腐烂病是由腐生型黑腐皮壳属真菌Valsa mali(Vm)和V.pyri(Vp)引起的植物病害,对中国乃至亚洲苹果和梨的生产发展形成威胁[1-2]。腐烂病导致的树体死亡、产量下降以及果实品质降低等问题,给果农和相关产业带来巨大的经济损失。病原菌通过因修剪、冻害、热损伤等操作引起的伤口侵入树体[3]。发病期间,果树的主枝上产生红褐色的水浸状病斑,病斑周围组织松软,伴有酒糟气味,逐渐失水干缩,颜色转为黑褐色,下陷形成溃疡。后期病斑表面出现黑色小粒点,并在潮湿时会溢出橘黄色的丝状物。当发病范围环绕枝干一周时,导致整株枯萎、死亡甚至破坏整个果园。目前,主要利用物理、生物和化学等方法来预防及控制腐烂病的传播[4]。然而,这些措施存在预防效果弱、运行成本高、环境污染风险高等问题[5]。抗性育种作为一种环境友好且可持续的防治手段,广受关注,但仍受到抗病机制研究不深入、单一和育种周期长的限制[6]。目前研究人员致力于筛选具有抗性的种质资源,如杜梨因深根系、萌发性强、耐修剪且抗病能力好,成为抗腐烂病研究的重要材料。通过现代生物技术手段,虽然已在抗病相关的主效基因挖掘和鉴定方面取得了一定进展,但与庞大的基因资源相比,仍显得不足。因此,迫切需要筛选出更多有助于抗病的主效基因,并系统地研究抗性机制,为腐烂病的抗性育种奠定基础。
在植物的整个生长发育周期内,会受到多种生物的侵害。有害生物凭借独特的感染机制,对植物的健康生长构成严重威胁。病害是农业生产及生态系统面临的重大挑战之一[7]。在病原体感染时,植物免疫系统被激活,细胞会转录重编程以激活免疫途径[8]。与此同时,植物也进化出复杂的调控机制用于对防御相关基因的转录过程加以调节。要实现基因表达的大规模转录重编程,需要转录因子(TF)之间的协同功能[9]。由于转录因子在调控抗性基因中的关键作用,筛选新的抗性基因显得尤为重要[10]。WRKY、bHLH、bZIP、C2H2 和NAC 等TF 家族已被证实与植物免疫相关[11-17]。NAC家族作为最大的植物特异性TF家族之一,参与各种生理过程和植物对环境胁迫的反应[18]。NAM、ATAF1/2、CUC1/2(NAC)转录因子于1996年首次被发现[19]。在典型的情况中,其N 端存在一个高度保守的NAC 结构域,包括约150个氨基酸残基,而C端是变化的转录调控区(TAR)[20]。到目前为止,已在多种植物中进行了系统研究,涵盖了从抗逆、抗病到生长发育等多个方面,包括拟南芥、烟草、水稻、玉米、大豆、番茄、苹果、葡萄、黄花蒿等[21-25]。NAC转录因子在调节细胞增殖、果实成熟、应激反应和程序性细胞死亡等方面取得了一系列重要进展[26-30];在植物激素信号传导方面也起着关键作用[31]。目前,NAC 转录因子在梨中的鉴定与对腐烂病抗性关系的分析鲜见报道。
本研究基于前期甘肃农业大学果树分子生物学实验室在杜梨基因组范围鉴定的转录因子NAC家族的基础上,筛选了一个差异表达的NAC 基因PbeNAC83(Chr9.g47397)并通过生物信息学分析、功能确认和表达分析进一步研究了该基因的功能。研究结果证实PbeNAC83 通过ROS(Reactive oxygen species)和植保素信号通路正向调节腐烂病抗性。
1 材料和方法
1.1 材料与处理
试验于2024—2025 年在甘肃农业大学园艺学院果树分子生物学实验室进行。黄冠梨和烟富3号苹果均购自甘肃农业大学校内超市。杜梨悬浮细胞Duli-G03 从幼嫩叶片中被诱导,并在MS 液体培养基中继代中[32]。梨腐烂病的病原菌株Vp-P-002由甘肃农业大学果树分子生物学实验室诱导和分离获得后,培养于马铃薯葡萄糖琼脂培养基(PDA:potato dextrose agar)[33]中。继代培养72 h 后,取直径为5 mm 的腐烂病菌饼15 个接种至150 mL 马铃薯葡萄糖液体培养基(PDB:potato dextrose broth)中,继续黑暗培养72 h,间隔12 h振荡1次。6000 r·min-1离心10 min,用0.22 μm过滤器和13 mm PES注射器过滤上清液,得到含有Vp次生代谢物(VpM:Valsa pyri Metabolite)的培养液。用上述VpM处理预先培养的细胞后,在0、1、3 和6 h 四个时间点收集样品,低温保存备用。PDA/PDB:马铃薯200 g,加水,煮沸,除去上清液,PDA加入20 g葡萄糖和10 g琼脂;PDB加入20 g葡萄糖,恒体积至1 L,煮沸,蒸压121 ℃20 min。
1.2 试验方法
1.2.1 PbeNAC83基因的生物信息学分析 PbeNAC83的编码区CDS 和蛋白序列从蔷薇科物种基因组数据库(GDR,https://www.rosaceae.org/)下载获得。基因组信息利用美国国家生物技术信息中心(NCBI,https://www.ncbi.nlm.nih.gov/)和拟南芥基因组数据库(TAIR,http://www.arabidopsis.org)检索获得,使用DNAMAN9.0[34]软件将PbeNAC83与多个物种进行氨基酸同源全长序列比对。在MEGA11[35]软件中通过邻接法(Neighbor-joining)构建PbeNAC83及其他物种同源基因蛋白序列的系统进化树。使用在线数据资源SMART(http://smart.embl-heidelberg.de/)对蛋白质的结构域进行预测,以e 值≤1e-5作为阈值。通过PlantCARE数据库(http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)对PbeNAC83启动子进行顺式作用元件预测[36],并利用TBtools进行可视化[37]。
1.2.2 表达模式分析“Duli-G03”悬浮细胞用20%的VpM 处理0、1、3和6 h后的表达数据从本课题组之前的转录组数据中检索获得。RNA 的提取使用RNAout 试剂盒(71203-50 天恩泽,北京),采用Evo M-MLV 反转录试剂盒(AG11728Accurate Biotechnology,湖南)去除基因组DNA,合成cDNA。使用Primer3 Input 0.4.0(http://bioinfo.ut.ee/primer3-0.4.0/)设计试验所需引物。在QuantStudio®5 实时PCR系统(Thermo Fisher Scientfc,美国)进行实时荧光定量PCR(quantitative real-time PCR,qRT-PCR),基因扩增引物及反应体系、程序均参考已发表的文献[38],计算相对表达量采用2-ΔΔCT法。
