叶绿素降解是果实成熟最明显的特征之一,也是植物代谢过程中的重要一环,促进营养从衰老器官和组织重新分配到正在生长的组织或贮存器官[1]。近年来,植物叶绿素主要降解途径已被阐明,由6 种叶绿素降解酶(chlorophyll catabolic enzyme,CCE)催化[2]。其中叶绿素酶(Chlorophyllase,CLH)一直被认为是叶绿素降解过程中的第一种酶。CLH也是研究比较广泛的叶绿素降解酶,目前已经在银杏(Ginkgo biloba L.)[3]、柑橘(Citrus reticulata Blanco)[4]等多种植物中纯化得到了CLH 蛋白。CLH 催化叶绿素a 脱植醇基形成脱植基叶绿素a(Chlorophyll a)[5]。有许多研究表明CLH 与叶绿素降解有直接联系,在衰老的叶片和成熟果实中可以检测到CLH 活性[6]。在南瓜(Cucurbita moschata Duch.)叶片和烟草(Nicotiana tabacum L.)叶片中过表达柑橘CLH 基因,将会加速叶片的叶绿素降解[7]。但有研究发现叶绿素酶活性的提高与叶绿素含量的下降并不完全一致。在菜豆突变体和野生型植株叶片上发现,新鲜叶片中CLH 酶活性高于衰老的叶片[8]。也有研究表明即使大部分叶绿素已经消失,苦楝(Melia azedarach L.)和旱金莲(Tropaeolum majus L.)在衰老过程中仍保持了较高的CLH 活性[9]。对拟南芥叶绿素酶双突变体的研究发现,叶绿素酶双突变体叶片衰老时叶绿素依旧可以降解,自此认为叶绿素酶可能与叶片衰老时的叶绿素降解没有关系[10]。最新的研究表明拟南芥中CLH1 参与了PSII 受损D1的降解,过表达CLH1 提高了幼苗对高光的耐受性,对AtCLH1 进行亚细胞定位发现,CLH1 定位在幼叶的叶绿体中,而在成熟和衰老叶片中定位在叶绿体外[11]。这说明CLH1 是幼叶中叶绿素降解的关键基因,而在老叶中不是叶绿素降解的关键因素。
但也有研究表明乙烯处理可使柑橘果皮中CLH 酶活性提高2.5~4.0 倍,导致叶绿素迅速降解,果皮明显褪绿[12]。青瓯柑和黄皮椪柑果实采后叶绿素降解过程中CLH 先上调后下调[13]。柑橘CLH 的N 段有一段转运肽序列,当该序列缺失时,成熟的叶绿素酶形成,引起柑橘中叶绿素的降解,说明CLH存在翻译后修饰,该过程对CLH 酶活性有重要的影响[14]。尽管CLH 在不同物种中的基因表达情况与酶活性规律陆续被发现,但CLH 在不同研究出现相互矛盾的结果,使得叶绿素酶在叶绿素降解过程中的功能一直是一个谜[15]。此外,当细胞膜解体时,CLH 降解叶绿素a产生脱植基叶绿素a,脱植基叶绿素a 对昆虫是有毒的,以此抵御昆虫对植物的侵害[16]。而且CLH 降解叶绿素a 产生脱植基叶绿素a及其衍生物也被证明具有抗病毒作用[17],AtCLH1 的下调会导致拟南芥对油菜芽孢杆菌的敏感性增加[18]。
CLH 在植物中可能具有更广泛的功能,在不同植物不同发育阶段中可能具有不同的作用,甚至在植物的不同器官也有着不同的基因表达情况,它参与的叶绿素降解过程可能与各种生物胁迫与非生物胁迫等多种生理现象相关。由于CLH 在光合生物中是保守的,因此通过生物信息学手段研究CLH 同系物在其他物种中的作用是有生物学意义的。
