA并进行反转录。用于qRT-PCR的基因特异性引物由Vector NTI 10软件设计,以CK处理的叶片作为参考样本,将基因的相对表达量设为1。引物设计序列如表1所示,以18sRNA为内参基因,相对表达量采用2-ΔΔCt法计算[29]。
表1 qRT-PCR基因所用引物
Table 1 Sequences of gene for qRT-PCR
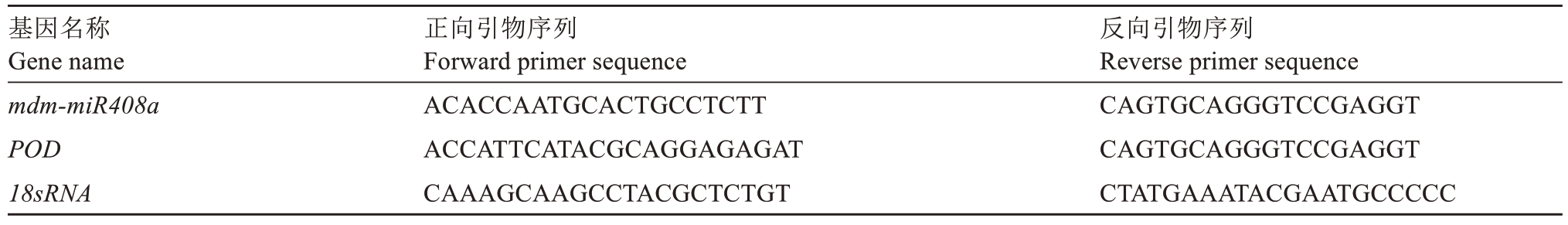
基因名称Gene name mdm-miR408a POD 18sRNA正向引物序列Forward primer sequence ACACCAATGCACTGCCTCTT ACCATTCATACGCAGGAGAGAT CAAAGCAAGCCTACGCTCTGT反向引物序列Reverse primer sequence CAGTGCAGGGTCCGAGGT CAGTGCAGGGTCCGAGGT CTATGAAATACGAATGCCCCC
1.8 统计分析
使用Microsoft Excel 2010、Origins 2018和SPSS 19.0(IBM)对试验数据进行统计和多重比较分析。图表中的数据以平均值±标准差表示,不同小写字母代表各处理间的差异显著(P<0.05)。
2 结果与分析
2.1 试验处理后酸枣幼苗表型
由图1-A~D可知,与CK相比,NaCl处理显著降低了酸枣幼苗的株高、根长、根表面积和根体积(51.63%、47.91%、35.75%和34.98%),单独外源喷施ALA处理对酸枣幼苗地上部生长和根系无显著影响;与NaCl处理相比,ALA+NaCl处理显著提高了酸枣幼苗的株高、根长、根表面积和根体积(36.54%、48.58%、17.07%和26.12%)。酸枣幼苗处理后形态图(图1-E)显示,与CK相比,外源喷施ALA酸枣幼苗的地上部无显著变化,根系侧根增加;NaCl处理后酸枣幼苗出现失水萎蔫的现象,根系形态受到显著抑制;与单独的NaCl处理相比,ALA+NaCl共同处理可缓解酸枣幼苗所受的损伤。由此可知,NaCl处理显著抑制了酸枣幼苗的生长和根系形态,外源喷施ALA能够显著地缓解NaCl胁迫对酸枣幼苗的伤害。

图1 胁迫处理后酸枣植株生长形态
Fig. 1 Growth pattern of treated wild jujube plants
2.2 酸枣sRNA文库测序与分析
为了鉴定能够响应盐胁迫的miRNAs,酸枣CK、ALA、NaCl及ALA+NaCl 4个处理共构建了12个sRNA文库,利用Illumina平台对文库进行测序分析。在所测12个文库中,CK-1、CK-2、CK-3、ALA-1、ALA-2、ALA-3、NaCl-1、NaCl-2、NaCl-3、ALA+NaCl-1、ALA+NaCl-2和ALA+NaCl-3分别产生的raw reads为:38.54 M、45.74 M、40.17 M、39.61 M、43.32 M、40.95 M、39.33 M、43.31 M、37.98 M、40.12 M、43.78 M 和39.86 M。经过杂质的过滤,去除低质量序列和polyA序列得到clean reads分别为38.21 M、45.51 M、39.97 M、39.36 M、43.09 M、40.79 M、38.98 M、43.07 M、37.8 M、39.94 M、43.55 M和39.45 M。各样品Q30均≥ 93%,将所有clean reads与参考基因组GCF_000826755.1_ZizJuj_1.1_genomic.fna进行比对,成功比对到基因的reads占clean reads的比例为86.46%~93.12%,数据质量符合分析要求,可进行后续分析(表2)。
表2 酸枣叶片测序质量控制
Table 2 Sequencing quality control of sour date leaves
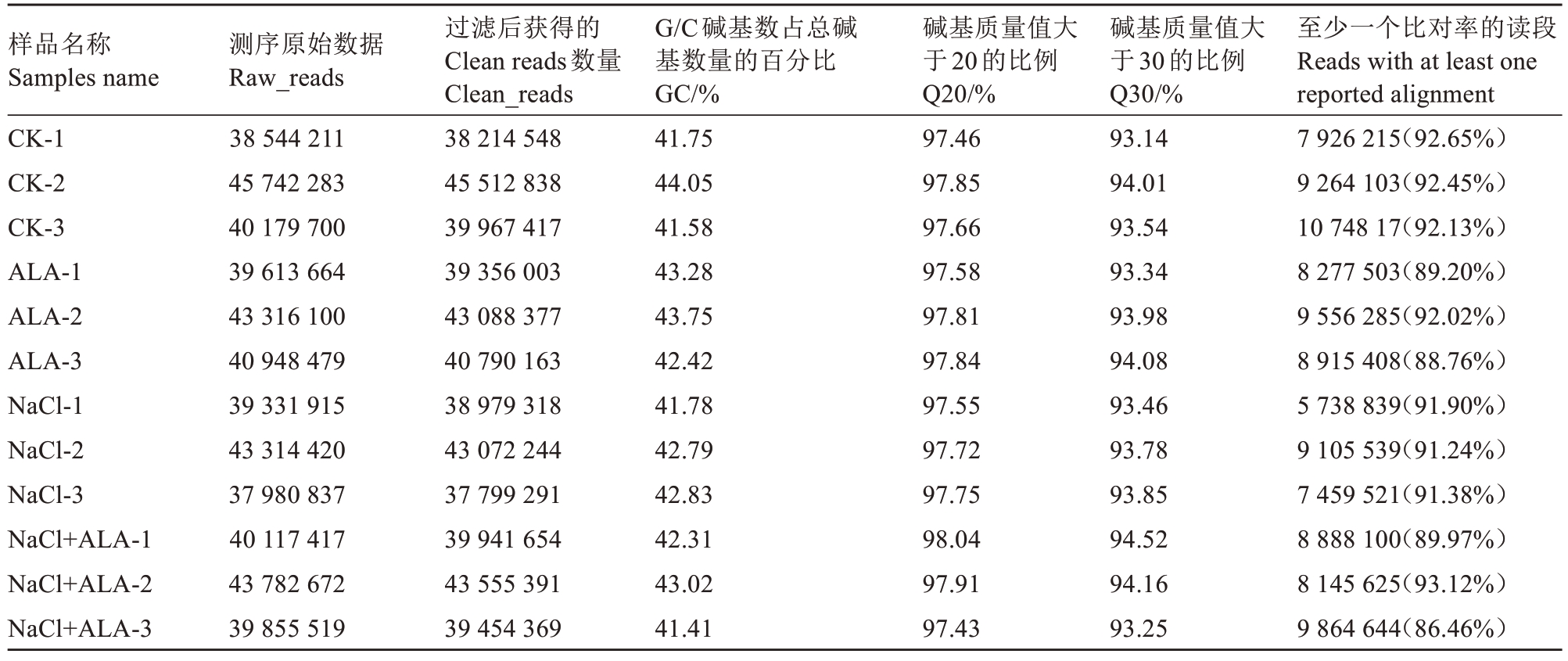