1.2.3 载体构建 设计上游引物F1(GAGGCGCGCCATGGAAAATATTAGAGAAAACTATG,含Asc Ⅰ酶切位点)和下游引物R1(CACCTAGGCTAAATATTAATCGGCATGCTAACACTG,含Avr Ⅱ酶切位点),克隆PbeNAC83 全长序列,连接19T 载体至DH5α,选取阳性单克隆进行测序。选择测序结果正确的单克隆进行扩繁,双酶切后连接过表达载体pFGC-5941 导入大肠杆菌DH5α。扩繁阳性菌落后提取质粒,取质粒利用冻融法转入农杆菌感受态细胞GV3101,经PCR 验证后用于后续试验。以pFGC-5941空载体为对照。
1.2.4 NAC 转录因子瞬时表达及病原菌接种 将pFGC-5941 空载体与携带目标基因的pFGC-5941-PbeNAC83农杆菌振荡12 h活化繁殖,8000 r·min-1、离心10 min 收集农杆菌,调节菌液OD600为0.4~0.6,重悬于MES-KOH 溶液,4 ℃静置3~4 h。用75%酒精擦拭清洁果实表皮,取0.2 mL菌液缓慢注射至果实,25 ℃浸润72 h 后,去除注射部位的边缘果皮后接种腐烂病病原菌,用游标卡尺分别在接种后36、48、60、72 h记录病斑的纵横直径,病斑直径=(纵径+横径)/2。病斑抑制率/%=[(对照组病斑直径-处理组病斑直径)/(对照组病斑直径)]×100。以空载体作为对照。以上试验设5次生物学重复。
1.2.5 杜梨Duli-G03 悬浮细胞的遗传转化 对预先培养的Duli-G03 悬浮细胞经过孔径为40~200 目的细胞过滤器后,在黑暗环境中以110 r·min-1的转速振荡培养72 h。在此期间,农杆菌经过活化后,重悬处理,调节其OD600值至约0.4。随后,用农杆菌悬浮液侵染杜梨悬浮细胞,在黑暗条件下,静置5 min,以促进细胞与农杆菌的接触。去除多余农杆菌,将细胞转移至MS 培养基中继续黑暗静置培养。48 h后,使用头孢菌素处理以杀灭残留的农杆菌,并将细胞转移至含有抗生素的MS培养基中继续培养。从生长良好的细胞团中筛选,并扩繁、转化细胞系,利用PCR 和qRT-PCR 技术对转化细胞系进行基因表达量检测[39]。
1.2.6 NAC 转录因子稳定表达及病原菌接种 Vp抗性分析采用方法:将细胞团均匀接种于MS 平板上再接种Vp菌饼。在接种后的36、48、60和72 h,使用游标卡尺测量病斑的纵横径。在接种72 h 后,使用2 mL MTT 染料对细胞染色,以观察细胞活性[40]。所有结果均通过拍照记录。VpM 抗性分析采用方法:细胞团浓度(φ)为20 μL(密实体积)·mL-1,20%浓度的VpM处理。于处理后1、3和6 h取样,用于细胞活性测定和基因表达量分析。
1.2.7 基因表达分析 依据田丹等[41]的方法,提取RNA 用于qRT-PCR 检测。选取与免疫反应信号相关的基因进行表达量测定,涉及模式触发免疫(PTI)、茉莉酸(JA)、植保素和活性氧(ROS)[42]。基因名称和引物序列参照Sun 等[40]的方法,具体信息见表1。基因的相对表达量采用2-ΔΔCT法计算[43-44]。
表1 qRT-PCR 相关基因及引物信息
Table 1 Genes and primer information associated to qRT-PCR
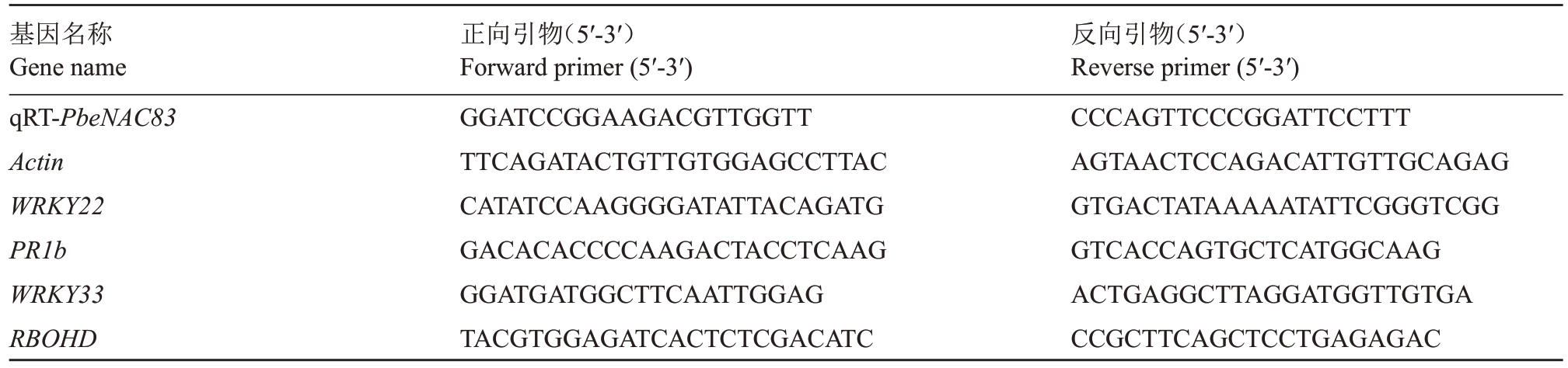
基因名称Gene name qRT-PbeNAC83 Actin WRKY22 PR1b WRKY33 RBOHD正向引物(5′-3′)Forward primer(5′-3′)GGATCCGGAAGACGTTGGTT TTCAGATACTGTTGTGGAGCCTTAC CATATCCAAGGGGATATTACAGATG GACACACCCCAAGACTACCTCAAG GGATGATGGCTTCAATTGGAG TACGTGGAGATCACTCTCGACATC反向引物(5′-3′)Reverse primer(5′-3′)CCCAGTTCCCGGATTCCTTT AGTAACTCCAGACATTGTTGCAGAG GTGACTATAAAAATATTCGGGTCGG GTCACCAGTGCTCATGGCAAG ACTGAGGCTTAGGATGGTTGTGA CCGCTTCAGCTCCTGAGAGAC
1.3 统计分析
通过Microsoft Excel(2016)软件进行数据的初步整理,采用t-test分析差异显著性(*P<0.05;**P<0.01)。所有数据以平均值±标准差的形式呈现。图标绘制利用OriginPro 9.0软件实现。
2 结果与分析
2.1 PbeNAC83生物信息学分析
将Chr9.g47397 蛋白序列提交至拟南芥基因组网站,发现其与NAC83(AT5G13180.1)序列高度同源,因此将其命名为PbeNAC83。为了更深入地了解进化关系,将其与10个物种中的13个同源基因构建了系统发育树,结果表明,PbeNAC83与白梨中的rna48275-v1.1-pbr基因的同源性最高(图1-A),表明他们可能具有相似的功能。进一步的结构域分析揭示,PbeNAC83 蛋白具有典型的NAM 结构域(图1-B),属于NAC 转录因子家族成员,具备调控基因表达的潜能。此外,对PbeNAC83 启动子区域的顺式作用元件预测分析显示,该区域富含与MeJA(茉莉酸甲酯)、ABA(脱落酸)信号通路、厌氧诱导相关的元件,以及与植物抗病相关的其他元件(图1-C),表明PbeNAC83 可能参与多种激素信号的调控。综上,PbeNAC83 为典型的NAC 转录因子,在进化上表现出保守性,并且可能响应多种激素和胁迫信号。
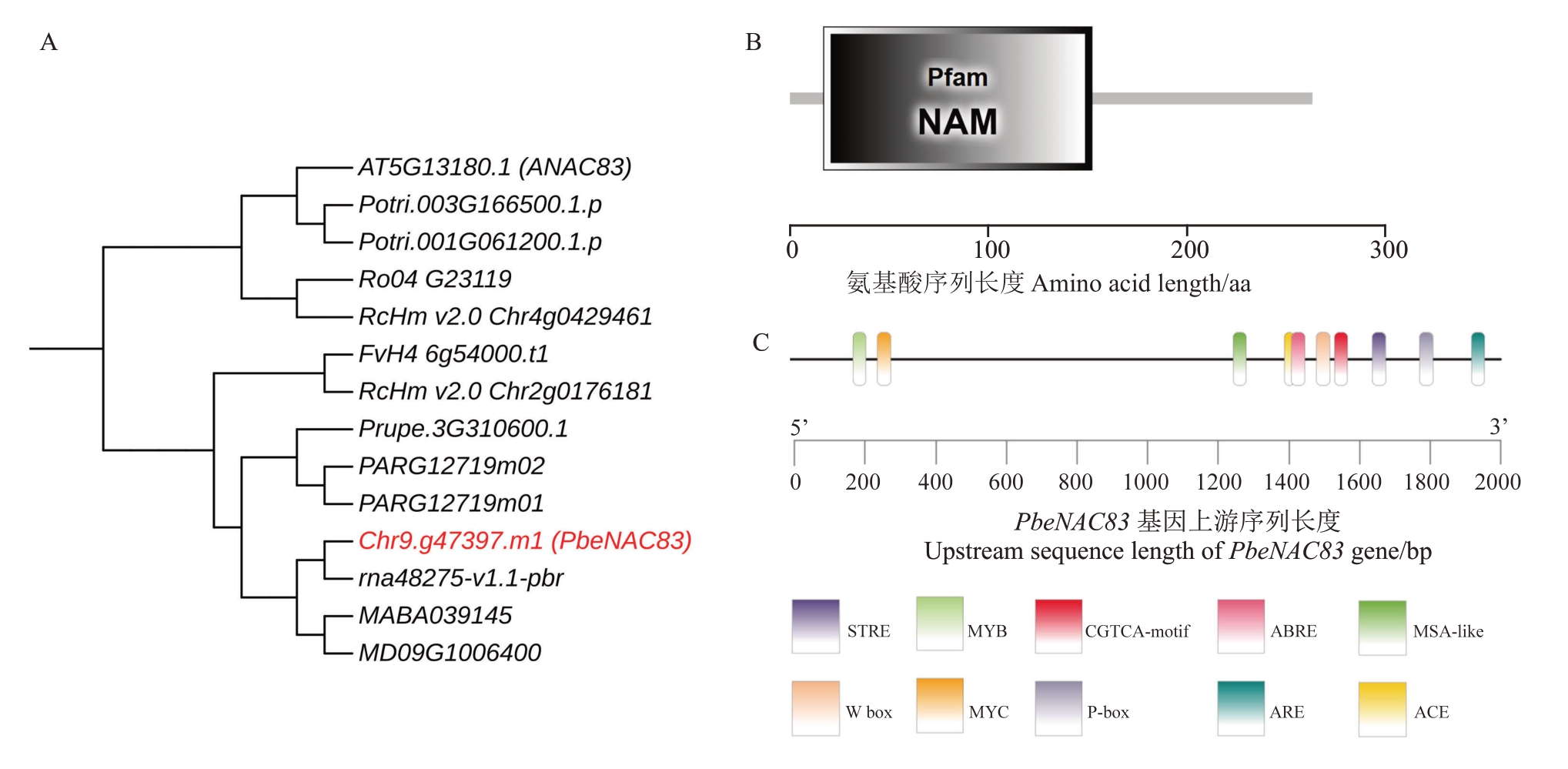
图1 PbeNAC83 基本参数及序列系统发育分析
Fig.1 Basic parameters and the sequence phylogenetic analysis of PbeNAC83
A.PbeNAC83 的系统发育分析,包括拟南芥、毛果杨、黑树莓、月季花、野草莓、桃、杏、白梨、山丁子、苹果;B.PbeNAC83 的保守结构域分析;C.PbeNAC83 的顺式作用元件预测。
A.Phylogenetic analysis of PbeNAC8 in:Arabidopsis thaliana、Populus trichocarpa、Rubus occidentalis、Rosa chinensis、Fragaria vesca、Prunus persica、Prunus armeniaca、Pyrus bretschneideri、Malus baccata、Malus×domestica.B.Conserved domain analysis of PbeNAC83.C.Cis-acting element prediction of PbeNAC83.