目前,国内外对叶绿素酶的研究主要集中在模式植物拟南芥上,而对蔷薇科植物CLH 的探索较少,PpCLH1 基因在桃中的功能尚未鉴定,其对桃果实叶绿素降解的影响也未得到深入研究。基于此,笔者在本研究中对CLH1 的蛋白结构特征进行了生物信息学分析,并对PpCLH1 的基因功能进行鉴定和研究,这对桃果实成熟过程中果皮叶绿素降解的研究具有一定的启发意义,为进一步研究PpCLH1基因在桃中的生物学功能奠定理论基础。
1 材料和方法
1.1 试验材料
供试材料为中蟠102,来自中国农业科学院郑州果树研究所桃品种圃内,株行距为1.0 m×4.0 m,2017 年定植,常规管理,郑州地区7 月上旬成熟。2023年6—7月,于成熟前30 d开始采摘树体外围中上部大小均匀、无病虫害、成熟度一致的果实30 个,带回实验室使用尼康D 700 相机于自然光下以黑色植绒布为背景拍照。以10个果实为一个样本,3次重复为1组,取样间隔7 d,在果实转色期每隔4 d取样1次,削取表皮进行液氮速冻,保存至-80 ℃冰箱备用。
1.2 RNA提取及反转录
总RNA 提取参照OMEGA Plant RNA Kit 说明书(广州飞扬生物工程有限公司,广州,中国)。利用1.5%琼脂糖凝胶电泳检测RNA质量和纯度,取1 μL RNA 利用微量紫外分光光度计NANO,DROP2000(Thermo Scientific,麻省)测定浓度。取5 μL RNA参照HiScript Ⅲ All-in-one RT SuperMix Perfect for qPCR第一链合成试剂盒说明书(南京诺唯赞生物科技股份有限公司,南京,中国),进行逆转录,-20 ℃冰箱保存进行后续的实时荧光定量PCR。
1.3 实时荧光定量检测
从桃基因组数据库Peach DataBase(http://www.peachmd.com/#/)下载相关基因的CDS(Coding sequence),利用primer primer 5.0 软件设计基因克隆引物,长度在20 bp 左右,G+C 含量在40%~60%,引物Tm值在57~63 ℃,引物避免发夹结构和引物二聚体,扩增片段大小为80~150 bp;PCR 体系为15 μL:cDNA 1 μL、Mix 7.5 μL、引物F 0.3 μL、引物R 0.3 μL、水6 μL。PCR 程序设置为:95 ℃预变性 5 min;95 ℃变性30 s、60 ℃退火30 s、72 ℃延伸30 s,扩增循环数为40,设置3 个技术重复。引物序列见表1。实时荧光定量PCR 所用的仪器为罗氏480,所用试剂盒为aq Pro Universal SYBR qPCR Master Mix(南京诺唯赞生物科技股份有限公司,南京,中国)。选取Actin(ppa007228mg)为桃内参基因[19],最后按照2-ΔΔCT方法计算结果[20]。
表1 实时定量PCR 反应中各基因的引物序列
Table 1 Primers sequence for real-time quantitative PCR reaction

基因Gene Actin登记号GenBank ppa007228m PpCLH1引物(5'-3')Primer (5'-3')GATTCCGGTGCCCAGAAGT CCAGCAGCTTCCATTCCAA CATGCCAAAACTGCCCTGTC AGGATATGGGGCCTGGTTCT ppa009825m