样品名称Samples name CK-1 CK-2 CK-3 ALA-1 ALA-2 ALA-3 NaCl-1 NaCl-2 NaCl-3 NaCl+ALA-1 NaCl+ALA-2 NaCl+ALA-3测序原始数据Raw_reads 38 544 211 45 742 283 40 179 700 39 613 664 43 316 100 40 948 479 39 331 915 43 314 420 37 980 837 40 117 417 43 782 672 39 855 519过滤后获得的Clean reads数量Clean_reads 38 214 548 45 512 838 39 967 417 39 356 003 43 088 377 40 790 163 38 979 318 43 072 244 37 799 291 39 941 654 43 555 391 39 454 369 G/C碱基数占总碱基数量的百分比GC/%41.75 44.05 41.58 43.28 43.75 42.42 41.78 42.79 42.83 42.31 43.02 41.41碱基质量值大于20的比例Q20/%97.46 97.85 97.66 97.58 97.81 97.84 97.55 97.72 97.75 98.04 97.91 97.43碱基质量值大于30的比例Q30/%93.14 94.01 93.54 93.34 93.98 94.08 93.46 93.78 93.85 94.52 94.16 93.25至少一个比对率的读段Reads with at least one reported alignment 7 926 215(92.65%)9 264 103(92.45%)10 748 17(92.13%)8 277 503(89.20%)9 556 285(92.02%)8 915 408(88.76%)5 738 839(91.90%)9 105 539(91.24%)7 459 521(91.38%)8 888 100(89.97%)8 145 625(93.12%)9 864 644(86.46%)
2.3 酸枣叶片miRNAs的鉴定
在这12个文库中,已知的miRNA表达水平也可以通过read content的频率来体现。根据reads content将不同处理组中已知miRNAs的表达水平分为8类:无表达(0 reads)、最低(>0~10 reads)、较低(>10~50 reads)、低(>50~100 reads)、中等(>100~500 reads)、高(>500~1000 reads)、较高(>1000~10 000 reads)和最高(>10 000 reads)。本研究结果显示,大多数文库中属于这8类的已知miRNAs的百分比高度相似。已知microRNA中最低(>0~10 reads)类别分布最多,在不同文库中的比例分别为18.3%、21.71%、23.49%。而在ALA vs CK、NaCl vs CK、ALA+NaCl vs CK中,最高(>10 000 reads)类别所占比例最小,分别为8.5%、8.55%、12.08%(图2-A)。为鉴定酸枣叶片中已知的miRNA,通过对miRBase18.0(http://www.mirbase.org)数据库进行比对,只考虑精确匹配的序列,共鉴定出149个已知miRNA,属于31个miRNA家族。其中,不同家族的miRNA成员数量相似,大多数保守的miRNA家族成员为1~6个。大多数已知miRNA家族都有多个成员,如miR156(26个成员)、miR167(10个成员)、miR171(11个成员)、miR171(11个成员)和miR395(9个成员)(图2-B)。sRNAs的长度主要在21~24nt之间,其中24nt sRNAs在12个文库中最丰富(图2-C)。使用miRdeep2从不同处理的酸枣叶片中鉴定出11个新的miRNA,新miRNA成熟序列的信息见表3,一些novel miRNA的二级结构如图2-D所示。
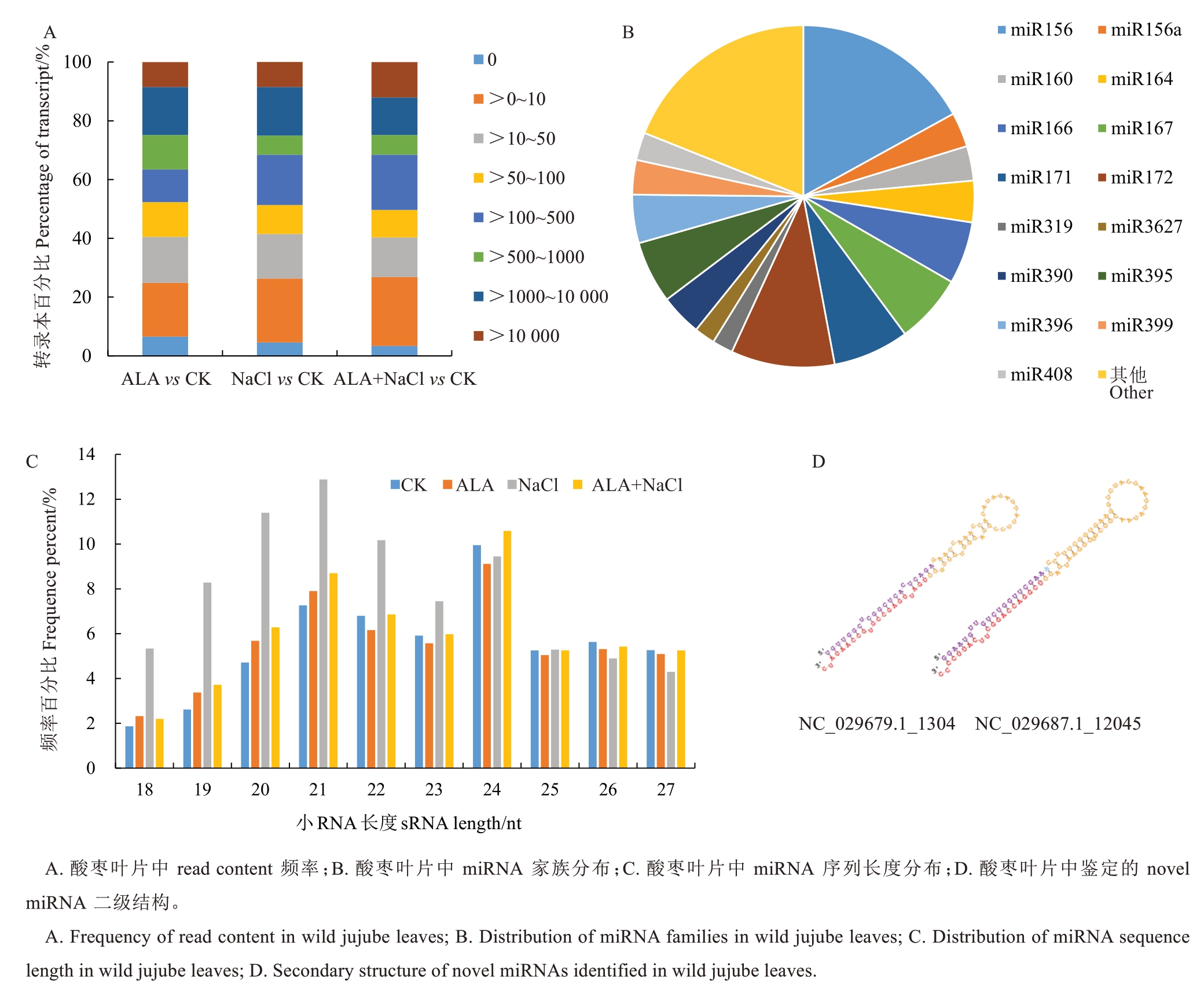
图2 酸枣叶片中miRNA家族鉴定
Fig. 2 Identification of miRNA families in wild jujube leaves