2.2 PbeNAC83表达模式分析
利用20%VpM 处理野生型杜梨悬浮细胞后的转录组数据,获得PbeNAC83 基因的FPKM 值(图2)。在20%VpM处理1 h后,PbeNAC83的FPKM值快速上升,由15.74 上升至173.38(图2-A)。进一步将杜梨的悬浮细胞用20%VpM 处理后,通过qRTPCR 技术分析其在1、3、6 h 后PbeNAC83 的表达量。结果显示,处理后PbeNAC83 的表达被立即激活,1 h 时由19.51 上升至211.2,上调约为10.82 倍(图2-B)。以上结果表明,PbeNAC83基因能够响应腐烂病信号,其表达水平的上调可能与植物抵御腐烂病的机制密切相关。
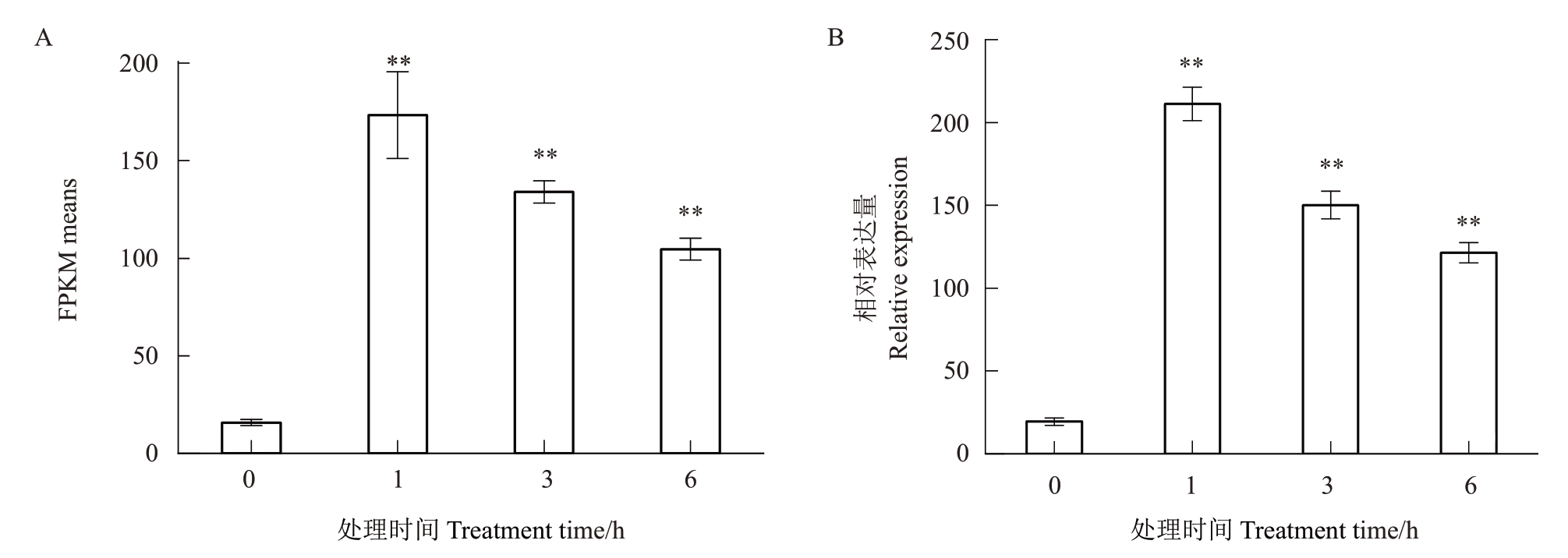
图2 PbeNAC83 对腐烂病信号的表达分析
Fig.2 The expression of PbeNAC83 in response to Valsa canker signals
A.PbeNAC83 响应腐烂病信号的表达分析:用20%Valsa pyri Metabolite(VpM)处理Pyrus betulifolia 悬浮细胞后通过RNA-seq 获得的基因FPKM 值;B.qRT-PCR。FPKM:每千个碱基的转录每百万映射读取的fragments。数据为平均值(±SD),n=3。*P<0.05,**P<0.01。下同。
A.The expression analysis of PbeNAC83 respond to Valsa canker signals:the FPKM value of Pyrus betulifolia suspension cells were treated with 20%Valsa pyri Metabolite(VpM)and detected by RNA-seq.B.qRT-PCR.FPKM(Fragments quantity per kilobase of the exon model for every million mapped fragments).The date were mean(±SD),n=3.*P<0.05,**P<0.01.The same below.