1.4 桃PpCLH1过表达载体的构建及转化
根据PpCLH1 目的基因编码区(CDS)的全长序列,结合PGreen 62-sk 载体上的Bam HⅠ和Eco RⅠ为酶切位点,利用primer 5.0软件设计基因克隆引物(表2),由上海生工公司进行引物的合成。以中蟠102 号的cDNA 为模板,利用合成的克隆引物扩出PpCLH1 目的基因CDS 区的全长序列,用限制性内切酶Bam HⅠ和Eco RⅠ对62-sk空载体进行双酶切以获得线性化载体,于水浴锅中37 ℃酶切30 min。利用1.5%琼脂糖凝胶电泳,切下带有目的基因条带的凝胶,参考凝胶回收试剂盒说明书FastPure Gel DNA Extraction Mini Kit(南京诺唯赞生物科技股份有限公司,南京,中国)进行目的DNA片段的回收及纯化,取1 μL DNA 利用微量紫外分光光度计NANO,DROP2000(Thermo Scientific,麻省)测定浓度。参考同源重组试剂盒说明书ClonExpress® ⅡOne Step Cloning Kit(南京诺唯赞生物科技股份有限公司,南京,中国),对PpCLH1 基因的纯化产物与线性化载体进行重组连接反应,构建PpCLH1 的过表达载体。转化至大肠杆菌感受态细胞DH5α,鉴定的含有基因PpCLH1 片段的阳性菌落送生工生物工程(上海)股份有限公司测序。选择测序正确的菌落采用质粒提取试剂盒纯化PpCLH1 质粒,用限制性内切酶Bam HⅠ对该质粒进行酶切1 h后,使用琼脂糖电泳检测。将PpCLH1 质粒转入农杆菌GV3101 感受态细胞,利用注射的方法瞬时浸染烟草叶片。
表2 构建PpCLH1 过表达载体的引物序列
Table 2 Construction primers sequence for PpCLH1 overexpression vector
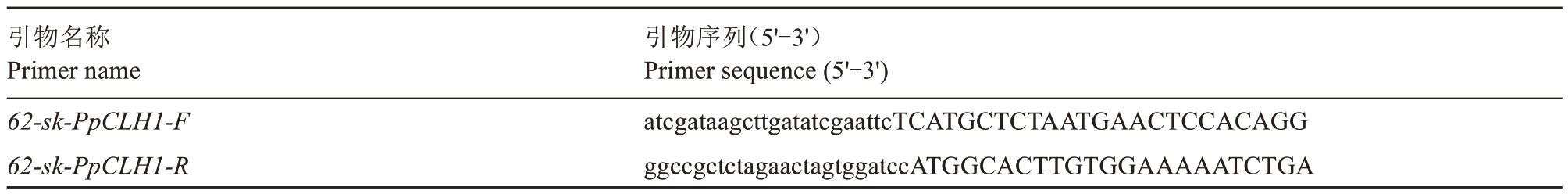
引物名称Primer name 62-sk-PpCLH1-F 62-sk-PpCLH1-R引物序列(5'-3')Primer sequence (5'-3')atcgataagcttgatatcgaattcTCATGCTCTAATGAACTCCACAGG ggccgctctagaactagtggatccATGGCACTTGTGGAAAAATCTGA
1.5 叶绿素含量
叶绿素含量测定参照李合生[21]的方法进行。
1.6 生物信息学分析
利用桃基因组数据库Peach DataBase(http://www.peachmd.com/#/)获得PpCLH1 基因序列,利用NCBI 数据库中BLASTp 在线分析软件对蛋白PpCLH1 进行同源性比对并下载其同源蛋白序列。通过DNAMAN 获得不同物种中CLH1 氨基酸序列比对图,并结合MEGA7.0 的Clustal W 进行聚类分析。使用Expasy(https://web.expasy.org/protparam/)在线分析,对PpCLH1 蛋白的分子质量、等电点、氨基酸数目、氨基酸组分、脂肪指数、亲水性等进行分析。用SOPMA(https://npsa prabi.ibcp.fr/)在线软件对PpCLH1 蛋白的二级结构进行预测;使用Swissmodel(https://swissmodel.expasy.org/)对PpCLH1 蛋白的三级结构进行预测。