表3 酸枣叶片中鉴定的新miRNAs
Table 3 Novel miRNAs identified in sour date leaves
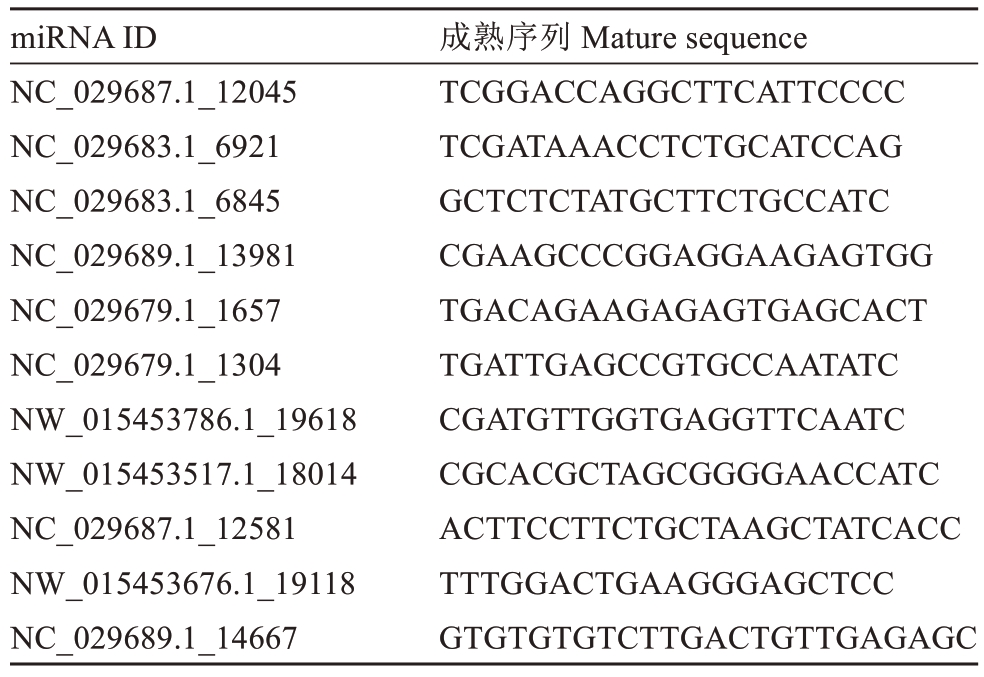
miRNA ID NC_029687.1_12045 NC_029683.1_6921 NC_029683.1_6845 NC_029689.1_13981 NC_029679.1_1657 NC_029679.1_1304 NW_015453786.1_19618 NW_015453517.1_18014 NC_029687.1_12581 NW_015453676.1_19118 NC_029689.1_14667成熟序列 Mature sequence TCGGACCAGGCTTCATTCCCC TCGATAAACCTCTGCATCCAG GCTCTCTATGCTTCTGCCATC CGAAGCCCGGAGGAAGAGTGG TGACAGAAGAGAGTGAGCACT TGATTGAGCCGTGCCAATATC CGATGTTGGTGAGGTTCAATC CGCACGCTAGCGGGGAACCATC ACTTCCTTCTGCTAAGCTATCACC TTTGGACTGAAGGGAGCTCC GTGTGTGTCTTGACTGTTGAGAGC
2.4 酸枣叶片中miRNAs表达定量分析
对各样本中已知和新miRNA进行表达量的统计,并用TPM进行表达量归一化处理。miRNA表达的分布描述了每个样品中总体miRNA表达模式。在每个样本中,miRNA的log10(TPM)+1主要在1~3之间(图3-A~C)。每个样品中TPM密度的分布如图3-D~F所示。样品3次重复中miRNA的表达水平趋于一致,表明生物学重现性是可靠的。
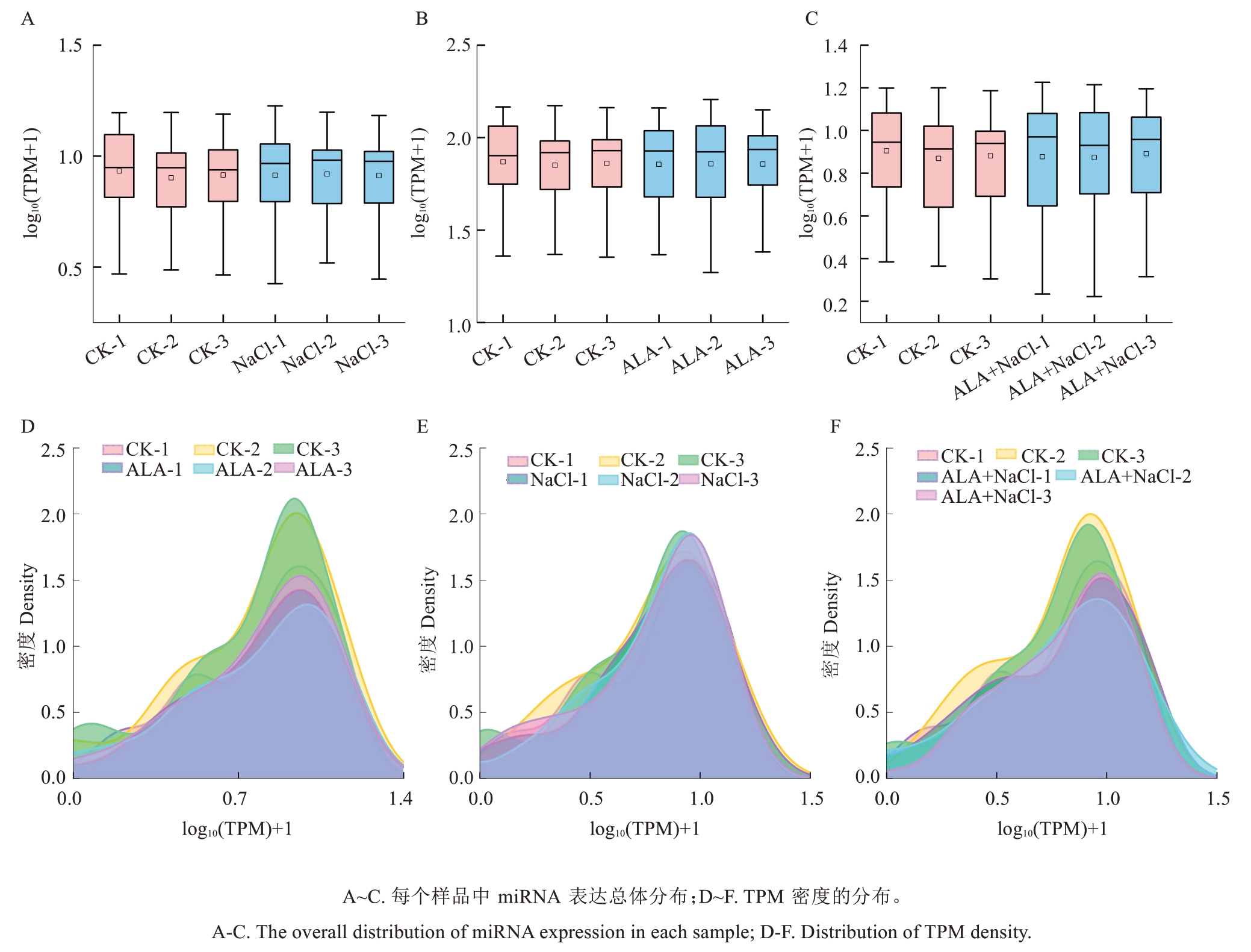
图3 每个样品中的miRNA表达状态
Fig. 3 miRNA expression status in each sample
2.5 差异miRNA分析
为鉴定CK、ALA、NaCl和ALA+NaCl处理组中差异表达的miRNAs,笔者对miRNAs的差异表达模式进行层次聚类分析和统计学比较分析。结果表明,与CK相比,ALA处理组中有153个miRNA差异表达,包括94个下调基因和59个上调基因;NaCl组中有152个miRNA差异表达,包括76个下调基因和76个上调基因;ALA+NaCl组中有149个miRNA差异表达,包括78个下调基因和71个上调基因(表4)。许多响应盐胁迫的miRNAs,如zju-miR172d、zju-miR395i和zju-miR172a均显著下调,表明这些miRNAs可能受盐胁迫抑制。进一步分析发现,ALA显著诱导zju-miR391表达,并抑制zju-miR408a、zju-miR398b的表达,进一步证明了miRNA在ALA缓解盐胁迫调控作用中的复杂性(图4)。
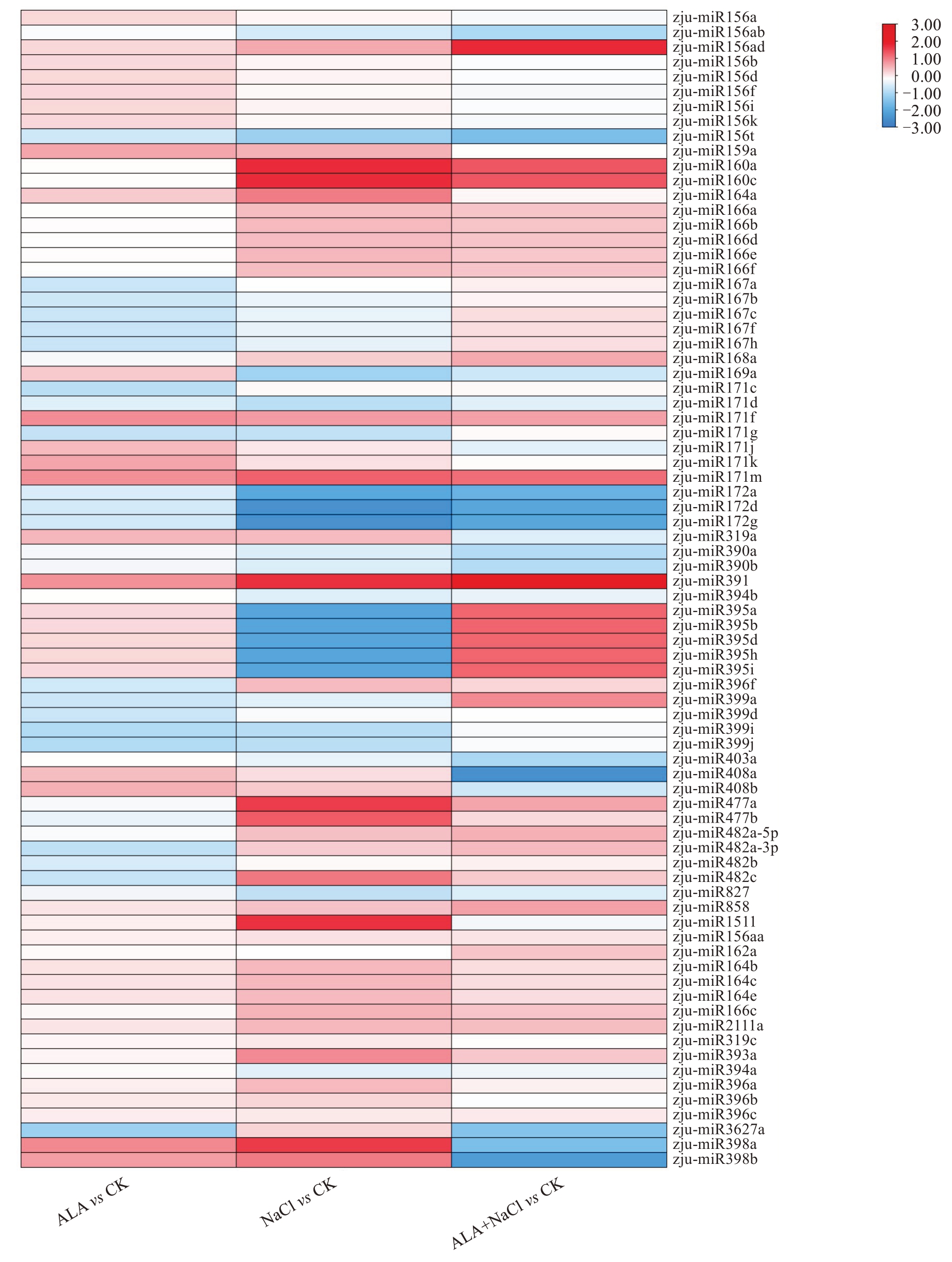
图4 酸枣叶片中各比较组中差异miRNA的表达水平
Fig. 4 Expression levels of differential miRNAs in each comparative group in leaves of wild jujube leaves
表4 叶片差异miRNA数量统计
Table 4 Number statistics of differential miRNA in leaves
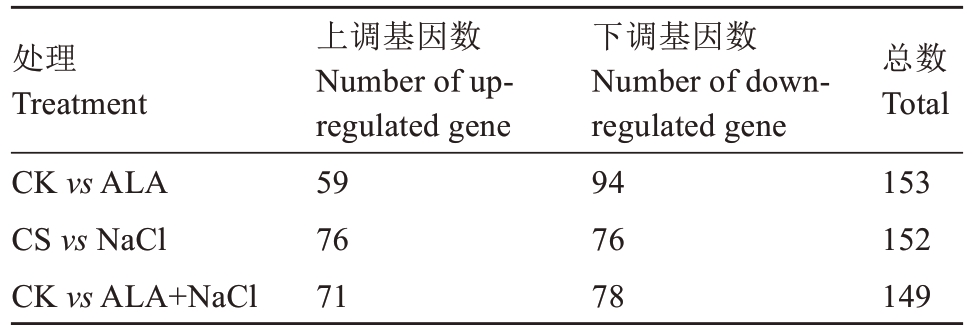
处理Treatment CK vs ALA CS vs NaCl CK vs ALA+NaCl上调基因数Number of upregulated gene 59 76 71下调基因数Number of downregulated gene 94 76 78总数Total 153 152 149
2.6 差异基因功能富集分析及响应NaCl胁迫的miRNA408a靶基因预测
