花色苷属于类黄酮类物质,具有典型的C6-C3-C6结构(两个苯环通过3个碳原子相连),由花色素与葡萄糖分子通过糖苷键连接而成[1]。花色苷是红葡萄品种及红葡萄酒中的呈色物质,直接影响了葡萄和葡萄酒的品质和经济价值,是酿酒葡萄最重要的经济性状之一。此外,花色苷还具有抗氧化、抗炎、抗肿瘤、保护视力、降血脂等生理功能,进而影响消费者的偏好和葡萄酒的市场价值[2-3]。因此花色苷是酿酒葡萄育种中的重点关注的目标之一。
欧亚种葡萄(Vitis vinifera L.)中主要存在5种花色素,包括花青素(Cyanidin)、花翠素(Delphinidin)、甲基花青素(Peonidin)、甲基花翠素(Petunidin)、二甲花翠素(Malvidin)[1]。这些花色素C3位上的羟基与葡萄糖C1 位上的羟基失去一分子水,形成花色素-3-O-葡萄糖苷。花色苷的生物合成在关键酶类黄酮-3’-羟基化酶(Flavonoid-3’-hydroxylase,F3’H)和类黄酮-3’,5’-羟基化酶(Flavonoid-3’5’-hydroxylase,F3’5’H)的作用下,形成两个分支,在F3’H分支下合成的花青素和甲基花青素被称为F3’H花色素,其在B 环上有两个取代基,而在F3’5’H 分支下合成的花翠素、甲基花翠素和二甲花翠素被称为F3’5’H 花色素,其在B 环上有3 个取代基[4]。自然条件下花色素很少以游离状态存在,在欧亚种葡萄中大多为C环上C3位羟基被糖苷化,并且糖基上的羟基还可以与脂肪酸(如乙酸)、羟基肉桂酸(如咖啡酸、对香豆酸、阿魏酸)相连形成酰化花色苷,进一步增强了花色苷在水溶液中的稳定性[5]。花色苷的组成与含量直接影响葡萄浆果的颜色。花色苷的呈色基团与B 环上的羟基和甲基数量有关,甲基数量越多时,花色苷越趋向于红色色调;而羟基数量越多,花色苷越表现出蓝紫色色调[6]。
由于葡萄是多年生果树,树体较大、世代周期长且高度杂合,而花色苷含量属于复杂的数量性状,使用传统育种方式进行颜色改良或不同果色创制难度较大,利用分子生物学技术进行葡萄基因组辅助育种显得尤为重要[7]。随着高通量测序技术和分析技术的发展,基于连锁不平衡(Linkage disequilibrium,LD)的关联映射(Association mapping,AM)可用于复杂性状的解析[8]。作为AM 方法之一的全基因组关联分析可以在全基因组范围内,检测群体的遗传变异多态性,与表型进行关联分析,最终筛选出与表型相关的位点和基因,是目前解析复杂数量性状的重要手段[9]。
近年来,研究者开始将全基因组关联分析(Genome-wide association analysis,GWAS)应用于葡萄遗传位点的鉴定,如抗性[10-12]、叶片形态[13-14]、香气物质[15-16]、浆果颜色[17]、浆果特性[18-20]等多个方面,但前人对葡萄果皮着色机制的研究主要集中在控制果皮颜色的有无或颜色分类的基因或位点上,未关注不同结构花色苷的遗传趋势及相关的SNP 位点及基因。基于不同结构花色苷呈色的差异,开展相关的遗传规律研究,对果实花色育种有重要的指导意义。
笔者在本研究中以欧亚种酿酒葡萄品种赤霞珠685和西拉100的杂交F1子代作为群体材料,利用高效液相色谱-串联三重四级杆质谱联用(High performance liquid chromatography triple quadrupole mass spectrometry,HPLC-QqQ-MS)技术检测花色苷的组成及含量,为全基因组关联分析提供表型数据。利用笔者课题组前期对该杂交F1群体全基因组重测序获得的大量SNP位点[21-22],进行GWAS分析,以期获得与葡萄果皮花色苷显著关联的SNP 位点和候选基因,用于开发分子标记,并为葡萄色泽的分子设计育种提供依据。
1 材料和方法
1.1 植物材料
以山西省农业科学院果树研究所培育的酿酒葡萄赤霞珠685(Vitis vinifera L.‘Cabernet Sauvignon 685’,CS 685)和西拉100(V. vinifera L.‘Syrah 100’,X 100)以及两者的正交和反交得到的F1代为试材,杂交在2013 年进行,在2018 年开始大量结果。这些子代均已通过杂交真实性鉴定。亲本及其子代均种植于山西农业科学院果树研究所酿酒葡萄育种圃(37°34′ N,112°49′ E),行向为南北行向,株行距为0.5 m×2.5 m。
试验所用的葡萄果实样品于2019 年采集。采样在葡萄果实总可溶性固形物(TSS,°Brix)含量约为21°Brix时进行,此时果实已成熟[23]。采得真实杂交子代样品共81份。