2 结果与分析
2.1 PpCLH1基因克隆
以中蟠102 号果皮cDNA 为模板,扩增PpCLH1目的基因CDS 区的全长序列,1%琼脂糖凝胶电泳检测,使用胶回收试剂盒(南京诺唯赞生物科技股份有限公司)进行目的DNA 片段的回收及纯化,用微量紫外分光光度计测定浓度。随后将克隆片段连接至PGreen 62-sk 载体中。阳性克隆测序结果表明中蟠102 PpCLH1 基因与UNIPORT 数据库(https://www.uniprot.org/)中桃基因组公布的PpCLH1 序列比对率为100%(图1)。

图1 PpCLH1 测序结果比对
Fig. 1 Alignment of PpCLH1 sequencing results
2.2 PpCLH1蛋白基本理化性质分析
PpCLH1 编码区全长为972 bp,编码323 个氨基酸。用Expasy 在线分析PpCLH1 蛋白的理化性质,该蛋白的分子式为C1568H2441N403O451S16,总原子数为4879,分子质量为34 667.02 Da,等电点为6.12,带正电荷的残基总数为26,带负电荷的残基总数为31,总平均亲水性为0.065,大于平均值,不稳定系数为44.56,脂肪族指数是88.76,并预测PpCLH1 蛋白是一个不稳定的蛋白。
2.3 PpCLH1蛋白二级结构、三级结构分析
利用Sopma 网站分析桃PpCLH1 的二级结构(图2),发现多个氨基酸参与α-螺旋、β-折叠等二级结构的形成,其中α-螺旋占比29.72%、β-折叠占比4.64%、延伸链占比16.10%、无规则卷曲占比49.54%。通过SWISS-MODEL网站预测PpCLH1蛋白的三维结构(图3),发现该蛋白无规卷曲占比最大,同时还存在延伸链和β-折叠等结构,该预测结果与二级结构预测结果一致。
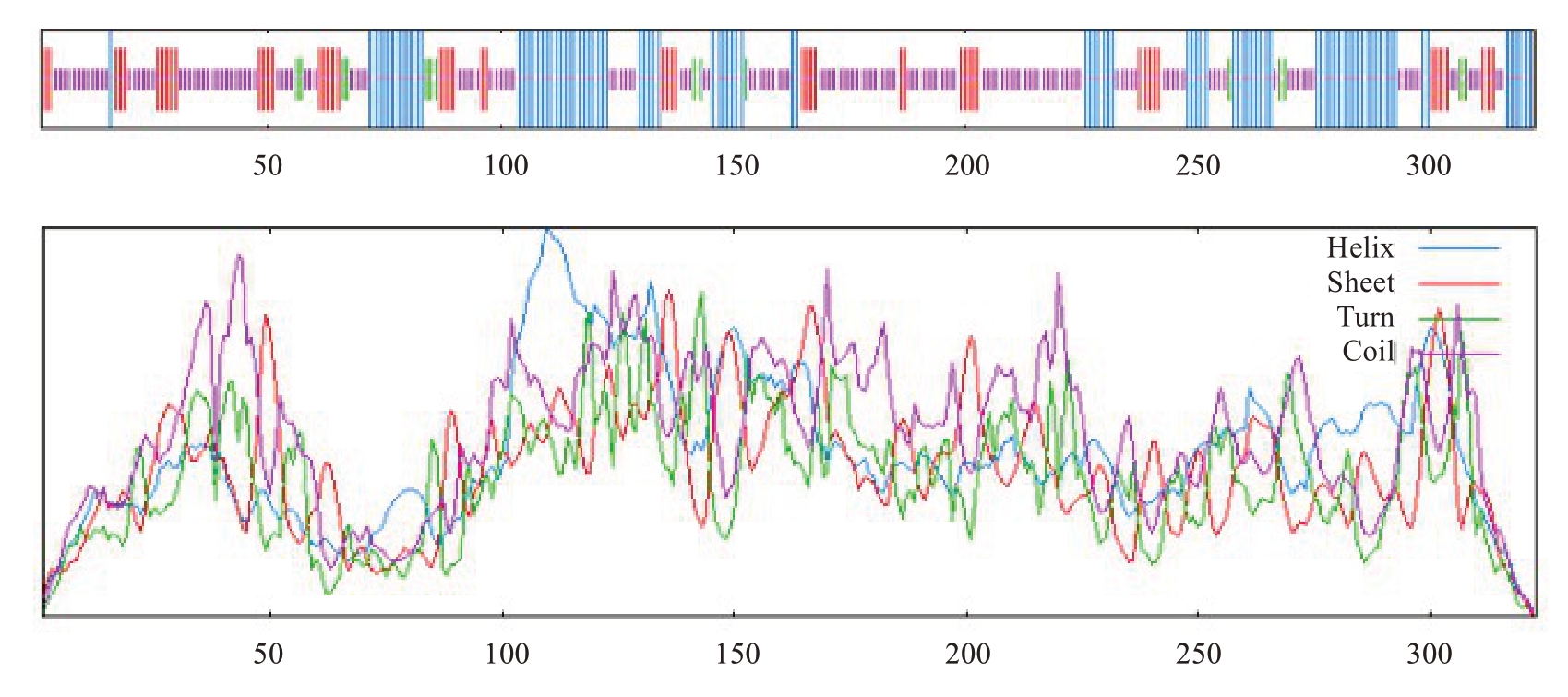
图2 PpCLH1 蛋白的二级结构
Fig. 2 Secondary structure for PpCLH1 protein
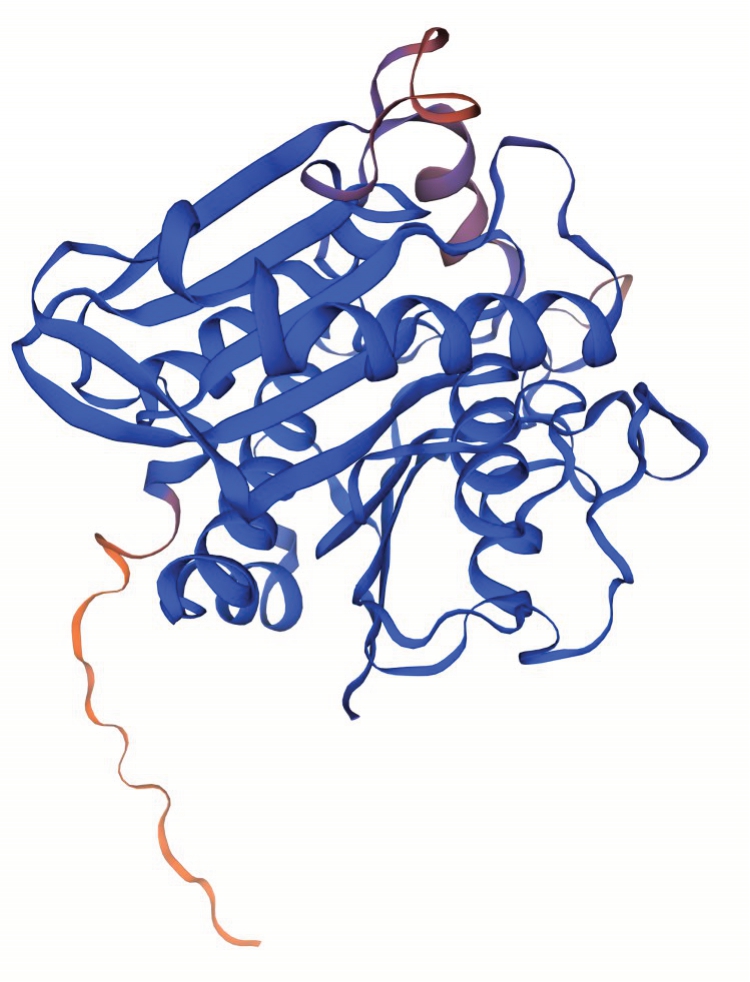
图3 PpCLH1 蛋白的三级结构预测
Fig. 3 3D structure prediction of PpCLH1 protein
2.4 多序列分析与CLH1蛋白系统进化树分析
不同物种CLH1氨基酸序列比对的结果显示(图4),桃PpCLH1蛋白与其他物种中CLH1蛋白具有较高的同源性。DNAMAN 显示桃PpCLH1、杏Pd-CLH1、樱桃PaCLH1、苹果MdCLH1、柑橘CsCLH1、葡萄VvCLH1、拟南芥AtCLH1、番茄SlCLH1、烟草NtCLH1 的一致性为67.48%。其中桃PpCLH1 与杏中PdCLH1 蛋白的同源性最高,为99.2%,这也证明了桃与杏的亲缘关系。樱桃PaCLH1 与PpCLH1 相似度也高达96.6%,说明CLH1在蔷薇科李属植物中有着高度的保守性。

图4 不同物种中CLH1 氨基酸序列比对
Fig. 4 Amino acid sequence comparison of SGR in different species