2.3 PbeNAC83正调控梨和苹果果实对Vp的抗性
为了探究基因是否在抗腐烂病中发挥作用,将PbeNAC83瞬时表达于黄冠梨和烟富3号苹果中,分析其对腐烂病的抗性(图3)。结果显示,PbeNAC83过表达的果实病斑直径小于空载体(图3-A),表明该基因可能增强了果实对腐烂病的抵抗力。随着接种部位逐渐发病,进一步测量在36、48、60和72 h的病斑直径,分析对照和过表达果实上病斑大小的变化(图3-B),结果表明,过表达PbeNAC83 可显著降低梨腐烂病菌(Vp)在黄冠梨和烟富3号苹果果实上的病斑直径,病斑抑制率分别为10.3%和14.9%。qRT-PCR 检测显示,与空载体对照相比,PbeNAC83在注射部位的表达水平显著上调(图3-C)。以上结果表明,PbeNAC83 的过表达正调控梨和苹果果实对腐烂病的抗性。
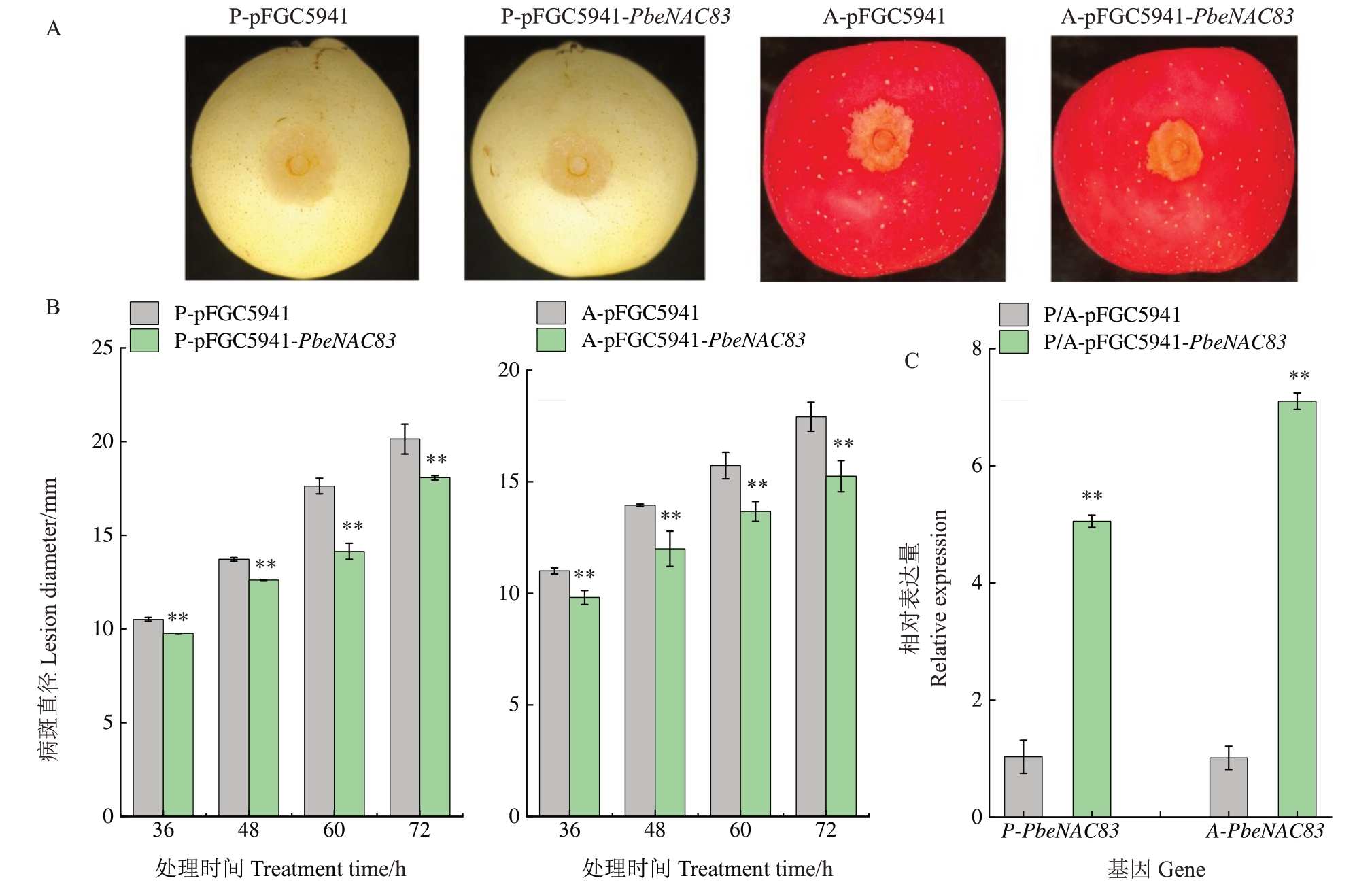
图3 PbeNAC83 在苹果和梨果实中的瞬时表达分析
Fig.3 Transient expression analysis of PbeNAC83 in apple and pear fruits
A.拍摄于果实接种腐烂病72 h 的病变表现。pFGC-5941 为空载体对照,pFGC-5941-PbeNAC83 过表达。P 是黄冠梨,A 是烟富3 号苹果;B.果实病斑大小测量数据;C.果实中PbeNAC83 的表达量测定。
A.Photographs taken 72 hours post-inoculation with Valsa canker show disease symptoms.pFGC-5941 served as the empty vector control,while pFGC-5941-PbeNAC83 was used for overexpression.P represents Huangguan pear,and A represents Yanfu No.3 apple.B.Lesion size measurement data on fruit.C.Expression of PbeNAC83 in fruits.
2.4 PbeNAC83 的过表达增强了Duli-G03 悬浮细胞对Vp的抗性
为了进一步证明PbeNAC83对腐烂病的调控作用,利用农杆菌介导的遗传转化法将PbeNAC83 转入Duli-G03,筛选出生长良好的过表达细胞系:PbeNAC83-OE1、PbeNAC83-OE2 和PbeNAC83-OE3(图4)。将3 个过表达细胞系分别铺板并接种Vp后,腐烂病的扩散速率受到显著抑制(图4-A)。处理至72 h的PbeNAC83-OE1细胞系的病斑直径较对照减少了73.1%(图4-B)。qRT-PCR检测显示,与对照相比,过表达细胞系中PbeNAC83 基因的表达水平显著上调(图4-C)。以上结果进一步表明,PbeNAC83的过表达显著增强了Duli-G03悬浮细胞对腐烂病感染的抗性。
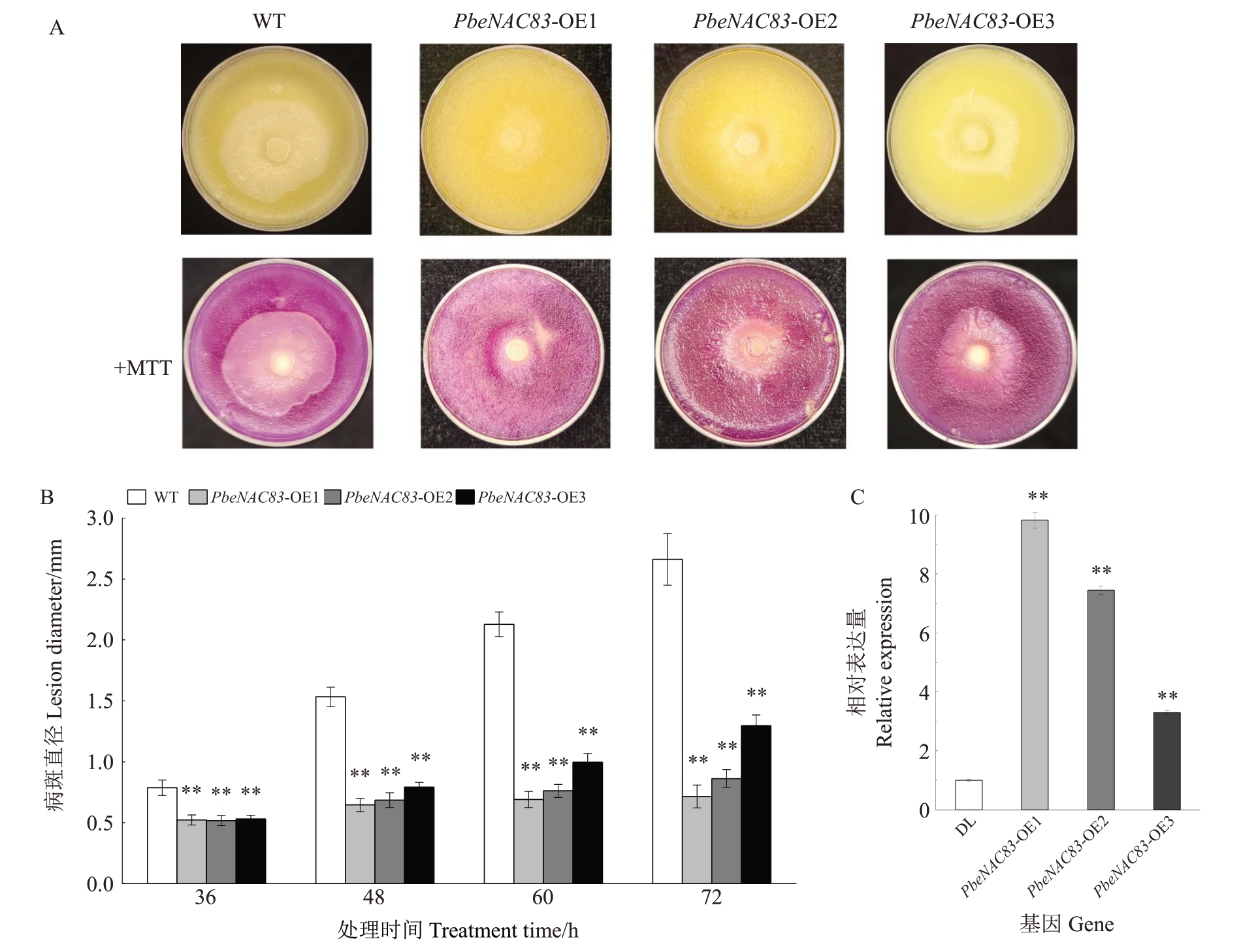
图4 PbeNAC83 过表达增强了Duli-G03 悬浮细胞对腐烂病菌的抗性
Fig.4 PbeNAC83 overexpression enhanced the resistance of Dali-G03 suspension cells to Valsa pyri
A.接种Vp 72 h 后,野生型细胞(WT)和3 个过表达转基因细胞系(PbeNAC83-OE1、2、3)在培养皿上的发病情况。活细胞鉴定:MTT 染为紫色;B.细胞接种Vp 后,各时间点下的病斑大小测量数据;C.以Actin 作参照,转基因株系中PbeNAC83 的表达水平。表达量是相对于WT的,其值设为1。
A.After inoculation with Vp for 72 hours,the disease incidence of wild-type cells(WT)and three overexpressing transgenic cell lines(PbeNAC83-OE1,OE2,and OE3)on petri dishes.Identification of viable cells:stained purple by MTT.B.Lesion size measurements at different time points after inoculation with Vp.C.Expression levels of PbeNAC83 in transgenic lines,normalized to Actin.Expression values are relative to WT,which is set as 1.