1.2 试剂与标准品
分析纯:甲醇,购自天津化工厂;色谱级:甲醇、甲酸、乙腈,购自美国Sigma-Aldrich 公司;标准品:二甲花翠素-3-O-葡萄糖苷购自美国Sigma-Aldrich公司。
1.3 主要仪器
高效液相色谱(Agilent1200 系列)串联三重四级杆质谱仪(HPLC-QqQ-MS)(美国Agilent 有限公司);Poroshell 120 EC-C18(150 mm×2.1 mm,2.7µm)色谱柱(美国Agilent有限公司)。
1.4 花色苷的提取与检测
参照已有花色苷提取与检测方法,并稍作修改[22]。
果皮中的花色苷提取:果实在液氮冷冻后剥皮,果皮在液氮环境下研磨成粉,真空冻干36 h。于2 mL离心管中精确称量0.100 g冻干粉末,加入1 mL 50%的甲醇水溶液,冰浴避光超声处理20 min后,在4 ℃下12 000 r·min-1离心5 min。收集上清液,对残渣重复提取1次,将2次上清液合并后在-40 ℃冰箱保存。
果皮中花色苷的检测:提取液测定前使用0.22 μm 聚四氟乙烯(PTFE)滤膜过滤,进样量5 μL。流动相A为0.1%甲酸的水溶液,B为含0.1%甲酸的50%的甲醇乙腈溶液。HPLC-QqQ-MS 的洗脱程序:90%~100%A,10%~100% B,持续15 min,后运行程序5 min。流动相流速为0.4 mL·min-1。柱温控制为55 ℃。质谱采用电喷雾离子源,正离子模式,离子源温度为150 ℃,干燥气温度为350 ℃,流量为12 L·min-1,喷雾电压为4 kV,雾化器压力为35 psi,检测器为多反应监测模式。
花色苷的定性依据为已建立的葡萄与葡萄酒酚类物质HPLC-UV-MS指纹谱库[24]。采用外标法定量,以二甲花翠素-3-O-葡萄糖苷为外标物,单位表示为mg·kg(-1以鲜质量计)。标线如下:y=0.000 013 07x+2.8(线性范围:12.13~125.49 mg·L-1,R2=0.991)。
1.5 全基因组关联分析
笔者课题组前期对杂交子代群体进行了全基因组重测序,使用SAMTOOLS 软件在群体中检测得到8 417 765个SNP位点,经过滤最终得到3 314 995个高质量SNPs,用于GWAS分析。
笔者在本研究中使用GEMMA 软件,采用单变量混合线性模型、以亲缘关系矩阵(kinship)作为随机效应,用GATA 1.92.4 进行主成分分析,将前3 个主成分作为协变量加入到模型中,以校正群体分层,对群体不同结构花色苷性状进行关联分析,通过关联的显著度(p<1e-6),筛选潜在的候选SNP位点。
y=Xβ+Pγ+Zu+ϵ。
该式中y为表型值的向量;X为固定效应的设计矩阵,用于表示固定效应的因子;β为固定效应的参数向量;P为包含了主成分作为协变量的设计矩阵;γ为与主成分相关的固定效应参数向量;Z为与随机效应相关的设计矩阵;u 为随机效应的向量;ϵ 为残差项。
1.6 候选基因分析
结合各性状对应的SNP 位点在葡萄参考基因组(http://genomes.cribi.unipd.it/grape/)上的物理位置,通过Cribi Genomics 网站(http://genomes.cribi.unipd.it/)对显著SNP 位点±100 kb 范围内的基因进行功能注释。基于基因功能注释,筛选与花色苷生物合成调控有关的基因,并将其中SNP位点位于基因内部的基因确定为候选基因。
1.7 统计分析
使用Microsoft Excel 2021对样本的花色苷含量进行统计分析,计算变异系数(Variation coefficient,CV)、广义遗传力(Broad sense heritability,H2),各指标的计算公式如下:
式中:SD为子代标准差,![]() 为子代均值。
为子代均值。
H2/%=(VP-VE)/VP×100,
式中:VP为表现型方差,VE为环境方差。
VE=(VP1-VP2)/2,
式中:VP1为母本的表型方差,VP2为父本的表型方差。
2 结果与分析
2.1 杂交F1群体内花色苷表型变异
81 份赤霞珠685 和西拉100 杂交子代中,有53个子代的果皮呈现明显红色,28个子代肉眼观察不到红色。如表1 所示,从有颜色的葡萄果皮中共检出20种花色苷,包括5种基本花色苷和15种乙酰化、香豆酰化和咖啡酰化形式,而乙酰化花青苷和香豆酰化甲基花青苷在所有未观察到红色的果皮中都未检出。F1子代之间花色苷组分的浓度有较大差别。
表1 杂交F1群体各个花色苷性状的遗传指标
Table 1 Genetic indicators of anthocyanins in the F1population