根据多个物种的CLH1 蛋白序列,利用MEGA 7.0 软件,采用邻接法构建了包括PpCLH1 等9 个CLH1 蛋白家族的系统进化树(图5)。发现桃PpCLH1、杏PdCLH1、樱桃PaCLH1位于进化树同一分支,亲缘关系最近,同时与苹果MdCLH1 蛋白的同源性也较高,这说明蔷薇科中CLH1 蛋白进化较为保守,但与番茄和烟草的CLH1 蛋白亲缘关系较远。
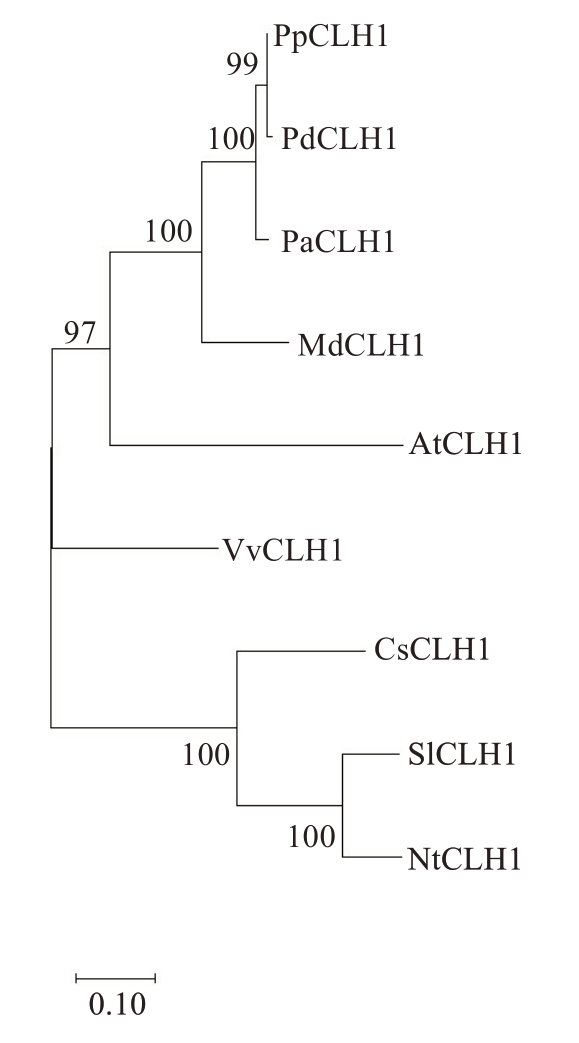
图5 桃PpCLH1 蛋白系统进化树(对比PpCLH1 蛋白序列)
Fig. 5 Phylogenetic tree of PpCLH1 protein in peach
2.5 PpCLH1在不同发育时期桃果实中的表达分析
叶绿素降解主要发生在果实完全成熟前的发育阶段中。因此,笔者选用中蟠102 果实成熟前30 d、23 d、16 d、12 d、8 d、4 d、0 d共7个时间点的桃果皮,进行PpCLH1转录水平分析。荧光定量PCR分析的结果显示(图6),在果实成熟前30 d,果实呈现明显绿色时,PpCLH1转录水平较低;随着果实逐步成熟,果皮褪绿,表达量不断升高。在果实成熟前16 d,PpCLH1 的表达量快速上升,并在果实成熟前12 d达到最高。基于以上结果,推测PpCLH1 基因可能是调控桃果实转色时褪绿的重要基因。
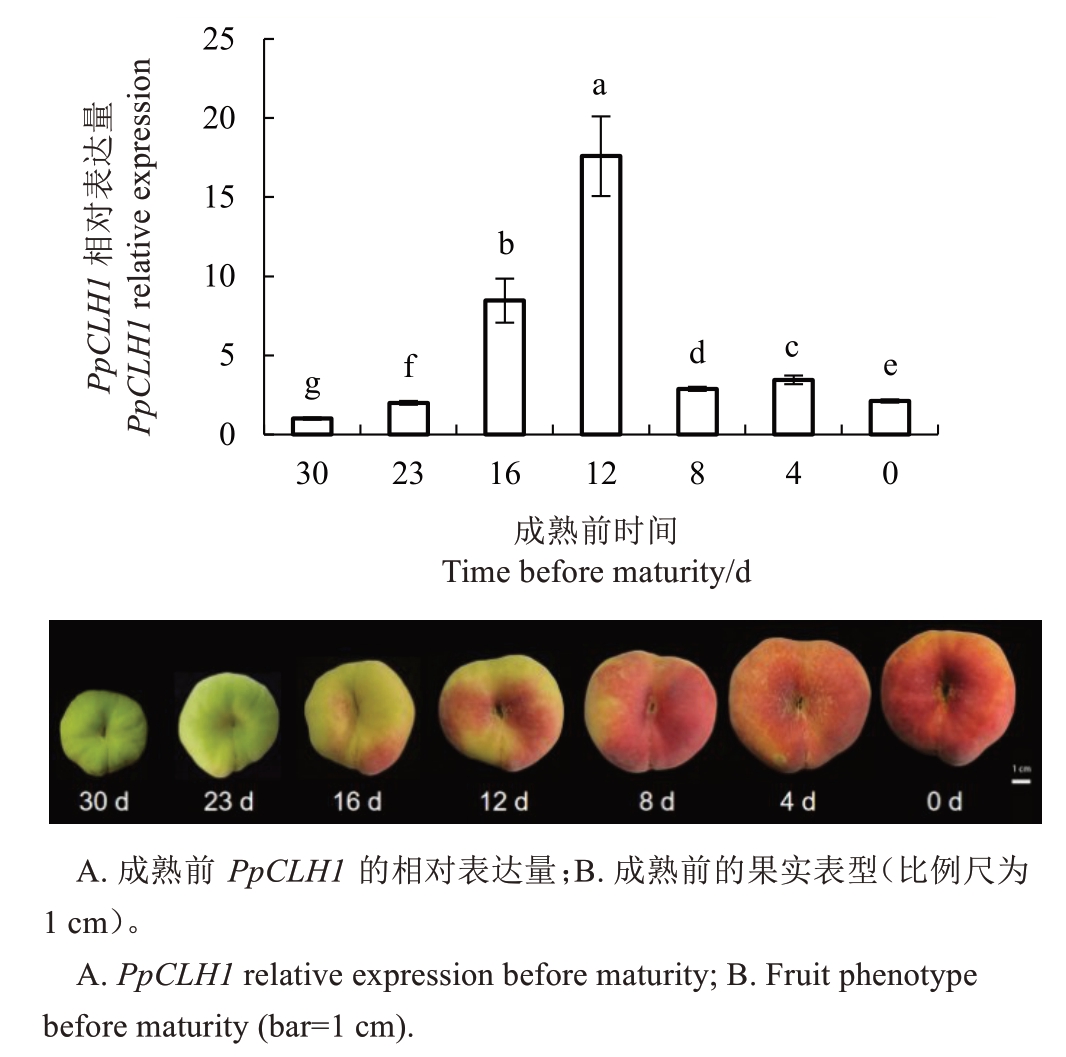
图6 桃果实不同发育时期中PpCLH1 基因的表达分析
Fig. 6 Expression analysis of PpCLH1 gene in peach fruit at different developmental stages
2.6 瞬时注射烟草叶片验证PpCLH1的功能
为了验证PpCLH1 基因的功能,利用瞬时注射烟草叶片以Pgreen 62-sk 空载作为对照,对PpCLH1基因的功能进行初步的验证。对烟草叶片表型分析,发现瞬转PpCLH1 烟草叶片颜色呈现淡绿色,而对照烟草叶片呈现深绿色(图7)。
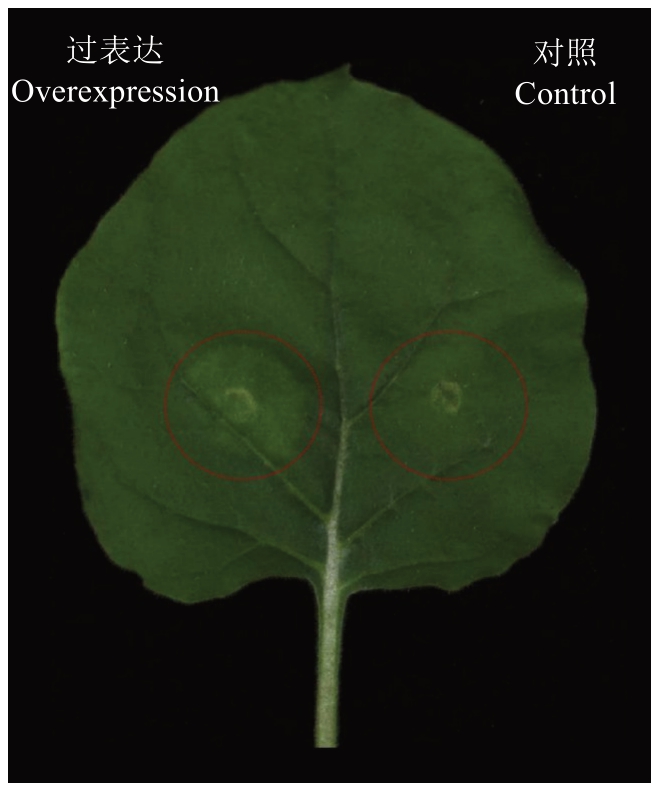
图7 瞬时注射烟草的表型
Fig. 7 Transient injection of tobacco phenotype
对叶片叶绿素含量进行测定,结果显示瞬转PpCLH1 烟草叶片叶绿素含量相较于对照烟草叶片显著降低(图8-A)。荧光定量结果显示在瞬时注射的烟草叶片中检测到了PpCLH1 的表达,与对照相比,PpCLH1 基因在烟草叶片中的表达量相较于野生型WT 上调了33 倍(图8-B)。上述结果表明,PpCLH1 具有促进叶绿素降解,进而导致烟草叶片叶绿素含量降低的功能。

图8 瞬时注射烟草叶绿素含量与荧光定量PCR 验证
Fig. 8 Chlorophyll content and qPCR validation in transiently injected tobacco leaves
3 讨 论