2.5 PbeNAC83 过表达增强了Duli-G03 悬浮细胞对腐烂病菌代谢物的抗性
采用MTT法对细胞活性跟踪检测,研究野生型(WT)和PbeNAC83 过表达细胞系对腐烂病菌代谢物(VpM)的耐受性(图5)。结果显示,与野生型相比,PbeNAC83 过表达细胞系(PbeNAC83-OE1)在20%VpM处理后的1、3和6 h,细胞活性均显著高于野生型。这一结果表明,PbeNAC83 的过表达显著增强了Duli-G03 悬浮细胞对腐烂病菌代谢物的耐受性。
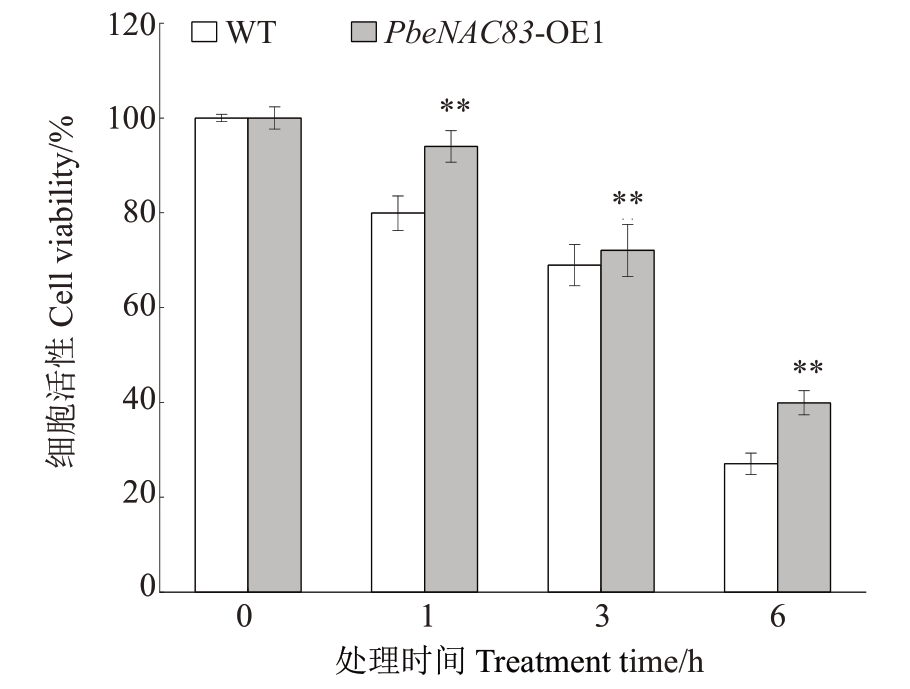
图5 过表达PbeNAC83 过表达增强了Duli-G03 悬浮细胞对腐烂病菌代谢物的耐受性
Fig.5 The overexpression of PbeNAC83 enhanced the tolerance of Duli-G03 suspension cells to VpM
20%VpM 处理后不同时间点Duli-G03 与PbeNAC83 转基因细胞系细胞的活性分析。
Analysis of the activity of Duli-G03 with cells from PbeNAC83 transgenic cell lines at different time points after 20%VpM treatment.
2.6 PbeNAC83的免疫机制分析
为了研究PbeNAC83 激活的信号通路,笔者检测了与植物免疫相关的关键基因的表达,包括PTI、JA、植保素和ROS信号通路基因(图6)。在VpM处理后,与野生型相比,PbeNAC83-OE1细胞中,PTI和JA 相关基因WRKY22 和PR1b 的表达水平上调(图6-A、B)。此外,植保素相关基因WRKY33在处理3 h后的表达量上调至对照的2.26 倍,ROS 相关基因RBOHD 的表达量上调至对照的1.96 倍(图6-C、D)。以上结果表明PbeNAC83的过表达可能激活了ROS和植保素信号通路来抵抗腐烂病。
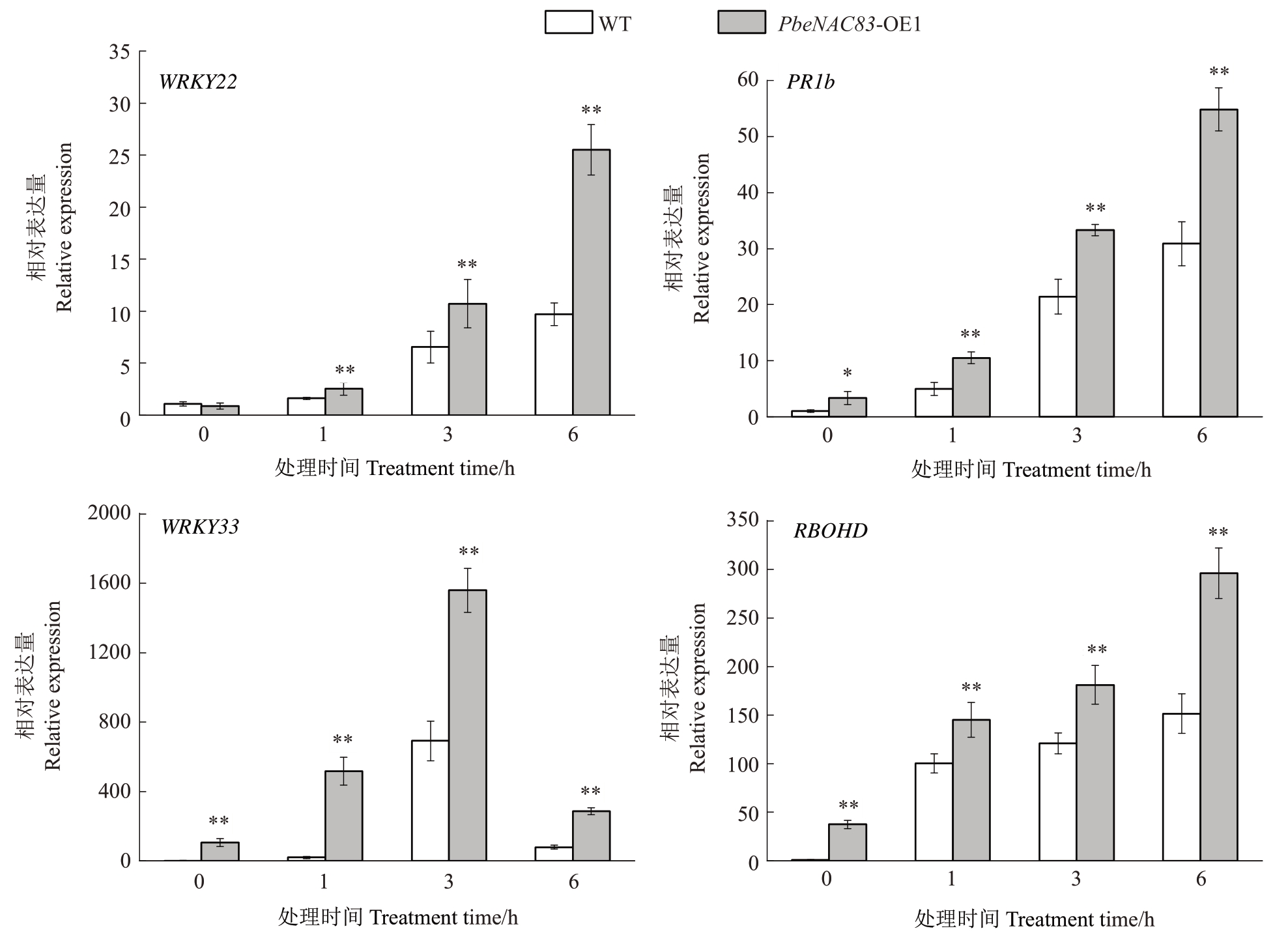
图6 PbeNAC83 激活多个免疫反应相关基因的表达
Fig.6 PbeNAC83 activates the expression of multiple immune response related genes
A.WRKY22 与PTI 信号通路相关;B.PR1b 与JA 信号通路相关;C.WRKY33 与植保素信号通路相关;D.RBOHD 与ROS 信号通路相关。
A.WRKY22 is associated with the PTI signaling pathway.B.PR1b is associated with the JA signaling pathway.C.WRKY33 is related to the phytoalexin signaling pathway.D.RBOHD is related to the ROS signaling pathway.