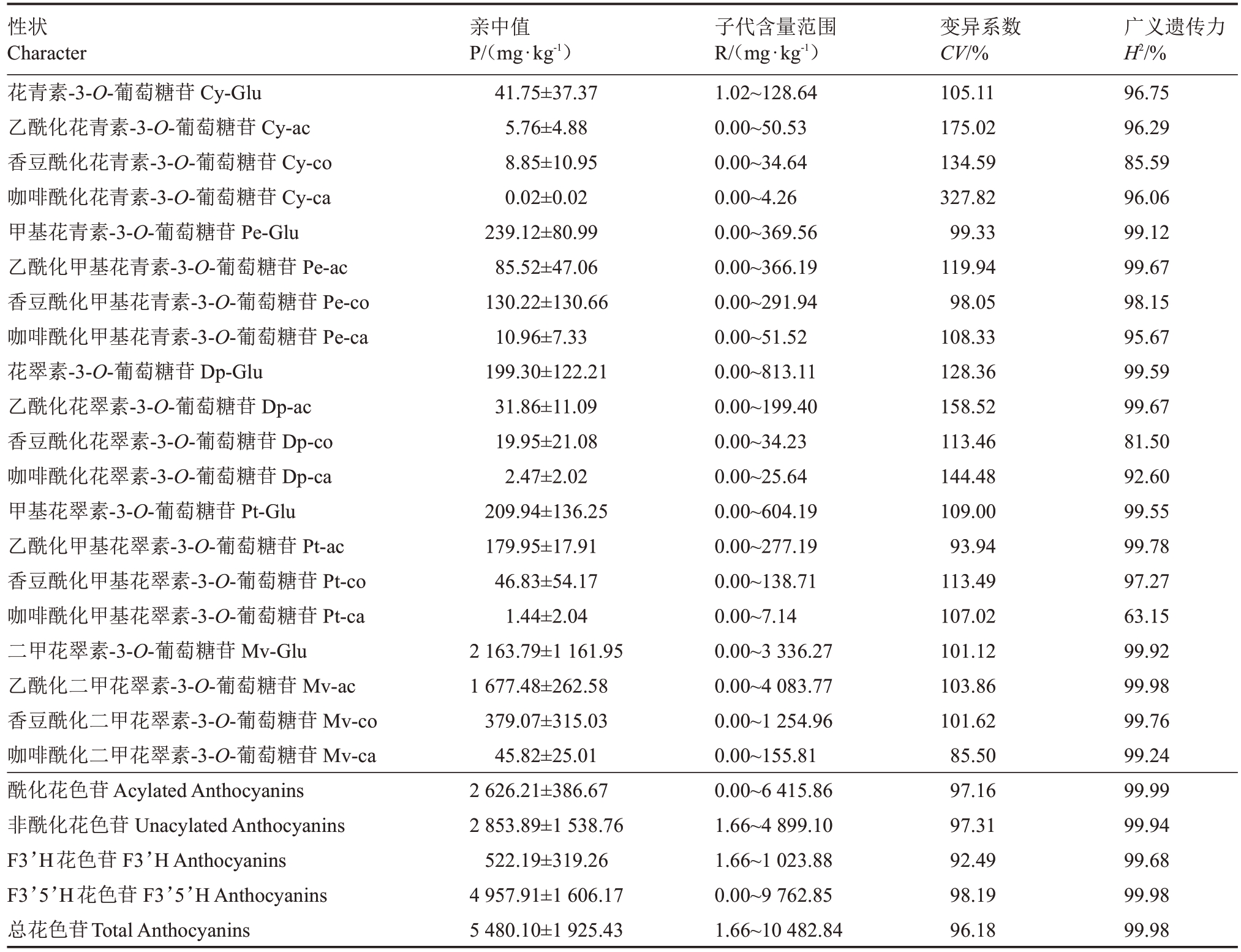
性状Character花青素-3-O-葡萄糖苷Cy-Glu乙酰化花青素-3-O-葡萄糖苷Cy-ac香豆酰化花青素-3-O-葡萄糖苷Cy-co咖啡酰化花青素-3-O-葡萄糖苷Cy-ca甲基花青素-3-O-葡萄糖苷Pe-Glu乙酰化甲基花青素-3-O-葡萄糖苷Pe-ac香豆酰化甲基花青素-3-O-葡萄糖苷Pe-co咖啡酰化甲基花青素-3-O-葡萄糖苷Pe-ca花翠素-3-O-葡萄糖苷Dp-Glu乙酰化花翠素-3-O-葡萄糖苷Dp-ac香豆酰化花翠素-3-O-葡萄糖苷Dp-co咖啡酰化花翠素-3-O-葡萄糖苷Dp-ca甲基花翠素-3-O-葡萄糖苷Pt-Glu乙酰化甲基花翠素-3-O-葡萄糖苷Pt-ac香豆酰化甲基花翠素-3-O-葡萄糖苷Pt-co咖啡酰化甲基花翠素-3-O-葡萄糖苷Pt-ca二甲花翠素-3-O-葡萄糖苷Mv-Glu乙酰化二甲花翠素-3-O-葡萄糖苷Mv-ac香豆酰化二甲花翠素-3-O-葡萄糖苷Mv-co咖啡酰化二甲花翠素-3-O-葡萄糖苷Mv-ca酰化花色苷Acylated Anthocyanins非酰化花色苷Unacylated Anthocyanins F3’H花色苷F3’H Anthocyanins F3’5’H花色苷F3’5’H Anthocyanins总花色苷Total Anthocyanins亲中值P/(mg·kg-1)41.75±37.37 5.76±4.88 8.85±10.95 0.02±0.02 239.12±80.99 85.52±47.06 130.22±130.66 10.96±7.33 199.30±122.21 31.86±11.09 19.95±21.08 2.47±2.02 209.94±136.25 179.95±17.91 46.83±54.17 1.44±2.04 2 163.79±1 161.95 1 677.48±262.58 379.07±315.03 45.82±25.01 2 626.21±386.67 2 853.89±1 538.76 522.19±319.26 4 957.91±1 606.17 5 480.10±1 925.43子代含量范围R/(mg·kg-1)1.02~128.64 0.00~50.53 0.00~34.64 0.00~4.26 0.00~369.56 0.00~366.19 0.00~291.94 0.00~51.52 0.00~813.11 0.00~199.40 0.00~34.23 0.00~25.64 0.00~604.19 0.00~277.19 0.00~138.71 0.00~7.14 0.00~3 336.27 0.00~4 083.77 0.00~1 254.96 0.00~155.81 0.00~6 415.86 1.66~4 899.10 1.66~1 023.88 0.00~9 762.85 1.66~10 482.84变异系数CV/%105.11 175.02 134.59 327.82 99.33 119.94 98.05 108.33 128.36 158.52 113.46 144.48 109.00 93.94 113.49 107.02 101.12 103.86 101.62 85.50 97.16 97.31 92.49 98.19 96.18广义遗传力H2/%96.75 96.29 85.59 96.06 99.12 99.67 98.15 95.67 99.59 99.67 81.50 92.60 99.55 99.78 97.27 63.15 99.92 99.98 99.76 99.24 99.99 99.94 99.68 99.98 99.98
为了解不同结构花色苷的遗传倾向,将在亲本和子代中检测到的花色苷按不同结构进行归类,将浓度进行累加,从而延伸出酰化花色苷和非酰化花色苷,以及F3’H 花色苷和F3’5’H 花色苷。将他们以及所有花色苷的总含量作为表型,共计25 个表型,对各表型进行统计分析(表1)。
广义遗传力代表了一个群体由基因型所决定变异的大小,在本研究中,除咖啡酰化甲基花翠苷外,其他24个表型的广义遗传力都在80%以上,其中15 种表型的广义遗传力达到99%,他们是甲基花青苷、乙酰化甲基花青苷、花翠苷、乙酰化花翠苷、甲基花翠苷、乙酰化甲基花翠苷、二甲花翠苷、乙酰化二甲花翠苷、香豆酰化二甲花翠苷、咖啡酰化二甲花翠苷、酰化和非酰化花色苷、F3’H 和F3’5’H花色苷、总花色苷,这些结果表明,各个花色苷表型均有较高的遗传效应。