在叶片衰老和果实成熟过程中叶绿素降解是标志性的信号之一[22]。在过量光照条件下,叶绿体中光系统产生大量ROS,而叶绿素参与光系统中能量的吸收与转移,对光保护和清除活性氧具有重要作用[23]。最近的研究表明,叶绿素酶参与叶绿素脱植基反应过程中产生的植醇基,可以作为α-生育酚合成的前体,说明叶绿素酶参与叶绿素降解在清除ROS 过程中具有重要作用[24]。樊艳燕等[25]发现Bo-CLH1 主要在青花菜初始衰老中起降解叶绿素的作用。但也有研究表明CLH 的表达与银杏叶片衰老或果实成熟过程均无关[6]。将匍匐剪股颖(Agrostis stolonifera)暴露于热胁迫温度下35 d,响应热胁迫后,其叶绿素酶活性增强[26]。在正常生长条件下,拟南芥CLH 的敲除系仍然能够以与野生型植物相同的速率降解叶绿素,拟南芥CLH 没有参与到衰老进程中叶绿素的降解代谢[27]。但CLH 突变体的幼苗在强光持续照射下,比野生型表现出更严重的光伤害,积累更多的光保护色素(花色素苷)以降低光伤害[28]。因此拟南芥叶绿素降解的另一条途径涉及脱酶叶绿素水解酶(pheophytin pheophorbide hydrolase,PPH),它是在拟南芥中催化叶绿素降解的必需酶[29],但在PPH番茄果实成熟过程中不起作用[30]。
因此,CLH 在果实发育和叶片中是否具有不同的叶绿素代谢作用呢?番茄(Solanum lycopersicum)中SlPPH 沉默番茄系叶绿素降解不正常,在叶片衰老期间积累脱镁叶绿素,但果实中只会短暂积累,最终这些果实能够像野生类型一样降解叶绿素。因此,番茄果实成熟过程中叶绿素降解还涉及其他叶绿素降解酶[30]。在番茄果实成熟过程中,CLH 有可能是参与叶绿素降解的酶,类似于柑橘CLH。多种植物的研究表明,CLH 对植物激素信号转导具有重要作用[31]。鲜枣色泽与叶绿素酶活性显著相关,叶绿素酶直接参与鲜枣的红变[32]。杧果成熟时叶绿素酶活性升高,叶绿素降解,但采前喷洒赤霉素会导致杧果叶绿素酶活性降低,从而延缓叶绿素的降解[33]。柑橘CLH 位于叶绿体中,可以响应乙烯,并参与果实成熟过程中叶绿素的降解[34]。在蜜柑(Citrus unshiu)中,乙烯能够增强叶绿素酶活性,从而加速叶绿素的降解[35]。将茉莉酸甲酯(Methyl jas monate,MeJA)作用于拟南芥植物时,CLH 基因的表达量达到最高[36]。因此,CLH 的功能可能因物种或植物器官而异。
尽管CLH 距离其被发现已经过了一个多世纪,并且在其功能和相关机制上在不同物种上也有了很多研究,但CLH 的功能还有许多未解答的问题[37]。由于CLH 在果实成熟与叶片衰老过程叶绿素降解中扮演的角色尚不够清晰,其功能可能因物种或植物器官而异。因此,进一步解析PpCLH1 是如何调控果实成熟过程与幼嫩叶片叶绿素降解的具有重要意义,这将为探索桃果实发育过程中叶绿素降解的分子机制提供参考。
4 结 论
PpCLH1 基因随着桃果实成熟转录水平逐渐上升,在果实成熟前16~12 d 时表达量升至最高,之后随着果实成熟表达量不断下降。烟草叶片瞬时表达结果表明,PpCLH1 具有促进叶片叶绿素降解的功能。基于本研究的结果,笔者初步推测PpCLH1 具有促进桃果皮叶绿素降解的功能,这对桃果实外观品质研究具有一定的启发意义,也为CLH1 基因转录水平和功能研究提供了一定的理论参考。
[1] LIM P O,KIM H J,GIL NAM H. Leaf senescence[J]. Annual Review of Plant Biology,2007,58:115-136.