3 讨 论
转录因子通过与靶基因启动子区域的顺式作用元件特异性地结合,从而调控基因的转录。一般来说,典型的转录因子结构域包含4 个主要功能:即DNA 结合区(DNA-binding domain,DBD)、转录调控区(transcription regulation domain,TRD)、寡聚化位点区(oligomerization site,OS)以及核定位信号区(nuclear localization signal,NLS)[45]。NAC(NAM、ATAF 和CUC)作为植物中最大的转录因子(TF)家族之一,在拟南芥和水稻中包含100 多个成员[46-47]。本研究通过进化分析比较了杜梨与其他物种中的NAC 转录因子,发现PbeNAC83 与拟南芥ANAC83具有较近的亲缘关系,推测二者可能具有相似的功能。另外,通过对PbeNAC83 启动子区域进行顺式作用元件分析,发现其表达调控可能受到多种激素协同作用,其中赤霉素与脱落酸可能在诱导PbeNAC83 基因表达过程中发挥关键作用。此外,启动子区域还包含与植物抗逆相关的元件,推测PbeNAC83 可能在植物的逆境响应中扮演重要角色。基于转录数据和荧光定量PCR 分析表明杜梨PbeNAC83基因在腐烂病菌处理后显著上调。
NAC 转录因子由广泛分布在各种高等植物物种中的众多成员组成,在植物的生长发育、器官建成、激素调控以及非生物胁迫响应过程中,发挥着至关重要的作用[48]。NAC 在病原体的免疫反应中起着至关重要的作用[49]。研究表明,许多NAC基因作为植物对寄生型、半寄生型或腐生型病原体免疫的正或负调节因子,调节超敏反应和ROS信号通路或作为病原体效应子的毒力靶标[50]。在拟南芥中,至少有7 种NAC 蛋白ATAF1、ATAF2、ANAC019、ANAC055、ANAC072、NTL9/CBNAC(钙调素结合NAC 蛋白)和ANAC042/JUB1(JUNGBRUNNEN1)参与调节植物对腐生型真菌病原体的免疫,包括灰霉病菌(Botrytis cinerea)、油菜链格孢菌(Alternaria brassicicola)和尖孢镰刀菌(Fusarium oxysporum)[51-53]。此外,在水稻的最新研究中发现NAC 的过表达,如OsNAC066、OsNAC096、OsNAC6 和Os-NAC111[54-57],增强了对稻瘟病的抗性。本文结合VpM处理悬浮细胞后差异基因的表达分析,筛选了NAC转录因子家族的1个基因进行克隆以及进一步的功能验证。结果表明,在果实和细胞过表达PbeNAC83 显著降低了病斑的扩散速率,表明PbeNAC83 正调控腐烂病抗性。未来可通过基因过表达,在抗病的基础上深入研究其他方面,为梨的抗性育种提供理论指导。
HR(Hypersensitive Response)诱导的细胞死亡可以防止或延缓病原体的进一步扩张,从而减轻对植物细胞的损伤。此过程中,这种效应通常伴随着ROS的爆发和积累。PbeNAC83通过调控ROS和植保素的合成,显著增强植物抗病性。具体而言,PbeNAC83 可能通过激活ROS 通路关键基因RBOHD的表达,促使细胞壁加固和活性氧爆发,直接抑制病原菌侵染与扩散。植保素作为一类低分子质量次生代谢物,在植物遭受病原菌侵染时被诱导合成,具备抗菌特性,在植物防范病原菌入侵进程中扮演关键角色。在健康植物中,植保素的含量通常很低,但细菌、真菌和卵菌等病原菌的入侵会显著诱导其合成[58]。研究发现,在接种梨腐烂病病菌后,PbeNAC83 过表达细胞中WRKY33 基因的表达被显著诱导。推测PbeNAC83的过表达通过激活植保素合成,增强植物对腐烂病的抗性,其独特的调控机制为抗病育种提供了新思路。其他抗病NAC 转录因子如TaNAC1调控细胞壁和ROS[59];HvNAC6调控水杨酸和茉莉酸[60];OsNAC4 调节细胞死亡[61];Os-NAC60 调节ROS 积累和胼胝质沉积[62]。笔者的研究结果表明,PbeNAC83 可能作为ROS 和植保素的关键调节因子,可以整合两个信号通路,协同调控抗病反应,实现高效抗病。
4 结 论
本研究从杜梨中筛选鉴定获得了一个响应腐烂病信号的NAC家族成员PbeNAC83。通过瞬时和稳定转化进行功能分析,明确了PbeNAC83 能够正向调控对腐烂病的抗性。进一步研究发现,PbeNAC83的过表达可激活植物体内的植保素和活性氧信号通路相关基因的表达,进而抑制腐烂病菌的入侵。以上发现为腐烂病的抗病分子育种提供依据,还为生产实践中有效防控腐烂病的发生提供新的思路和方法。通过识别关键抗病基因并研究机制,可加快抗病育种进程,为培育抗腐烂病新品种提供支持。
[1] MENG X L,YANG R,LIU A T,HU T L,WANG Y N,CAO K Q,WANG S T.The influence of lower temperature induction of Valsa mali on the infection of apple trees[J]. Plant Disease,2021,105(10):2776-2780.
[2] FENG H,WANG C L,HE Y T,TANG L,HAN P L,LIANG J H,HUANG L L.Apple Valsa canker:Insights into pathogenesis and disease control[J].Phytopathology Research,2023,5(1):45.
[3] WANG S T,HU T L,WANG Y N,LUO Y,MICHAILIDES T J,CAO K Q. New understanding on infection processes of Valsa canker of apple in China[J]. European Journal of Plant Pathology,2016,146(3):531-540.
[4] 邱德文.生物农药的发展现状与趋势分析[J].中国生物防治学报,2015,31(5):679-684.QIU Dewen.Analysis of the development situation and trends of biological pesticides in China[J]. Chinese Journal of Biological Control,2015,31(5):679-684.
[5] 王晓焕,潘彤彤,练森,王彩霞,李保华.环境因子对腐烂病菌在苹果枝条木质部内生长扩展的影响[J]. 中国农业科学,2018,51(17):3291-3301.WANG Xiaohuan,PAN Tongtong,LIAN Sen,WANG Caixia,LI Baohua. Effects of environmental factors on the growth and extension of Valsa mali in the xylem of apple branches[J].Scientia Agricultura Sinica,2018,51(17):3291-3301.
[6] ABE K,KOTODA N,KATO H,SOEJIMA J I. Genetic studies on resistance to Valsa canker in apple:genetic variance and breeding values estimated from intra- and inter-specific hybrid progeny populations[J]. Tree Genetics & Genomes,2011,7(2):363-372.
[7] 孙娥.苹果类受体激酶基因MdLecRK-S.4.3 调控腐烂病的功能研究[D].兰州:甘肃农业大学,2023.SUN E. Functional analysis of apple receptor like kinase gene MdLecRK-S.4.3 in regulating Valsa canker[D]. Lanzhou:Gansu Agricultural University,2023.
[8] DUPLAN V,RIVAS S.E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity[J].Frontiers in Plant Science,2014,5:42.
[9] 张娜.小麦与叶锈菌互作过程中TaNAC35 转录因子的特征分析[D].保定:河北农业大学,2019.ZHANG Na. Characterization analysis of transcription Factor TaNAC35 in the wheat-Puccinia triticina interaction[D]. Baoding:Hebei Agricultural University,2019.