对比亲中值与子代含量范围,可以发现,子代呈现明显的分离,各表型变异系数均超过85%,其中咖啡酰化花青苷的变异系数最高,达到327.82%。图1 直观展示了花色苷的含量分布,大部分表型在杂交子代群体中都呈现出广泛且连续的分布特征。

图1 在杂交F1群体中花色苷含量的分布频率
Fig.1 Distribution frequency of anthocyanin concentrations in hybrid F1 population
2.2 各个花色苷组分含量的相关性分析
基于不同花色苷在含量上的分布规律极为相似,笔者进一步采用Pearson 相关性分析评估不同表型之间的相关性。结果如图2 所示,除花青苷含量和咖啡酰化花青苷含量与其他花色苷含量之间有较低的相关性之外,其他花色苷含量之间均存在显著的正相关。这些结果表明,不同花色苷性状可能受类似机制的调控,其调控位点可能位于花色苷生物合成途径的主干路径基因或是调控该途径的转录因子。
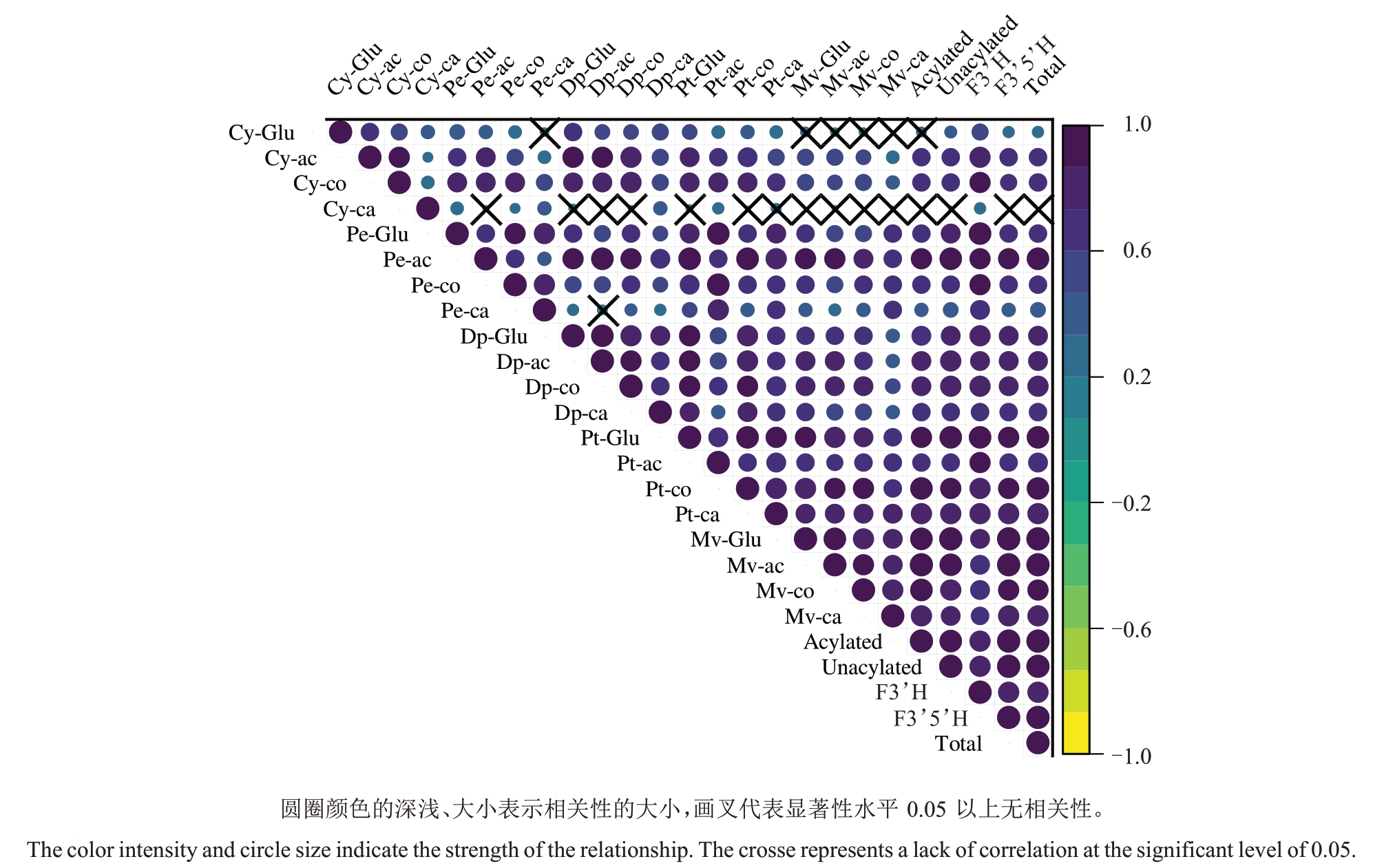
图2 杂交F1群体中各个花色苷性状含量的相关性分析
Fig.2 Correlations among the concentrations of various anthocyanins in the F1 population
2.3 花色苷含量的全基因组关联分析
将25 个花色苷含量性状数据分别与3 314 995个高质量SNP 位点进行全基因组关联分析,图3 展示了花翠苷、甲基花翠苷、二甲花翠苷、乙酰化二甲花翠苷和香豆酰化二甲花翠苷花色苷含量全基因组关联分析得到的曼哈顿图和Q-Q 图。以p<1e-6为筛选阈值,超过阈值线的即为与性状显著关联的SNP位点。Q-Q图反映了有较好的GWAS质控。

图3 花色苷全基因组关联分析曼哈顿图、Q-Q 图
Fig.3 Genome-wide association analysis of anthocyanin Manhattan plot,Q-Q plot
GWAS 结果显示,各表型合计定位到17 382 个显著SNP 位点,每个表型均关联到显著SNP 位点,这些位点绝大多数都分布于2号染色体上。大部分表型曼哈顿图峰型非常相似,在2 号染色体处有显著SNP簇,这表明与葡萄花色苷合成相关的位点在染色体上集中分布。图4显示了各表型显著SNP位点的交集情况,其中香豆酰化花翠苷、咖啡酰化甲基花翠苷、乙酰化甲基花青苷、香豆酰化甲基花青苷、乙酰化花翠苷、甲基花翠苷、乙酰化甲基花翠苷、香豆酰化甲基花翠苷、F3’H 花色苷、乙酰化二甲花翠苷、咖啡酰化二甲花翠苷、二甲花翠苷、非酰化花色苷、F3’5’H 花色苷、酰化花色苷、总花色苷、香豆酰化二甲花翠苷交集数量最多,存在1275个共同的显著SNP 位点。此外,各集合也存在大量交集,表明不同花色苷之间受相同位点调控,存在共同的遗传基础。
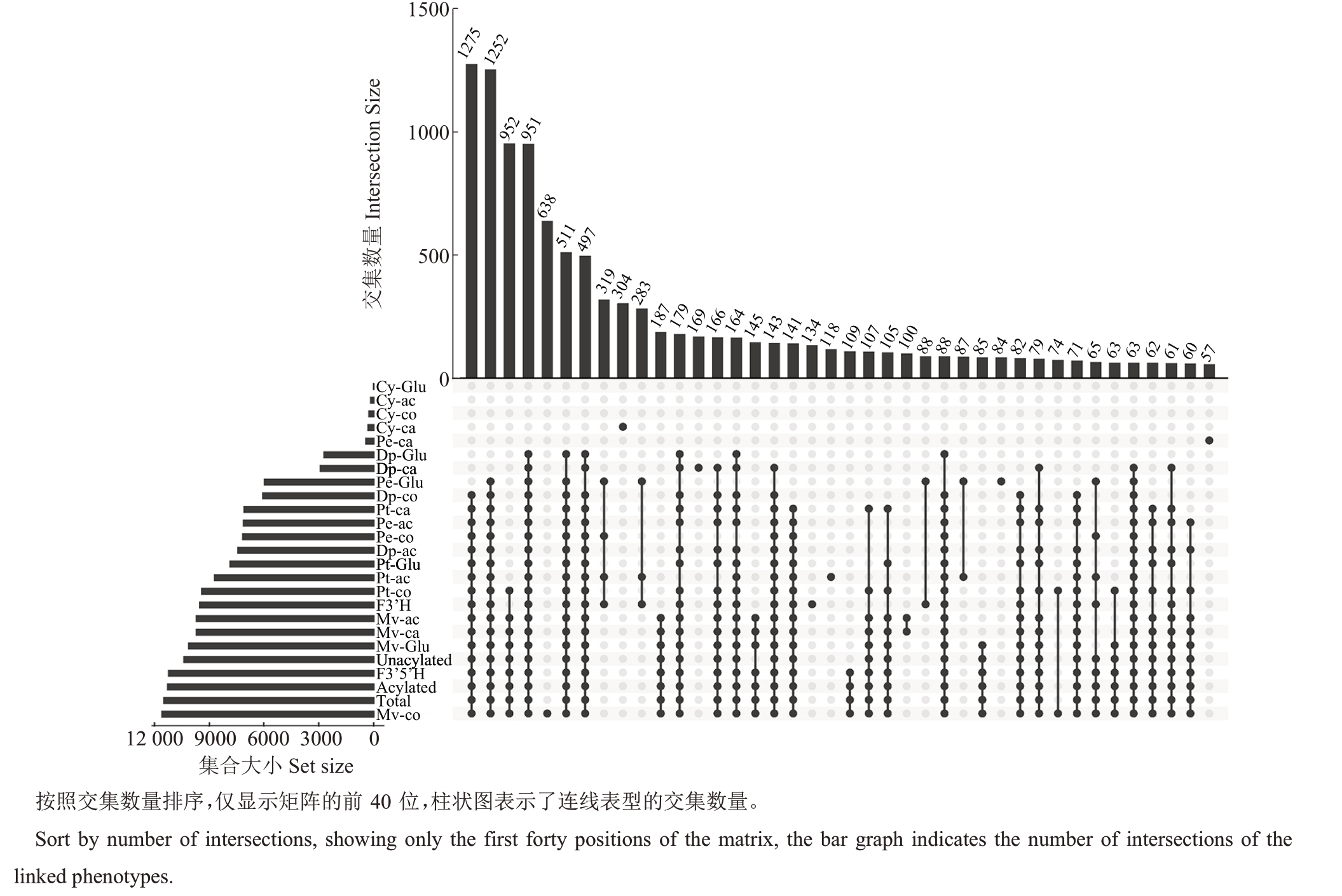
图4 花色苷显著SNP 位点交集
Fig.4 Significant SNP site intersection for anthocyanins
2.4 候选基因与SNP位点的挖掘
利用已公布的葡萄(Vitis vinifera‘Pinot Noir’PN40024 v2.1)全基因组,将与显著关联的SNP位点定位在葡萄基因组上,以挖掘SNP 位点±100 kb 范围内的基因。结果显示,所有显著SNP位点共对应7127个基因。基于基因功能注释进行筛选,重点关注位于基因编码区内的SNP 位点及其所对应的基因,得到3个候选基因及其9个位于基因编码区内的SNP位点(表2,图5)。以上筛选出SNP位点合计关联到除花青苷与咖啡酰化花青苷外的23个表型,且大多数表型关联到的SNP位点和基因都彼此重复,可能由于花青苷与咖啡酰化花青苷在2号染色体上SNP 位点密度较低,未筛选出与之相关的SNP 位点。

图5 候选SNP 位点不同基因型个体在杂交F1群体中的含量分布箱线图
Fig.5 Boxplot of the distribution of the content of individuals of different genotypes of candidate SNP loci in F1
表2 基因编码区内显著SNP 位点信息及其基因注释
Table 2 Information of significant SNP loci within genes CDS and their gene annotation.