[2] HÖRTENSTEINER S. Update on the biochemistry of chlorophyll breakdown[J]. Plant Molecular Biology,2013,82(6):505-517.
[3] OKAZAWA A,TANGO L,ITOH Y,FUKUSAKI E,KOBAYASHI A. Characterization and subcellular localization of chlorophyllase from Ginkgo biloba[J]. Zeitschrift Fur Naturforschung. C,Journal of Biosciences,2006,61(1/2):111-117.
[4] JACOB-WILK D,HOLLAND D,GOLDSCHMIDT E E,RIOV J,EYAL Y. Chlorophyll breakdown by chlorophyllase:Isolation and functional expression of the Chlase1 gene from ethylenetreated Citrus fruit and its regulation during development[J].The Plant Journal,1999,20(6):653-661.
[5] CHOU Y L,LEE Y L,YEN C C,CHEN L F O,LEE L C,SHAW J F. A novel recombinant chlorophyllase from Cyanobacterium cyanothece sp. ATCC 51142 for the production of bacteriochlorophyllide a[J]. Biotechnology and Applied Biochemistry,2016,63(3):371-377.
[6] TANG L,OKAZAWA A,ITOH Y,FUKUSAKI E I,KOBAYASHI A. Expression of chlorophyllase is not induced during autumnal yellowing in Ginkgo biloba[J]. Zeitschrift Fur Naturforschung. C,Journal of Biosciences,2004,59(5/6):415-420.
[7] HARPAZ-SAAD S,AZOULAY T,ARAZI T,BEN-YAAKOV E,METT A,SHIBOLETH Y M,HÖRTENSTEINER S,GIDONI D,GAL-ON A,GOLDSCHMIDT E E,EYAL Y. Chlorophyllase is a rate-limiting enzyme in chlorophyll catabolism and is posttranslationally regulated[J]. The Plant Cell,2007,19(3):1007-1022.
[8] MAJUMDAR S,GHOSH S,GLICK B R,DUMBROFF E B.Activities of chlorophyllase,phosphoenolpyruvate carboxylase and ribulose-1,5-bisphosphate carboxylase in the primary leaves of soybean during senescence and drought[J]. Physiologia Plantarum,1991,81(4):473-480.
[9] BEN-YAAKOV E,HARPAZ-SAAD S,GALILI D,EYAL Y,GOLDSCHMIDT E. The relationship between chlorophyllase activity and chlorophyll degradation during the course of leaf senescence in various plant species[J]. Israel Journal of Plant Sciences,2006,54(2):129-135.
[10] SCHENK N,SCHELBERT S,KANWISCHER M,GOLDSCHMIDT E E,DÖRMANN P,HÖRTENSTEINER S. The chlorophyllases AtCLH1 and AtCLH2 are not essential for senescence-related chlorophyll breakdown in Arabidopsis thaliana[J].FEBS Letters,2007,581(28):5517-5525.
[11] TIAN Y N,ZHONG R H,WEI J B,LUO H H,EYAL Y,JIN H L,WU L J,LIANG K Y,LI Y M,CHEN S Z,ZHANG Z Q,PANG X Q. Arabidopsis CHLOROPHYLLASE 1 protects young leaves from long-term photodamage by facilitating FtsHmediated D1 degradation in photosystem II repair[J]. Molecular Plant,2021,14(7):1149-1167.
[12] TREBITSH T,GOLDSCHMIDT E E,RIOV J. Ethylene induces de novo synthesis of chlorophyllase,a chlorophyll degrading enzyme,in citrus fruit peel[J]. Proceedings of the National Academy of Sciences of the United States of America,1993,90(20):9441-9445.
[13] 罗焘. 青瓯柑和黄皮椪柑果实采后色泽变化及调控机理研究[D]. 武汉:华中农业大学,2016.LUO Tao. Characteristics and regulatory mechanism of pigmentation in Postharvest Green Ougan and Yellowish Ponkan[D].Wuhan:Huazhong Agricultural University,2016.
[14] AZOULAY SHEMER T,HARPAZ-SAAD S,BELAUSOV E,LOVAT N,KROKHIN O,SPICER V,STANDING K G,GOLDSCHMIDT E E,EYAL Y. Citrus chlorophyllase dynamics at ethylene-induced fruit color-break:A study of chlorophyllase expression,posttranslational processing kinetics,and in situ intracellular localization[J]. Plant Physiology,2008,148(1):108-118.
[15] CHRIST B,HÖRTENSTEINER S. Mechanism and significance of chlorophyll breakdown[J]. Journal of Plant Growth Regulation,2014,33(1):4-20.
[16] HU X Y,MAKITA S,SCHELBERT S,SANO S,OCHIAI M,TSUCHIYA T,HASEGAWA S F,HÖRTENSTEINER S,TANAKA A,TANAKA R. Reexamination of chlorophyllase function implies its involvement in defense against chewing herbivores[J]. Plant Physiology,2015,167(3):660-670.
[17] GUO H T,PAN X B,MAO R C,ZHANG X C,WANG L J,LU X Y,CHANG J H,GUO J T,PASSIC S,KREBS F C,WIGDAHL B,WARREN T K,RETTERER C J,BAVARI S,XU X D,CUCONATI A,BLOCK T M. Alkylated porphyrins have broad antiviral activity against hepadnaviruses,flaviviruses,filoviruses,and arenaviruses[J]. Antimicrobial Agents and Chemotherapy,2011,55(2):478-486.
[18] KARIOLA T,BRADER G,LI J,PALVA E T. Chlorophyllase 1,a damage control enzyme,affects the balance between defense pathways in plants[J]. The Plant Cell,2005,17(1):282-294.