差异miRNA靶基因进行GO富集分析,结果表明,miRNA靶基因参与多种生物学过程,包括生物调节、细胞过程、代谢过程、信号转导和其他生物学过程。此外,miRNAs靶基因涉及分子功能包括转录因子活性和催化活性。KEGG数据库分析发现,靶基因参与了代谢、遗传信息过程、组织系统、环境信息过程和细胞过程5个不同的功能。在这些靶基因中,发现了与其他代谢途径有关的靶基因,包括TMV抗性蛋白、含SPX结构域的膜蛋白、钠钾转运蛋白、核糖体RNA处理蛋白的转录因子,如生长素应答因子(ARF)家族、MYB转录因子、WRKY转录因子(图5)。

图5 zju-miR408a靶基因富集分析
Fig. 5 zju-miR408a target gene enrichment analysis
对差异表达miRNA分析发现,无论是否经过NaCl胁迫处理,ALA处理均能显著下调zjumiR408a的表达水平。这表明酸枣zju-miR408a可能参与了ALA介导的缓解NaCl胁迫的过程。为进一步了解miR408a在NaCl胁迫中的功能,对zjumiR408进行靶基因预测(表5),发现酸枣中zjumiR408a靶基因并不相同,zju-miR408a的靶基因包括TMV抗性蛋白N样、原卟啉原氧化酶,线粒体、过氧化物酶4、木质素形成阴离子过氧化物酶、钙依赖性蛋白激酶1、漆酶和过氧化物酶等。因此,笔者推测ALA通过介导zju-miR408a对POD活性进行调控,增强植物耐盐性,从而缓解NaCl胁迫的影响。
表5 zju-miR408a靶基因预测结果
Table 5 zju-miR408a target gene prediction results

基因ID Gene ID LOC107403307 LOC107404868 LOC107405097 LOC107405832 LOC107406893 LOC107407356 LOC107407364 LOC107408008 LOC107410427 LOC107410617 LOC107410668 LOC107411129 LOC107412498 LOC107415375 LOC107415564 LOC107416117 LOC107416183 LOC107417746 LOC107419839 LOC107421706 LOC107423464 LOC107425191 LOC107426494 LOC107426842 LOC107427104 LOC107429130 LOC107430444 LOC107430555 LOC107430628 LOC107431265 LOC107431646 LOC107431819 LOC107432145 LOC107433279 LOC107434080 LOC107434313 LOC107435508注释结果Annotation results TMV抗性蛋白N样 TMV resistance protein N-like原卟啉原氧化酶,线粒体 Protoporphyrinogen oxidase, mitochondrial过氧化物酶4 Peroxidase 4木质素形成阴离子过氧化物酶 Lignin-forming anionic peroxidase Tubby样F-box蛋白8 Tubby-like F-box protein 8谷胱甘肽-C9 Glutaredoxin-C9可能的转录因子WRKY 51 Probable WRKY transcription factor 51转录因子MYB3R-1 Transcription factor MYB3R-1 La相关蛋白1A La-related protein 1A L-抗坏血酸氧化酶同源物 L-ascorbate oxidase homolog过氧化物酶 P7 Peroxidase P7可能的LRR受体样丝氨酸/苏氨酸蛋白激酶 At5g45780 Probable LRR receptor-like serine/threonine-protein kinase At5g45780 F-box/kelch-repeat 蛋白 At3g06240 F-box/kelch-repeat protein At3g06240-like钾转运蛋白8 