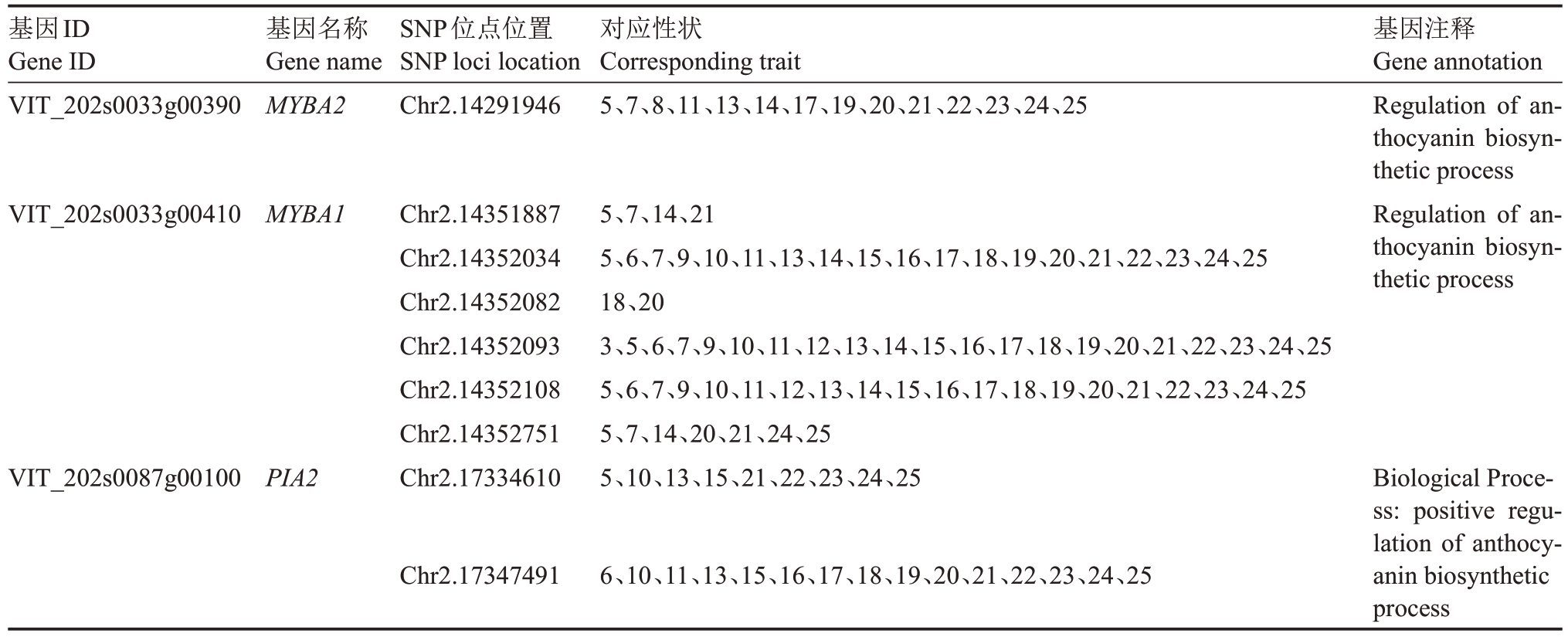
注:对应性状的序号:1.Cy-Glu;2.Cy-ac;3.Cy-co;4.Cy-ca;5.Pe-Glu;6.Pe-ac;7.Pe-co;8.Pe-ca;9.Dp-Glu;10.Dp-ac;11.Dp-co;12.Dp-ca;13.Pt-Glu;14.Pt-ac;15.Pt-co;16.Pt-ca;17.Mv-Glu;18.Mv-ac;19.Mv-co;20.Mv-ca;21.酰化花色苷;22.非酰化花色苷;23.F3’H 花色苷;24.F3’5’H 花色苷;25.总花色苷。
Note:Corresponding trait numbers:1.Cy-Glu;2.Cy-ac;3.Cy-co;4.Cy-ca;5.Pe-Glu;6.Pe-ac;7.Pe-co;8.Pe-ca;9.Dp-Glu;10.Dp-ac;11.Dpco;12.Dp-ca;13.Pt-Glu;14.Pt-ac;15.Pt-co;16.Pt-ca;17.Mv-glu;18.Mv-ac;19.Mv-co;20.Mv-ca;21.Acylated anthocyanin;22.Unacylated anthocyanin;23.F3’H anthocyanin;24.F3’5’H anthocyanin;25.Total anthocyanin.