[19] BRANDI F,BAR E,MOURGUES F,HORVÁTH G,TURCSI E,GIULIANO G,LIVERANI A,TARTARINI S,LEWINSOHN E,ROSATI C. Study of ‘Redhaven’ peach and its white-fleshed mutant suggests a key role of CCD4 carotenoid dioxygenase in carotenoid and norisoprenoid volatile metabolism[J]. BMC Plant Biology,2011,11:24.
[20] SCHMITTGEN T D,LIVAK K J. Analyzing real-time PCR data by the comparative C(T) method[J]. Nature Protocols,2008,3(6):1101-1108.
[21] 李合生. 植物生理生化实验原理和技术[M]. 北京:高等教育出版社,2000.LI Hesheng. Principles and techniques of plant physiological biochemical experiment[M]. Beijing:Higher Education Press,2000.
[22] TANAKA A,TANAKA R. Chlorophyll metabolism[J]. Current Opinion in Plant Biology,2006,9(3):248-255.
[23] MUÑOZ P,MUNNÉ-BOSCH S. Vitamin E in plants:Biosynthesis,transport,and function[J]. Trends in Plant Science,2019,24(11):1040-1051.
[24] VOM DORP K,HÖLZL G,PLOHMANN C,EISENHUT M,ABRAHAM M,WEBER A P M,HANSON A D,DÖRMANN P. Remobilization of phytol from chlorophyll degradation is essential for tocopherol synthesis and growth of Arabidopsis[J].The Plant Cell,2015,27(10):2846-2859.
[25] 樊艳燕,刘玉梅,李占省,方智远,杨丽梅,庄木,张扬勇,孙培田. 青花菜衰老过程中叶绿素降解相关基因的表达分析[J].园艺学报,2015,42(7):1338-1346.FAN Yanyan,LIU Yumei,LI Zhansheng,FANG Zhiyuan,YANG Limei,ZHUANG Mu,ZHANG Yangyong,SUN Peitian.Analysis of the expression of chlorophyll degrading genes during senescence of broccoli[J]. Acta Horticulturae Sinica,2015,42(7):1338-1346.
[26] ROSSI S,CHAPMAN C,HUANG B R. Suppression of heat-induced leaf senescence by γ-aminobutyric acid,proline,and ammonium nitrate through regulation of chlorophyll degradation in creeping bentgrass[J]. Environmental and Experimental Botany,2020,177:104116.
[27] HU X Y,JIA T,HÖRTENSTEINER S,TANAKA A,TANAKA R. Subcellular localization of chlorophyllase2 reveals it is not involved in chlorophyll degradation during senescence in Arabidopsis thaliana[J]. Plant Science,2020,290:110314.
[28] 田亚楠. 拟南芥叶绿素酶1 在幼叶中光保护作用的分子机制[D]. 广州:华南农业大学,2020.TIAN Yanan. Molecular mechanisms of photo-protective role of chlorophyllase 1 in young leaves of Arabidopsis thaliana[D].Guangzhou:South China Agricultural University,2020.
[29] SCHELBERT S,AUBRY S,BURLA B,AGNE B,KESSLER F,KRUPINSKA K,HÖRTENSTEINER S. Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis[J]. The Plant Cell,2009,21(3):767-785.
[30] GUYER L,HOFSTETTER S S,CHRIST B,LIRA B S,ROSSI M,HÖRTENSTEINER S. Different mechanisms are responsible for chlorophyll dephytylation during fruit ripening and leaf senescence in tomato[J]. Plant Physiology,2014,166(1):44-56.
[31] SHARAFI E,DEHESTANI A,FARMANI J,PAKDIN-PARIZI A. Bioinformatics evaluation of plant chlorophyllase,the key enzyme in chlorophyll degradation[J]. Applied Food Biotechnology,2017,4(3):167-178.
[32] 纵伟,张沙沙,赵光远,张华. 热处理下鲜枣红变与叶绿素酶活性相关性的研究[J]. 食品科技,2012,37(7):44-46.ZONG Wei,ZHANG Shasha,ZHAO Guangyuan,ZHANG Hua.Correlation between the going-red of fresh jujube and the chlorophyllase activity[J]. Food Science and Technology,2012,37(7):44-46.
[33] 董真真,曾凤,徐孝兰,李雯. 采前喷洒赤霉素对‘红贵妃’芒果色泽及相关酶活性的影响[J]. 热带生物学报,2017,8(2):178-184.DONG Zhenzhen,ZENG Feng,XU Xiaolan,LI Wen. Effect of pre-harvest spraying with gibberellic acid on coloration and related enzyme activities of ‘Hongguifei’ mango fruits[J]. Journal of Tropical Biology,2017,8(2):178-184.
[34] Jia Z, Zhang H, Zhang X, Zhang J, Xu C. Transcriptome analysis reveals candidate genes involved in ethylene-induced color break of Satsuma mandarin (Citrus unshiu Marc.) [J]. Scientia Horticulturae, 2018, 237:43-51.
[35] SHIMOKAWA K. Purification of ethylene-enhanced chlorophyllase from Citrus unshiu fruits[J]. Agricultural and Biological Chemistry,1981,45(10):2357-2359.
[36] TSUCHIYA T,OHTA H,OKAWA K,IWAMATSU A,SHIMADA H,MASUDA T,TAKAMIYA K. Cloning of chlorophyllase,the key enzyme in chlorophyll degradation:Finding of a lipase motif and the induction by methyl jasmonate[J]. Proceedings of the National Academy of Sciences of the United States of America,1999,96(26):15362-15367.
[37] HU X Y,KHAN I,JIAO Q S,ZADA A,JIA T. Chlorophyllase,a common plant hydrolase enzyme with a long history,is still a puzzle[J]. Genes,2021,12(12):1871.