Potassium transporter 8依赖ATP的锌金属蛋白酶 FTSH 6,叶绿体 ATP-dependent zinc metalloprotease FTSH 6, chloroplastic双孔钾通道3 Two-pore potassium channel 3 ALA-相互作用亚基1 ALA-interacting subunit 1过氧化物酶47 Peroxidase 47过氧化氢酶同工酶1 Catalase isozyme 1 NAD依赖性苹果酸酶62 kDa同工酶,线粒体 NAD-dependent malic enzyme 62 kDa isoform, mitochondrial F-box蛋白SKP2A F-box protein SKP2A钙调蛋白结合转录激活因子3 Calmodulin-binding transcription activator 3 LRR类受体丝氨酸/苏氨酸蛋白激酶RGI2 LRR receptor-like serine/threonine-protein kinase RGI2钠转运体HKT1 Sodium transporter HKT1 F-box蛋白At1g10780 F-box protein At1g10780热休克蛋白90-5,叶绿体 Heat shock protein 90-5, chloroplastic钙依赖性蛋白激酶1 Calcium-dependent protein kinase 1漆酶17 Laccase-17-like漆酶2 Laccase-2过氧化物酶P7 Peroxidase P7 NADPH 依赖性硫氧还原酶3 NADPH-dependent thioredoxin reductase 3过氧化物酶体脂肪酸β-氧化多功能蛋白AIM1 Peroxisomal fatty acid beta-oxidation multifunctional protein AIM1 LRR受体样丝氨酸/苏氨酸蛋白激酶FEI 1 LRR receptor-like serine/threonine protein kinase FEI 1谷胱甘肽S转移酶F13 Glutathione S-transferase F13抗病蛋白RUN1 Disease resistance protein RUN1 NADH脱氢酶 NADH dehydrogenase可能的转录因子WRKY20 Probable WRKY transcription factor 20
2.7 酸枣叶片POD酶活性及zju-miRNA408a和靶基因qRT-PCR验证
为验证测序结果中zju-miRNA408a对ALA和NaCl胁迫的动态响应在生物学层面的一致性,本研究采用qRT-PCR技术分析了ALA处理及NaCl胁迫条件下该miRNA的表达水平。测序数据与qRTPCR结果呈现出高度一致的表达变化趋势,证实了miRNA-Seq检测结果的可靠性,且与预期试验假设完全吻合(图6)。靶基因POD的表达量及POD酶活性进一步证实了ALA可能通过下调zju-miRNA408a的表达来上调POD基因的表达量从而提高POD酶活性来增强酸枣耐盐能力。
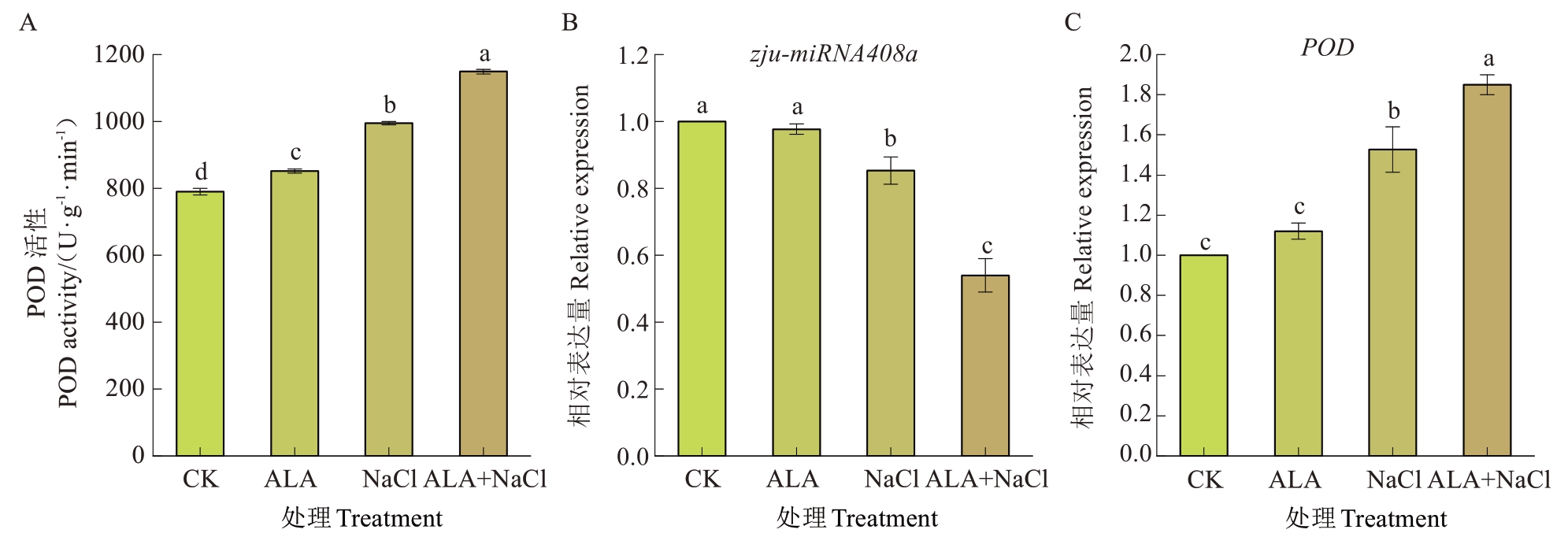
图6 酸枣叶片POD酶活性(A)和zju-miRNA408a(B)、靶基因POD(C)的qRT-PCR验证
Fig. 6 POD enzyme activity (A) and qRT-PCR validation of zju-miRNA408a (B), target genes POD (C)in wild jujube leaves
3 讨 论
盐胁迫是一种在自然环境中广泛存在的非生物胁迫形式。由于不合理的灌溉、施肥和气候变化,受盐胁迫影响的土地面积正在增加。长期的盐胁迫会抑制植物生长,降低作物产量,最终降低农产品的经济效益和土地利用效率[30]。幼苗期作为植物生长发育的起点,盐胁迫对其的影响程度决定了植物能否顺利度过苗期。周琦等[31]研究发现鹅耳枥在盐胁迫下随着盐浓度的增加,生长势受到抑制,这一结果与施娴等[32]和于伟[33]的研究结果一致,表明酸枣在盐胁迫下生长受到抑制,在喷施ALA后抑制作用得到缓解。