基因ID Gene ID VIT_202s0033g00390基因名称Gene name MYBA2 SNP位点位置SNP loci location Chr2.14291946对应性状Corresponding trait 5、7、8、11、13、14、17、19、20、21、22、23、24、25 VIT_202s0033g00410MYBA1基因注释Gene annotation Regulation of anthocyanin biosynthetic process Regulation of anthocyanin biosynthetic process VIT_202s0087g00100Biological Process: positive regulation of anthocyanin biosynthetic process PIA2 Chr2.14351887 Chr2.14352034 Chr2.14352082 Chr2.14352093 Chr2.14352108 Chr2.14352751 Chr2.17334610 5、7、14、21 5、6、7、9、10、11、13、14、15、16、17、18、19、20、21、22、23、24、25 18、20 3、5、6、7、9、10、11、12、13、14、15、16、17、18、19、20、21、22、23、24、25 5、6、7、9、10、11、12、13、14、15、16、17、18、19、20、21、22、23、24、25 5、7、14、20、21、24、25 5、10、13、15、21、22、23、24、25 Chr2.173474916、10、11、13、15、16、17、18、19、20、21、22、23、24、25
基因VIT_202s0033g00390 和VIT_202s0033g0-0410 分别被注释为VvMYBA2 和VvMYBA1,它们均是已报道的调控花色苷合成的转录因子[25-27],这些也证实了本研究结果的可靠性。 基因VIT_202s0087g00100 在拟南芥中的同源基因被注释为光敏色素互作蛋白2(PIA2),PIA2 是拟南芥中花色苷积累的正调节因子[28],但在葡萄中其功能未被证实。
2.5 三个转录因子关联的SNP 位点基因型与花色苷含量的关系
由于密码子的简并性,位于基因编码区内的SNP位点突变可能不会引起非同义突变。在本研究中,位于MYBA2 基因编码区内的Chr2.14291946 和位于MYBA1 基因编码区内的Chr2.14351887、Chr2.14352034、 Chr2.14352082、 Chr2.14352108、Chr2.14352751 以及PIA2 基因内的Chr2.17334610均发生了非同义突变,其余2 个SNP 位点为同义突变(表3)。
表3 候选SNP 位点的变化信息
Table 3 Candidate SNPs mutation information

注:氨基酸缩写:D.天冬氨酸;E.谷氨酸;V.缬氨酸;A.丙氨酸;Q.谷氨酰胺;P.脯氨酸;R.精氨酸;T.苏氨酸;S.丝氨酸;I.异亮氨酸;L.亮氨酸。
Note:Amino acid abbreviations:D.Aspartic acid;E.Glutamic acid;V.Valine;A.Alanine;Q.Glutamine;P.Proline;R.Arginine;T.Threonine;S.Serine;I.Isoleucine;L.Leucine.
基因Gene MYBA2 SNP位点位置SNP loci location Chr2.14291946氨基酸突变Amino acid mutation E→D MYBA1Chr2.14351887V→A Chr2.14352034Q→P Chr2.14352082R→P Chr2.14352093T Chr2.14352108R→S Chr2.14352751V→I PIA2Chr2.17334610L→I Chr2.17347491 SNP位点突变信息SNP loci mutation information G→C(低→高)G→C(Low-High)A→G(低→高)A→G(Low-High)T→G(低→高)T→G(Low-High)G→C(低→高)G→C(Low-High)A→C(低→高)A→C(Low-High)C→A(低→高)C→A(Low-High)C→T(低→高)C→T(Low-High)G→T(低→高)G→T(Low-High)T→C(低→高)T→C(Low-High)突变类型Mutation type非同义突变Non-synonymous mutations非同义突变Non-synonymous mutations非同义突变Non-synonymous mutations非同义突变Non-synonymous mutations同义突变Synonymous mutations非同义突变Non-synonymous mutations非同义突变Non-synonymous mutations非同义突变Non-synonymous mutations同义突变Synonymous mutations S
在杂交F1群体中大部分SNP位点存在3种基因型,且不同基因型对应的花色苷含量具有明显差异,基因型与表型含量的变化关系见表3。由于突变引起的表型变异规律相似,笔者在本研究中重点关注了与酰化花色苷、非酰化花色苷、F3’H花色苷、F3’5’H花色苷相关的SNP位点的基因分型结果(图5),如与酰化花色苷、非酰化花色苷、F3’H 花色苷、F3’5’H花色苷均显著关联的SNP位点Chr2.14291946,位于MYBA2基因的CDS区域,该位点处基因型为G/C的子代含量均值高于基因型为G/G 的子代含量均值,且当该位点处核苷酸由G 突变为C 时,引起了非同义突变,导致该基因氨基酸序列中的缬氨酸(E)被谷氨酸(D)取代,相应的各表型的含量增加。
3 讨 论
尽管葡萄果皮花色苷含量受多种因素影响,但其组成和相对含量主要由遗传因素决定[29-31]。全基因组关联分析是解析这一复杂数量性状遗传结构的有效方法。花色苷的生物合成过程中涉及多个基因的相互作用和调控网络,目前,花色苷的生物合成途径已经明确,途径中合成关键酶的结构基因功能大多保守,一些转录因子是导致花色苷差异积累的主要因素[32]。
前人研究表明,位于Chr2上的相邻基因VvMYBA1和VvMYBA2是连锁的,可将二者视为一个单倍型,两基因可以独立调控VvUFGT基因的表达,决定果皮是否呈色,也就是说红葡萄品种至少含有一种类型的功能性等位基因,且等位基因组成不同的单倍型产生的葡萄果皮颜色也不同[33-35]。有研究表明,VvMYBA1 启动子区域的SNP 位点突变导致了红色果皮的Benitaka 到黑色果皮的Brazil 的芽变[36]。笔者在本研究中也关联到了VvMYBA1和VvMYBA2基因编码区的SNP 位点,在前人的研究中未被报道,将为探究葡萄果皮颜色调控机制提供靶标。VIT_202s0087g00100 在拟南芥中的同源基因为PIA2,以往有关PIA2 的报道较少,在拟南芥幼苗中PIA2 被发现可通过激活UFGT基因表达参与调控花色苷的生物合成,PIA2也可以抑制光敏色素A介导的PIF3快速磷酸化,防止磷酸化后的PIF3被泛素-蛋白酶体途径降解[37-39]。转录因子PIF3属于bHLH家族,是花色苷合成路径中关键结构基因CHS 的正调控因子[40]。综合来看,PIA2 在调控植物花色苷合成途径中发挥着多重作用。笔者在本研究中首次发现PIA2可能参与了葡萄果皮花色苷合成的调控,其机制将有待深入探究。
在本研究中,大多数表型含量之间显著正相关,且关联到的SNP 位点和基因都彼此重复。共筛选到9个显著SNP位点,关联到3个转录因子,包括已验证参与葡萄花色苷合成的MYBA2、MYBA1以及尚未报道与葡萄花色苷路径有关的基因PIA2。有7个显著SNP位点发生非同义突变,突变后对应的花色苷含量显著上升,具有开发为分子标记的潜力。对于2 个发生同义突变的位点,突变后对应花色苷含量同样显著上升,可能由于同义突变在转录水平调控、翻译效率等层面对表型仍能对表型产生影响。笔者在本研究中筛选与花色苷合成相关的关键基因和位点,可为深入研究葡萄花色苷合成的转录调控机制提供参考。并可通过挖掘花色苷遗传调控位点开发分子标记,可用于葡萄育种的早期筛选,加快分子育种进程。
4 结 论
不同结构花色苷有相似的遗传趋势,控制不同结构花色苷合成的遗传位点高度相似,且主要位于2 号染色体上。首次在酿酒葡萄杂交F1群体中,利用GWAS分析筛选到调控花色苷生物合成的9个遗传位点和所对应的3 个候选基因,并发现PIA2(VIT_202s0087g00100)与葡萄花色苷生物合成相关联,这些发现对葡萄生产和育种工作具有重要意义。
[1] 张欣珂,赵旭,成池芳,齐梦瑶,石英. 葡萄酒中的酚类物质Ⅰ:种类、结构及其检测方法研究进展[J].食品科学,2019,40(15):255-268.ZHANG Xinke,ZHAO Xu,CHENG Chifang,QI Mengyao,SHI Ying. Phenolics in wines Ⅰ:A review of categories,structures and detection methods[J]. Food Science,2019,40(15):255-268.