[10] ZHANG H,LV S K,WANG C Y,JI W Q.The role of transcription factor in wheat defense against pathogen and its prospect in breeding[J]. Journal of Plant Biology and Crop Research,2018,1(1):1005.
[11] REN C X,CHEN S Y,HE Y H,XU Y P,YANG J,CAI X Z.Fine-tuning of the dual-role transcription factor WRKY8 via differential phosphorylation for robust broad-spectrum plant immunity[J].Plant Communications,2024,5(12):101072.
[12] ZHANG Q L,WANG J B,LI Y Y,TUNG J,DENG Y T,BAKER B,DINESH-KUMAR S P,LI F.Conserved transcription factors NRZ1 and NRM1 regulate NLR receptor-mediated immunity[J].Plant Physiology,2024,195(1):832-849.
[13] ZHU X Y,ZHAO Y D,SHI C M,XU G J,WANG N N,ZUO S M,NING Y S,KANG H X,LIU W D,WANG R Y,YAN S Y,WANG G L,WANG X L.Antagonistic control of rice immunity against distinct pathogens by the two transcription modules via salicylic acid and jasmonic acid pathways[J]. Developmental Cell,2024,59(12):1609-1622.e4.
[14] LI W T,ZHU Z W,CHERN M,YIN J J,YANG C,RAN L,CHENG M P,HE M,WANG K,WANG J,ZHOU X G,ZHU X B,CHEN Z X,WANG J C,ZHAO W,MA B T,QIN P,CHEN W L,WANG Y P,LIU J L,WANG W M,WU X J,LI P,WANG J R,ZHU L H,LI S G,CHEN X W.A natural allele of a transcription factor in rice confers broad-spectrum blast resistance[J].Cell,2017,170(1):114-126.e15.
[15] MAO G H,MENG X Z,LIU Y D,ZHENG Z Y,CHEN Z X,ZHANG S Q. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis[J].The Plant Cell,2011,23(4):1639-1653.
[16] HUYSMANS M,BUONO R A,SKORZINSKI N,RADIO M C,DE WINTER F,PARIZOT B,MERTENS J,KARIMI M,FENDRYCH M,NOWACK M K. NAC transcription factors ANAC087 and ANAC046 control distinct aspects of programmed cell death in the Arabidopsis columella and lateral root cap[J].The Plant Cell,2018,30(9):2197-2213.
[17] WANG N,FAN X,HE M Y,HU Z Y,TANG C L,ZHANG S,LIN D X,GAN P F,WANG J F,HUANG X L,GAO C X,KANG Z S,WANG X J. Transcriptional repression of TaNOX10 by TaWRKY19 compromises ROS generation and enhances wheat susceptibility to stripe rust[J]. The Plant Cell,2022,34(5):1784-1803.
[18] XU B,OHTANI M,YAMAGUCHI M,TOYOOKA K,WAKAZAKI M,SATO M,KUBO M,NAKANO Y,SANO R,HIWATASHI Y,MURATA T,KURATA T,YONEDA A,KATO K,HASEBE M,DEMURA T. Contribution of NAC transcription factors to plant adaptation to land[J]. Science,2014,343(6178):1505-1508.
[19] SOUER E,VAN HOUWELINGEN A,KLOOS D,MOL J,KOES R. The No apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries[J]. Cell,1996,85(2):159-170.
[20] AIDA M,ISHIDA T,FUKAKI H,FUJISAWA H,TASAKA M.Genes involved in organ separation in Arabidopsis:An analysis of the cup-shaped cotyledon mutant[J]. The Plant Cell,1997,9(6):841-857.
[21] NURUZZAMAN M,MANIMEKALAI R,SHARONI A M,SATOH K,KONDOH H,OOKA H,KIKUCHI S. Genome-wide analysis of NAC transcription factor family in rice[J]. Gene,2010,465(1/2):30-44.
[22] OOKA H,SATOH K,DOI K,NAGATA T,OTOMO Y,MURAKAMI K,MATSUBARA K,OSATO N,JUN K W,CARNINCI P,HAYASHIZAKI Y,SUZUKI K,KOJIMA K,TAKAHARA Y,YAMAMOTO K,KIKUCHI S.Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana[J].DNA Research,2003,10(6):239-247.
[23] KIKUCHI K,UEGUCHI-TANAKA M,YOSHIDA K T,NAGATO Y,MATSUSOKA M,HIRANO H Y. Molecular analysis of the NAC gene family in rice[J].Molecular and General Genetics MGG,2000,262(6):1047-1051.
[24] SU H Y,ZHANG S Z,YUAN X W,CHEN C T,WANG X F,HAO Y J. Genome-wide analysis and identification of stress-responsive genes of the NAM-ATAF1 2-CUC2 transcription factor family in apple[J]. Plant Physiology and Biochemistry,2013,71:11-21.
[25] WANG N,ZHENG Y,XIN H P,FANG L C,LI S H. Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera[J].Plant Cell Reports,2013,32(1):61-75.
[26] HENDELMAN A,STAV R,ZEMACH H,ARAZI T.The tomato NAC transcription factor SlNAM2 is involved in flowerboundary morphogenesis[J]. Journal of Experimental Botany,2013,64(18):5497-5507.
[27] ZHANG Q J,LI T,ZHANG L J,DONG W X,WANG A D.Expression analysis of NAC genes during the growth and ripening of apples[J].Horticultural Science,2018,45(1):1-10.
[28] LI W H,LI H W,WEI Y F,HAN J X,WANG Y,LI X G,ZHANG L H,HAN D G. Overexpression of a Fragaria vesca NAM ATAF and CUC (NAC) transcription factor gene(FvNAC29) increases salt and cold tolerance in Arabidopsis thaliana[J]. International Journal of Molecular Sciences,2024,25(7):4088.
[29] XU W B,GUO Q H,LIU P,DAI S,WU C G,YANG G D,HUANG J G,ZHANG S Z,SONG J M,ZHENG C C,YAN K.A long non-coding RNA functions as a competitive endogenous RNA to modulate TaNAC018 by acting as a decoy for taemiR6206[J].Plant Molecular Biology,2024,114(3):36.
[30] LIU Y M,YOU H G,LI H P,ZHANG C J,GUO H,HUANG X L,ZHANG Q,ZHANG X Y,MA C,WANG Y J,LI T D,JI W Q,KANG Z S,ZHANG H.TaNAC1 boosts powdery mildew resistance by phosphorylation-dependent regulation of TaSec1a and TaCAMTA4 via PP2Ac/CDPK20[J]. New Phytologist,2024,244(2):635-653.
[31] BIAN Z Y,GAO H H,WANG C Y. NAC transcription factors as positive or negative regulators during ongoing battle between pathogens and our food crops[J].International Journal of Molecular Sciences,2021,22(1):81.
[32] ZHAO D,TIAN Y Z,YU H Q,MAO X,WANG C,DUO H,SUN E,ZUO C W.Establishment of the“Valsa pyri metabolites(VpM)-suspension cell”-based system to study the response of pears to VpM[J]. Physiological and Molecular Plant Pathology,2022,120:101850.
[33] ZUO C W,MAO J,CHEN Z J,CHU M Y,DUO H,CHEN B H.RNA sequencing analysis provides new insights into dynamic molecular responses to Valsa mali pathogenicity in apple‘Changfu No.2’[J].Tree Genetics&Genomes,2018,14(5):75.
[34] SOLOVYEV V,KOSAREV P,SELEDSOV I,VOROBYEV D.Automatic annotation of eukaryotic genes,pseudogenes and promoters[J].Genome Biology,2006,7(1):S10.
[35] TAMURA K,STECHER G,KUMAR S. MEGA11:Molecular evolutionary genetics analysis version 11[J]. Molecular Biology and Evolution,2021,38(7):3022-3027.
[36] LESCOT M,DÉHAIS P,THIJS G,MARCHAL K,MOREAU Y,VAN DE PEER Y,ROUZÉ P,ROMBAUTS S.PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J].Nucleic Acids Research,2002,30(1):325-327.
[37] CHEN C J,CHEN H,ZHANG Y,THOMAS H R,FRANK M H,HE Y H,XIA R. TBtools:An integrative toolkit developed for interactive analyses of big biological data[J]. Molecular Plant,2020,13(8):1194-1202.
[38] 郑艳.MdMDS1 调控腐烂病抗性的功能研究[D].兰州:甘肃农业大学,2024.ZHENG Yan.Functional analysis of MdMDS1 in regulating valsa canker resistance[D]. Lanzhou:Gansu Agricultural University,2024.