miRNA及其靶基因在植物抵御盐胁迫过程中可通过调控种子的萌发、根系生长的改变、生长周期变化、活性氧清除等过程,缓解胁迫对植物的损伤[34]。植物体内叶绿素生物合成的前体物质是ALA,外源施加ALA可以提高植物抗氧化能力,降低膜脂过氧化的损害程度,提高植物对非生物胁迫的抵抗能力[35]。Kanto等[36]推测5-ALA可作为抗氧化酶活性的启动子,促进抗氧化酶活性升高。在本试验中,ALA处理后酸枣幼苗叶片的POD酶活性均高于CK,外源喷施ALA后可进一步提高酸枣幼苗叶片的POD酶活性,这与王亚新等[37]的研究结果一致。ALA还可以提高植物保护酶活性,减轻膜脂质过氧化程度,保持膜系统的完整性,增强植株抵御逆境的能力。
本研究探讨了ALA通过调控miRNAs表达在增强酸枣盐胁迫耐受性中的潜在作用。对以CK、ALA处理、NaCl处理和ALA+NaCl处理的试验材料测定了miRNA的表达情况,鉴定出149个已知miRNA(属于31个miRNA家族)和11个预测的novel miRNA。获得的这些miRNAs的序列符合miRNAs的二级结构标准。12个文库中sRNAs的长度主要在21~24nt之间,其中24nt sRNAs在12个文库中最丰富,推测他们可能在盐胁迫缓解效应中发挥关键作用。通过更深入地研究sRNA转录组,发现sRNA的大小分布与已发表的植物非常相似,如拟南芥[38],辣椒[39]和三叶橙[40],比例最高的是21-24nt sRNA。此外,已知的149个miRNA属于31个保守miRNA家族,这些miRNAs广泛分布于双子和单子叶植物中[41]。这些已知的miRNAs家族成员不止一个,如miR156(26个成员)、miR167(10个成员)miR171(11个成员)和miR395(9个成员)。
miRNA主要通过抑制靶基因的翻译或者对靶基因特异性切割来行使其功能。miR408是一类高度保守、古老的基因家族,在植物应对非生物胁迫时发挥着重要的作用[42]。现有研究中,miR408主要靶向车前花青素、漆酶(LAC)、AFG1和TaCLP1等合成基因[43]。研究表明,拟南芥中,AGO1和AGO2参与调控miR408介导的花青素合成[44];在蒺藜状苜蓿中miR408靶向质体蓝素蛋白响应水分胁迫[45]。笔者发现ALA显著抑制了zju-miR408a的表达,以酸枣转录组unigene为靶标预测的数据库,预测到酸枣zju-miR408家族的靶基因包括原卟啉原氧化酶、过氧化物酶4和漆酶等。qPCR结果也显示,zjumiR408a和靶基因在4个处理中的表达量均有显著差异。因此,笔者推测ALA可能通过抑制zjumiR408a的表达来正向调控POD活性,从而增强植物对盐胁迫的耐受性。该研究结果为进一步解析酸枣miR408在盐胁迫缓解中的功能机制奠定了基础。
4 结 论
笔者在本研究中使用高通量测序构建了不同处理(对照、ALA、NaCl和ALA+NaCl)下酸枣叶片的小RNA文库。对文库数据分析,鉴定到149个已知的miRNA和11个新的miRNA。许多响应盐胁迫的miRNA,如zju-miR172d、zju-miR395i和zju-miR172a,表达显著下调,表明它们可能被盐胁迫抑制。进一步分析发现,ALA显著抑制了zju-miR408a的表达,其靶基因主要包括过氧化物酶和原卟啉原氧化酶。基于酸枣叶片POD酶活性、zju-miRNA408a及其靶基因POD的qRT-PCR验证结果,表明ALA通过下调zju-miR408a表达来提高POD酶活性,进而清除活性氧(ROS),最终增强酸枣的盐胁迫耐受性。
[1] 刘凤之,王海波,李莉,宣景宏. 我国设施果树产业现状、存在问题与发展对策[J] . 中国果树,2021(11):1-4.LIU Fengzhi,WANG Haibo,LI Li,XUAN Jinghong. Current situation,issue and suggestion of the protected fruit industry in China[J] . China Fruits,2021(11):1-4.