[2] 张淑淑,吕想,刘伟,张菊华.果蔬花色苷的理化特性、提取技术及功能活性研究进展[J].食品与发酵工业,2024,50(2):360-371.ZHANG Shushu,LÜ Xiang,LIU Wei,ZHANG Juhua. Research progress on physical and chemical characteristics,extraction technology,and functional activity of anthocyanins from fruits and vegetables[J]. Food and Fermentation Industries,2024,50(2):360-371.
[3] 崔云斌,李嘉钰,刘钰,李苏辉,蒲旭欣,才让卓玛,王世光,邹登朗.植物花色苷及其生物合成与药理的研究进展[J].青海草业,2022,31(3):71-75.CUI Yunbin,LI Jiayu,LIU Yu,LI Suhui,PU Xuxin,CAI Rangzhuoma,WANG Shiguang,ZOU Denglang. Advances in the study of plant anthocyanins and their biosynthesis and pharmacology[J].Qinghai Prataculture,2022,31(3):71-75.
[4] 张龙,李卫华,姜淑梅,朱根发,王碧青,李洪清.花色素苷生物合成与分子调控研究进展[J]. 园艺学报,2008,35(6):909-916.ZHANG Long,LI Weihua,JIANG Shumei,ZHU Genfa,WANG Biqing,LI Hongqing.Progress of molecular basis of biosynthesis and transcriptional regulation of anthocyanins[J].Acta Horticulturae Sinica,2008,35(6):909-916.
[5] NAKAYAMA T,SUZUKI H,NISHINO T. Anthocyanin acyltransferases:Specificities,mechanism,phylogenetics,and applications[J]. Journal of Molecular Catalysis B:Enzymatic,2003,23(2/3/4/5/6):117-132.
[6] HE F,MU L,YAN G L,LIANG N N,PAN Q H,WANG J,REEVES M J,DUAN C Q. Biosynthesis of anthocyanins and their regulation in colored grapes[J]. Molecules,2010,15(12):9057-9091.
[7] 倪海枝,王引,颜帮国,陈方永.果树基因组辅助育种技术研究现状与展望[J].分子植物育种,2023,21(5):1535-1550.NI Haizhi,WANG Yin,YAN Bangguo,CHEN Fangyong. Research status and prospects of genomics-assisted breeding technology in fruit tree[J]. Molecular Plant Breeding,2023,21(5):1535-1550.
[8] TELLO J,IBÁÑEZ J. Review:Status and prospects of association mapping in grapevine[J].Plant Science,2023,327:111539.
[9] KORTE A,FARLOW A. The advantages and limitations of trait analysis with GWAS:A review[J].Plant Methods,2013,9:29.
[10] WANG Y,XIN H P,FAN P G,ZHANG J S,LIU Y B,DONG Y,WANG Z M,YANG Y Z,ZHANG Q,MING R,ZHONG G Y,LI S H,LIANG Z C.The genome of Shanputao(Vitis amurensis) provides a new insight into cold tolerance of grapevine[J].The Plant Journal,2021,105(6):1495-1506.
[11] TRENTI M,LORENZI S,BIANCHEDI P L,GROSSI D,FAILLA O,GRANDO M S,EMANUELLI F. Candidate genes and SNPs associated with stomatal conductance under drought stress in Vitis[J].BMC Plant Biology,2021,21(1):7.
[12] ZHANG Y,FAN X C,LI Y F,SUN H S,JIANG J F,LIU C H.Restriction site-associated DNA sequencing reveals the molecular genetic diversity of grapevine and genes related to white rot disease[J].Scientia Horticulturae,2020,261:108907.
[13] LAPLANTE E R,FLEMING M B,MIGICOVSKY Z,WEBER M G. Genome-wide association study reveals a genomic region associated with mite-recruitment phenotypes in the domesticated grapevine(Vitis vinifera)[J].Genes,2021,12(7):1013.
[14] CHITWOOD D H,RANJAN A,MARTINEZ C C,HEADLAND L R,THIEM T,KUMAR R,COVINGTON M F,HATCHER T,NAYLOR D T,ZIMMERMAN S,DOWNS N,RAYMUNDO N,BUCKLER E S,MALOOF J N,ARADHYA M,PRINS B,LI L,MYLES S,SINHA N R.A modern ampelography:A genetic basis for leaf shape and venation patterning in grape[J].Plant Physiology,2014,164(1):259-272.
[15] YANG X X,GUO Y S,ZHU J C,NIU Z Z,SHI G L,LIU Z D,LI K,GUO X W.Genetic diversity and association study of aromatics in grapevine[J].Journal of the American Society for Horticultural Science,2017,142(3):225-231.
[16] SUN Q,HE L,SUN L,XU H Y,FU Y Q,SUN Z Y,ZHU B Q,DUAN C Q,PAN Q H.Identification of SNP loci and candidate genes genetically controlling norisoprenoids in grape berry based on genome-wide association study[J]. Frontiers in Plant Science,2023,14:1142139.
[17] CARDOSO S,LAU W,EIRAS DIAS J,FEVEREIRO P,MANIATIS N.A candidate-gene association study for berry colour and anthocyanin content in Vitis vinifera L.[J].PLoS One,2012,7(9):e46021.
[18] ZHANG C,CUI L W,FANG J G.Genome-wide association study of the candidate genes for grape berry shape-related traits[J].BMC Plant Biology,2022,22(1):42.
[19] LIANG Z C,DUAN S C,SHENG J,ZHU S S,NI X M,SHAO J H,LIU C H,NICK P,DU F,FAN P G,MAO R Z,ZHU Y F,DENG W P,YANG M,HUANG H C,LIU Y X,DING Y Q,LIU X J,JIANG J F,ZHU Y Y,LI S H,HE X H,CHEN W,DONG Y. Whole-genome resequencing of 472 Vitis accessions for grapevine diversity and demographic history analyses[J].Nature Communications,2019,10(1):1190.