[39] MAO X,WANG C,LV Q Q,TIAN Y Z,WANG D D,CHEN B H,MAO J,LI W F,CHU M Y,ZUO C W. Cyclic nucleotide gated channel genes(CNGCs)in Rosaceae:Genome-wide annotation,evolution and the roles on Valsa canker resistance[J].Plant Cell Reports,2021,40(12):2369-2382.
[40] SUN E,YU H Q,CHEN Z J,CAI M R,MAO X,LI Y Y,ZUO C W. Enhanced Valsa canker resistance conferred by expression of MdLecRK-S.4.3 in Pyrus betulifolia is largely suppressed by PbePUB36[J]. Journal of Experimental Botany,2023,74(14):3998-4013.
[41] 田丹,朵虎,刘河,吕前前,左存武.苹果L-LEC-RLK 基因家族鉴定及响应病原真菌信号的表达分析[J].园艺学报,2019,46(3):421-432.TIAN Dan,DUO Hu,LIU He,LÜ Qianqian,ZUO Cunwu. Genome wide identification and expression patterns in response to signals from fungal pathogens of L-LEC-RLK gene family in apple[J].Acta Horticulturae Sinica,2019,46(3):421-432.
[42] 孙娥,闫文萍,余宏强,赵丹,朵虎,左存武.苹果和梨抗病相关基因的诱导与表达分析[J]. 西北植物学报,2022,42(9):1468-1476.SUN E,YAN Wenping,YU Hongqiang,ZHAO Dan,DUO Hu,ZUO Cunwu. Induction and expression analysis of the disease resistance related genes in apple and pear[J].Acta Botanica Boreali-Occidentalia Sinica,2022,42(9):1468-1476.
[43] 余宏强.蔷薇科植物模式识别受体鉴定及杜梨抗腐烂病成员筛选[D].兰州:甘肃农业大学,2023.YU Hongqiang. Identification of pattern recognition receptors in Rosaceae and screening of members of Pyrus betulifolia for resistance to valsa canker[D].Lanzhou:Gansu Agricultural University,2023.
[44] LIVAK K J,SCHMITTGEN T D.Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method[J].Methods,2001,25(4):402-408.
[45] 潘炬忠,韦萍,朱德平,邵胜雪,陈珊珊,韦雅倩,高维维.水稻转录因子OsERF104 的克隆和功能研究[J].作物学报,2025:1- 15(2025- 02- 07). https://link.cnki.net/urlid/11.1809.S.20250207.1715.006.PAN Juzhong,WEI Ping,ZHU Deping,SHAO Shengxue,CHEN Shanshan,WEI Yaqian,GAO Weiwei.Cloning and functional analysis of OsERF104 transcription factor in rice[J].Acta Agronomica Sinica,2025:1-15(2025-02-07). https://link.cnki.net/urlid/11.1809.S.20250207.1715.006.
[46] FANG Y J,YOU J,XIE K B,XIE W B,XIONG L Z.Systematic sequence analysis and identification of tissue-specific or stressresponsive genes of NAC transcription factor family in rice[J].Molecular Genetics and Genomics,2008,280(6):547-563.
[47] HAN K,ZHAO Y,LIU J,TIAN Y,EL-KASSABY Y A,QI Y,KE M,SUN Y,LI Y. Genome-wide investigation and analysis of NAC transcription factor family in Populus tomentosa and expression analysis under salt stress[J].Plant Biology,2024,26(5):764-776.
[48] NOMAN A,LIU Z Q,AQEEL M,ZAINAB M,KHAN M I,HUSSAIN A,ASHRAF M F,LI X,WENG Y H,HE S L.Basic leucine zipper domain transcription factors:The vanguards in plant immunity[J]. Biotechnology Letters,2017,39(12):1779-1791.
[49] ZANETTI M E,RÍPODAS C,NIEBEL A.Plant NF-Y transcription factors:Key players in plant-microbe interactions,root development and adaptation to stress[J]. Biochimica et Biophysica Acta (BBA)- Gene Regulatory Mechanisms,2017,1860(5):645-654.
[50] DONG B X,LIU Y,HUANG G,SONG A P,CHEN S M,JIANG J F,CHEN F D,FANG W M.Plant NAC transcription factors in the battle against pathogens[J]. BMC Plant Biology,2024,24(1):958.
[51] BU Q Y,JIANG H L,LI C B,ZHAI Q Z,ZHANG J,WU X Y,SUN J Q,XIE Q,LI C Y.Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses[J]. Cell Research,2008,18(7):756-767.
[52] WANG X E,BASNAYAKE B M V S,ZHANG H J,LI G J,LI W,VIRK N,MENGISTE T,SONG F M. The Arabidopsis ATAF1,a NAC transcription factor,is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens[J]. Molecular Plant-Microbe Interactions,2009,22(10):1227-1238.
[53] HE X,ZHU L F,XU L,GUO W F,ZHANG X L. GhATAF1,a NAC transcription factor,confers abiotic and biotic stress responses by regulating phytohormonal signaling networks[J].Plant Cell Reports,2016,35(10):2167-2179.
[54] LIU Q,YAN S J,HUANG W J,YANG J Y,DONG J F,ZHANG S H,ZHAO J L,YANG T F,MAO X X,ZHU X Y,LIU B. NAC transcription factor ONAC066 positively regulates disease resistance by suppressing the ABA signaling pathway in rice[J].Plant Molecular Biology,2018,98(4):289-302.
[55] WANG H,BI Y,GAO Y Z,YAN Y Q,YUAN X,XIONG X H,WANG J J,LIANG J Y,LI D Y,SONG F M.A Pathogen-Inducible rice NAC transcription factor ONAC096 contributes to immunity against Magnaprothe oryzae and Xanthomonas oryzae pv. oryzae by direct binding to the promoters of OsRap2.6,Os-WRKY62,and OsPAL1[J]. Frontiers in Plant Science,2021,12:802758.
[56] NAKASHIMA K,TRAN L P,VAN NGUYEN D,FUJITA M,MARUYAMA K,TODAKA D,ITO Y,HAYASHI N,SHINOZAKI K,YAMAGUCHI-SHINOZAKI K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress- responsive gene expression in rice[J]. The Plant Journal,2007,51(4):617-630.
[57] YOKOTANI N,TSUCHIDA- MAYAMA T,ICHIKAWA H,MITSUDA N,OHME-TAKAGI M,KAKU H,MINAMI E,NISHIZAWA Y. OsNAC111,a blast disease-responsive transcription factor in rice,positively regulates the expression of defense-related genes[J]. Molecular Plant-Microbe Interactions,2014,27(10):1027-1034.
[58] ZHOU J G,MU Q,WANG X Y,ZHANG J,YU H Z,HUANG T Z,HE Y X,DAI S J,MENG X Z. Multilayered synergistic regulation of phytoalexin biosynthesis by ethylene,jasmonate,and MAPK signaling pathways in Arabidopsis[J]. The Plant Cell,2022,34(8):3066-3087.
[59] WANG F T,LIN R M,FENG J,CHEN W Q,QIU D W,XU S C. TaNAC1 acts as a negative regulator of stripe rust resistance in wheat,enhances susceptibility to Pseudomonas syringae,and promotes lateral root development in transgenic Arabidopsis thaliana[J].Frontiers in Plant Science,2015,6:108.
[60] DALMAN K,WIND J J,NEMESIO-GORRIZ M,HAMMERBACHER A,LUNDÉN K,EZCURRA I,ELFSTRAND M.Overexpression of PaNAC03,a stress induced NAC gene family transcription factor in Norway spruce leads to reduced flavonol biosynthesis and aberrant embryo development[J]. BMC Plant Biology,2017,17(1):6.
[61] KANEDA T,TAGA Y R,TAKAI R,IWANO M,MATSUI H,TAKAYAMA S,ISOGAI A,CHE F S. The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death[J].The EMBO Journal,2009,28(7):926-936.
[62] WANG Z Y,XIA Y Q,LIN S Y,WANG Y R,GUO B H,SONG X N,DING S C,ZHENG L Y,FENG R Y,CHEN S L,BAO Y L,SHENG C,ZHANG X,WU J G,NIU D D,JIN H L,ZHAO H W. Osa-miR164a targets OsNAC60 and negatively regulates rice immunity against the blast fungus Magnaporthe oryzae[J].The Plant Journal,2018,95(4):584-597.