[2] 李会军,李萍,余国奠. 酸枣的研究进展及开发前景[J] . 中国野生植物资源,1999,18(3):15-19.LI Huijun,LI Ping,YU Guodian. Advance of research and prospect of development of Zizyphus jujuba var. spinosa (Bunge) Hu ex H F Chow[J] . Chinese Wild Plant Resources,1999,18(3):15-19.
[3] 马进杰,刘萍,马百平. 酸枣仁化学成分及其镇静催眠作用研究进展[J] . 国际药学研究杂志,2011,38(3):206-211.MA Jinjie,LIU Ping,MA Baiping. The chemical constituents of Semen Zizyphi Spinosae and pharmacologic mechanism of its sedative and hypnotic effects:Research advances[J] . Journal of International Pharmaceutical Research,2011,38(3):206-211.
[4] 赵天娇. 圣冠枣接穗对酸枣实生砧木根系生长及生理特性的影响[D] . 长沙:中南林业科技大学,2022.ZHAO Tianjiao. Scion of Ziziphus spina-christi effects on growth and physiological characteristics of seedling stock roots in Ziziphus acidojujuba[D] . Changsha:Central South University of Forestry & Technology,2022.
[5] WU Y,LIAO W B,DAWUDA M M,HU L L,YU J H. 5-Aminolevulinic acid (ALA) biosynthetic and metabolic pathways and its role in higher plants:A review[J] . Plant Growth Regulation,2019,87(2):357-374.
[6] AKRAM N A,ASHRAF M,AL-QURAINY F. Aminolevulinic acid-induced changes in some key physiological attributes and activities of antioxidant enzymes in sunflower (Helianthus ann‐uus L.) plants under saline regimes[J] . Scientia Horticulturae,2012,142:143-148.
[7] MEMON S A,HOU X L,WANG L J,LI Y. Promotive effect of 5-aminolevulinic acid on chlorophyll,antioxidative enzymes and photosynthesis of pakchoi (Brassica campestris ssp. chinen‐sis var. communis Tsen et Lee)[J] . Acta Physiologiae Plantarum,2009,31(1):51-57.
[8] XU F,CHANG J,CHENG S Y,ZHU J,LI L L,WANG Y,CHENG H. Promotive effect of 5-aminolevulinic acid on the antioxidant system in Ginkgo biloba leaves[J] . African Journal of Biotechnology,2009,8(16):3769-3776.
[9] BARTEL D P. MicroRNAs:Genomics,biogenesis,mechanism,and function[J] . Cell,2004,116(2):281-297.
[10] VOINNET O. Origin,biogenesis,and activity of plant micro-RNAs[J] . Cell,2009,136(4):669-687.
[11] ZHANG B H. MicroRNA:A new target for improving plant tolerance to abiotic stress[J] . Journal of Experimental Botany,2015,66(7):1749-1761.
[12] SUNKAR R,LI Y F,JAGADEESWARAN G. Functions of microRNAs in plant stress responses[J] . Trends in Plant Science,2012,17(4):196-203.
[13] JYOTHI M N,RAI D V,NAGESH BABU R. Identification and characterization of high temperature stress responsive novel miRNAs in French bean (Phaseolus vulgaris)[J] . Applied Biochemistry and Biotechnology,2015,176(3):835-849.
[14] BARRERA-FIGUEROA B E,GAO L,DIOP N N,WU Z G,EHLERS J D,ROBERTS P A,CLOSE T J,ZHU J K,LIU R Y.Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes[J] . BMC Plant Biology,2011,11:127.
[15] LIU H H,TIAN X,LI Y J,WU C G,ZHENG C C. Microarraybased analysis of stress-regulated microRNAs in Arabidopsis thaliana[J] . RNA,2008,14(5):836-843.
[16] MORO B,CHOROSTECKI U,ARIKIT S,SUAREZ I P,HÖBARTNER C,RASIA R M,MEYERS B C,PALATNIK J F.Efficiency and precision of microRNA biogenesis modes in plants[J] . Nucleic Acids Research,2018,46(20):10709-10723.
[17] NISHIHARA E,KONDO K,PARVEZ M M,TAKAHASHI K,WATANABE K,TANAKA K. Role of 5-aminolevulinic acid(ALA) on active oxygen-scavenging system in NaCl-treated spinach (Spinacia oleracea)[J] . Journal of Plant Physiology,2003,160(9):1085-1091.
[18] SUN Y P,ZHANG Z P,WANG L J. Promotion of 5-aminolevulinic acid treatment on leaf photosynthesis is related with increase of antioxidant enzyme activity in watermelon seedlings grown under shade condition[J] . Photosynthetica,2009,47(3):347-354.
[19] NAEEM M S,RASHEED M,LIU D,JIN Z L,MING D F,YONEYAMA K,TAKEUCHI Y,ZHOU W J. 5-Aminolevulinic acid ameliorates salinity-induced metabolic,water-related and biochemical changes in Brassica napus L.[J] . Acta Physiologiae Plantarum,2011,33(2):517-528.
[20] 常心怡,孙军利,赵宝龙,李芳芳,刘连玲,何旺. 外源ALA对NaCl胁迫下酸枣幼苗光合特性与膜脂过氧化的影响[J] . 新疆农业科学,2019,56(9):1635-1644.CHANG Xinyi,SUN Junli,ZHAO Baolong,LI Fangfang,LIU Lianling,HE Wang. Effects of exogenous ALA on photosynthesis and membrane peroxidation in the leaves of jujube seedlings under NaCl treatment[J] . Xinjiang Agricultural Sciences,2019,56(9):1635-1644.
[21] ZHU C M,LIU Z Y,CHANG X Y,ZHANG Z J,SHI W C,ZHANG Z R,ZHAO B L,SUN J L. Analysis of the alternative splicing events of exogenous δ-aminolevulinic acid under NaCl stress in wild jujube seedlings[J] . Forests,2022,13(12):2076.
[22] LANGMEAD B,TRAPNELL C,POP M,SALZBERG S L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome[J] . Genome Biology,2009,10(3):R25.