[20] MIGICOVSKY Z,SAWLER J,GARDNER K M,ARADHYA M K,PRINS B H,SCHWANINGER H R,BUSTAMANTE C D,BUCKLER E S,ZHONG G Y,BROWN P J,MYLES S.Patterns of genomic and phenomic diversity in wine and table grapes[J].Horticulture Research,2017,4:17035.
[21] 刘敖一,孙琪,董志刚,刘镇海,段长青,潘秋红.‘赤霞珠’与‘西拉’杂交群体内种子单宁的遗传趋势分析[J].中外葡萄与葡萄酒,2022(4):8-13.LIU Aoyi,SUN Qi,DONG Zhigang,LIU Zhenhai,DUAN Changqing,PAN Qiuhong. Inheritance trend of seed condensed tannins in F1 hybrids of‘Cabernet Sauvignon’and‘Syrah’of Vitis vinifera[J]. Sino-Overseas Grapevine & Wine,2022(4):8-13.
[22] 卢浩成,魏巍,成池芳,陈武,李树德,王晓军,刘宗昭,王军.5个调色葡萄品种酚类物质轮廓分析[J]. 食品科学,2021,42(16):145-154.LU Haocheng,WEI Wei,CHENG Chifang,CHEN Wu,LI Shude,WANG Xiaojun,LIU Zongzhao,WANG Jun. Analysis of the phenolic profiles of five blending grape varieties[J]. Food Science,2021,42(16):145-154.
[23] 钟海霞,丁祥,周晓明,潘明启,张付春,韩守安,张雯,谢辉,王敏,艾尔买克·才卡斯木,伍新宇. 新疆10 个酿酒葡萄品种(系)可溶性糖组分及含量分析[J].新疆农业科学,2021,58(7):1323-1331.ZHONG Haixia,DING Xiang,ZHOU Xiaoming,PAN Mingqi,ZHANG Fuchun,HAN Shouan,ZHANG Wen,XIE Hui,WANG Min,Ermaik·Caikasim,WU Xinyu.Analysis of soluble sugar components and contents in 10 mature grape varieties(lines) in Xinjiang[J]. Xinjiang Agricultural Sciences,2021,58(7):1323-1331.
[24] LIANG N N,PAN Q H,HE F,WANG J,REEVES M J,DUAN C Q. Phenolic profiles of Vitis davidii and Vitis quinquangularis species native to China[J]. Journal of Agricultural and Food Chemistry,2013,61(25):6016-6027.
[25] MUÑOZ C,FANZONE M,LIJAVETZKY D. Transcriptional regulation of the anthocyanin biosynthesis pathway in developing grapevine berries in cultivar‘Malbec’by putative R2R3 MYB negative regulators[J]. Scientia Horticulturae,2019,257:108663.
[26] KOBAYASHI S,GOTO-YAMAMOTO N,HIROCHIKA H.Retrotransposon-induced mutations in grape skin color[J]. Science,2004,304(5673):982.
[27] WALKER A R,LEE E,ROBINSON S P. Two new grape cultivars,bud sports of Cabernet Sauvignon bearing pale-coloured berries,are the result of deletion of two regulatory genes of the berry colour locus[J]. Plant Molecular Biology,2006,62(4/5):623-635.
[28] SHIN J,PARK E,CHOI G.PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis[J].The Plant Journal,2007,49(6):981-994.
[29] 付东艳,汪蕾,张龙生.遮阳对马瑟兰葡萄花色苷组分及合成相关基因表达的影响[J].果树学报,2025,42(1):30-47.FU Dongyan,WANG Lei,ZHANG Longsheng. Effects of Sun shading on anthocyanin components and the expression of genes related to anthocyanin synthesis in Marselan grape[J].Journal of Fruit Science,2025,42(1):30-47.
[30] 薛晓斌,李栋梅,张艳霞,王振平.水分胁迫对马瑟兰葡萄果实品质及花色苷合成代谢的影响[J].果树学报,2023,40(5):919-931.XUE Xiaobin,LI Dongmei,ZHANG Yanxia,WANG Zhenping.Effects of water stress on berry quality and anthocyanin metabolism in Marselan grape[J].Journal of Fruit Science,2023,40(5):919-931.
[31] 邢婷婷,杨航宇,王雯染,杨晓慧,王军.14 个欧亚种红色酿酒葡萄品种(品系)的花色苷组成和含量分析[J]. 果树学报,2018,35(2):147-157.XING Tingting,YANG Hangyu,WANG Wenran,YANG Xiaohui,WANG Jun.The compositions and contents of anthocyanins of 14 red wine grape varieties or clones (Vitis vinifera)[J]. Journal of Fruit Science,2018,35(2):147-157.
[32] ZHAO W,LIU Y H,LI L,MENG H J,YANG Y,DONG Z B,WANG L,WU G L. Genome-wide identification and characterization of bHLH transcription factors related to anthocyanin biosynthesis in red walnut(Juglans regia L.)[J].Frontiers in Genetics,2021,12:632509.
[33] WALKER A R,LEE E,BOGS J,MCDAVID D A J,THOMAS M R,ROBINSON S P.White grapes arose through the mutation of two similar and adjacent regulatory genes[J]. The Plant Journal,2007,49(5):772-785.
[34] AZUMA A,KOBAYASHI S,MITANI N,SHIRAISHI M,YAMADA M,UENO T,KONO A,YAKUSHIJI H,KOSHITA Y.Genomic and genetic analysis of Myb-related genes that regulate anthocyanin biosynthesis in grape berry skin[J]. Theoretical and Applied Genetics,2008,117(6):1009-1019.
[35] AZUMA A,UDO Y,SATO A,MITANI N,KONO A,BAN Y,YAKUSHIJI H,KOSHITA Y,KOBAYASHI S. Haplotype composition at the color locus is a major genetic determinant of skin color variation in Vitis × labruscana grapes[J]. Theoretical and Applied Genetics,2011,122(7):1427-1438.
[36] XU Y S,JIANG N,ZHANG Y G,WANG M,REN J P,TAO J M. A SNP in the promoter region of the VvmybA1 gene is responsible for differences in grape berry color between two related bud sports of grape[J].Plant Growth Regulation,2017,82(3):457-465.
[37] YOO J,SHIN D H,CHO M H,KIM T L,BHOO S H,HAHN T R.An ankyrin repeat protein is involved in anthocyanin biosynthesis in Arabidopsis[J]. Physiologia Plantarum,2011,142(4):314-325.
[38] YOO J,CHO M H,LEE S W,BHOO S H. Phytochrome-interacting ankyrin repeat protein 2 modulates phytochrome A-mediated PIF3 phosphorylation in light signal transduction[J]. The Journal of Biochemistry,2016,160(4):243-249.
[39] HENRIQUES R,JANG I C,CHUA N H. Regulated proteolysis in light-related signaling pathways[J]. Current Opinion in Plant Biology,2009,12(1):49-56.
[40] KIM J,YI H,CHOI G,SHIN B,SONG P S,CHOI G.Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction[J]. The Plant Cell,2003,15(10):2399-2407.