[23] FRIEDLÄNDER M R,MACKOWIAK S D,LI N,CHEN W,RAJEWSKY N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades[J] . Nucleic Acids Research,2012,40(1):37-52.
[24] WANG L K,FENG Z X,WANG X,WANG X W,ZHANG X G. DEGseq:An R package for identifying differentially expressed genes from RNA-seq data[J] . Bioinformatics,2010,26(1):136-138.
[25] YANG Y H,DUDOIT S,LUU P,LIN D M,PENG V,NGAI J,SPEED T P. Normalization for cDNA microarray data:A robust composite method addressing single and multiple slide systematic variation[J] . Nucleic Acids Research,2002,30(4):e15.
[26] ALLEN E,XIE Z X,GUSTAFSON A M,CARRINGTON J C.microRNA-directed phasing during trans-acting siRNA biogenesis in plants[J] . Cell,2005,121(2):207-221.
[27] ASHBURNER M,BALL C A,BLAKE J A,BOTSTEIN D,BUTLER H,CHERRY J M,DAVIS A P,DOLINSKI K,DWIGHT S S,EPPIG J T,HARRIS M A,HILL D P,ISSELTARVER L,KASARSKIS A,LEWIS S,MATESE J C,RICHARDSON J E,RINGWALD M,RUBIN G M,SHERLOCK G.Gene Ontology:Tool for the unification of biology[J] . Nature Genetics,2000,25(1):25-29.
[28] KANEHISA M,GOTO S,KAWASHIMA S,OKUNO Y,HATTORI M. The KEGG resource for deciphering the genome[J] .Nucleic Acids Research,2004,32(Database issue):D277-D280.
[29] LIVAK K J,SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J] . Methods,2001,25(4):402-408.
[30] MUNNS R,TESTER M. Mechanisms of salinity tolerance[J] .Annual Review of Plant Biology,2008,59:651-681.
[31] 周琦,祝遵凌,施曼. 盐胁迫对鹅耳枥生长及生理生化特性的影响[J] . 南京林业大学学报(自然科学版),2015,39(6):56-60.ZHOU Qi,ZHU Zunling,SHI Man. Effects of salt stress on growth,physiological and biochemical characteristics of Carpi‐nus turczaninowii seedlings[J] . Journal of Nanjing Forestry University (Natural Sciences Edition),2015,39(6):56-60.
[32] 施娴,李洪有,卢丙越,周云,赵继菊,赵孟丽,梁京,孟衡玲. 3个苦荞品种对盐胁迫的生理响应及耐受性评价[J] . 作物杂志,2022(3):149-154.SHI Xian,LI Hongyou,LU Bingyue,ZHOU Yun,ZHAO Jiju,ZHAO Mengli,LIANG Jing,MENG Hengling. Physiological responses of three tartary buckwheat varieties to salt stress and evaluation of salt tolerance[J] . Crops,2022(3):149-154.
[33] 于伟. 水杨酸和亚精胺缓解茄子幼苗盐胁迫的生理机制[D] .南京:南京农业大学,2015.YU Wei. Physiological mechanism of SA and Spd alleviating salt stress in eggplant seedling (Solaunm melongena L.)[D] .Nanjing:Nanjing Agricultural University,2015.
[34] 刘晓威,杨秀艳,刘正祥,武海雯,张华新,朱建峰. MicroRNA在植物抵御盐胁迫过程中的作用[J] . 生物技术通报,2017,33(12):12-21.LIU Xiaowei,YANG Xiuyan,LIU Zhengxiang,WU Haiwen,ZHANG Huaxin,ZHU Jianfeng. Role of microRNA in plant resistance to salt stress[J] . Biotechnology Bulletin,2017,33(12):12-21.
[35] HE S S,YANG H,CAO R Q,TANG Q,AN Y Y,WANG L J. 5-Aminolevulinic acid-induced salt tolerance in strawberry (cv.‘Benihoppe’):Possible role of nitric oxide on interception of salt ions in roots[J] . Scientia Horticulturae,2022,304:111294.
[36] KANTO U,JUTAMANEE K,OSOTSAPAR Y,CHAI-ARREE W,JATTUPORNPONG S. Promotive effect of priming with 5-aminolevulinic acid on seed germination capacity,seedling growth and antioxidant enzyme activity in rice subjected to accelerated ageing treatment[J] . Plant Production Science,2015,18(4):443-454.
[37] 王亚新,冯乃杰,赵黎明,郑殿峰,沈雪峰,刘美玲,杜有为. 植物生长调节剂与氮肥对盐胁迫下水稻幼苗生理特性的影响[J] .核农学报,2024,38(3):561-573.WANG Yaxin,FENG Naijie,ZHAO Liming,ZHENG Dianfeng,SHEN Xuefeng,LIU Meiling,DU Youwei. Effects of plant growth regulators and nitrogen fertiliser on the physiological characteristics of rice seedlings under salt stress[J] . Journal of Nuclear Agricultural Sciences,2024,38(3):561-573.
[38] JATAN R,CHAUHAN P S,LATA C. High-throughput sequencing and expression analysis suggest the involvement of Pseudo‐monas putida RA-responsive microRNAs in growth and development of Arabidopsis[J] . International Journal of Molecular Sciences,2020,21(15):5468.
[39] HWANG D G,PARK J H,LIM J Y,KIM D,CHOI Y,KIM S,REEVES G,YEOM S I,LEE J S,PARK M,KIM S,CHOI I Y,CHOI D,SHIN C. The hot pepper (Capsicum annuum) microRNA transcriptome reveals novel and conserved targets:A foundation for understanding microRNA functional roles in hot pepper[J] . PLoS One,2013,8(5):e64238.
[40] SONG C N,WANG C,ZHANG C Q,KORIR N K,YU H P,MA Z Q,FANG J G. Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata)[J] .BMC Genomics,2010,11:431.
[41] LIN S,SANTI C,JOBET E,LACUT E,EL KHOLTI N,KARLOWSKI W M,VERDEIL J L,BREITLER J C,PÉRIN C,KO S S,GUIDERDONI E,CHIOU T J,ECHEVERRIA M. Complex regulation of two target genes encoding SPX-MFS proteins by rice miR827 in response to phosphate starvation[J] . Plant &Cell Physiology,2010,51(12):2119-2131.
[42] AXTELL M J,BOWMAN J L. Evolution of plant microRNAs and their targets[J] . Trends in Plant Science,2008,13(7):343-349.
[43] ZHAO X Y,HONG P,WU J Y,CHEN X B,YE X G,PAN Y Y,WANG J,ZHANG X S. The tae-miR408-mediated control of Ta‐TOC1 genes transcription is required for the regulation of heading time in wheat[J] . Plant Physiology,2016,170(3):1578-1594.
[44] MAUNOURY N,VAUCHERET H. AGO1 and AGO2 act redundantly in miR408-mediated plantacyanin regulation[J] . PLoS One,2011,6(12):e28729.
[45] TRINDADE I,CAPITÃO C,DALMAY T,FEVEREIRO M P,DOS SANTOS D M. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula[J] . Planta,2010,231(3):705-716.