自交不亲和性(Self-incompatibility,SI)是显花植物普遍存在的一种遗传控制机制,即当自交不亲和植物自花授粉时,雌蕊花柱可以识别并拒绝自己的花粉,使自己的花粉不能正常萌发生长,从而促进异交,提高异交重组率,避免近交衰退,保持物种的遗传多样性[1]。早期遗传学发现,自交不亲和性通常由一个单一的复等位S 位点(S-locus)所调控[2]。S位点内通常包含着两类紧密连锁的基因,即花粉S基因和花柱S 基因,这两类基因构成的遗传单元的不同变体称为S 单倍体型,它们之间的相互作用可以调控自交不亲和性。自交不亲和可以分为孢子体自交不亲和与配子体自交不亲和。绝大部分栽培果树,包括梨属均为配子体自交不亲和[3],表现为自花授粉或相同S 基因型品种间异花授粉不结实,其机制在于花柱内的S 基因产物为具有核酸酶活性的糖蛋白S核酸酶(S-RNase),能够特异地控制花粉和雌蕊识别过程[4]。因此,确定梨品种的S基因型可为大田科学配置授粉树、杂交亲本选配提供重要依据。
在梨的自交不亲和性研究及S 基因型鉴定方面,早期日本学者采用田间授粉试验等传统方法鉴定S 基因型[5],随后建立PCR-RFLP 系统首先对亚洲梨自交不亲和基因开展了研究并分离鉴定出9 个S等位基因[6-7]。国内,最早是乌云塔娜[8]在中国白梨中分离鉴定7 个新的S 基因;随后Tan 等[9]、张妤艳等[10-11]、衡伟等[12-13]和陈慧等[14]利用PCR-RFLP 检测技术和核苷酸序列分析等方法分离鉴定出一些亚洲梨品种的S 基因型及大量新的S 基因。我国的梨自交不亲和基因的分离鉴定虽开展得相对较晚,但取得了很大的成绩。截至目前,我国梨品种中已分离鉴定出来的S 基因有60 个[15],极大丰富了梨的S 基因型信息库。
目前,梨品种S 基因型的鉴定主要是根据雌性决定因子S 核酸酶高变区(hyper variable region,HV)序列的差异来判别不同的S等位基因,采用梨S基因特异性引物FTQQYQ 和anti-IWPNV[16-17]进行PCR 扩增。但随着种质资源的挖掘利用,经典的S等位基因引物FTQQYQ和anti-IIWPNV并不完全适用于所有梨品种S 基因型的鉴定。笔者在本研究中根据重测序数据序列多态性比对结果,设计新的S基因简并引物FTQQYQ-B 和anti-GIIWPN。新的特异引物能够扩增出更为丰富的特异性S 单倍型序列,能够准确鉴定出丹霞红等23 个新品种梨的S 基因型,从而更有效地配置授粉品种,优化授粉策略,提高坐果率和果实品质,为品种遗传改良和推广应用奠定理论基础。
1 材料和方法
1.1 试验材料
供试梨新品种为红香酥,中梨1 号,红酥宝,丹霞红,红酥蜜,早红玉,红玛瑙,中梨碧玉,中梨秋香,中梨蜜脆,中梨291,阳光蜜露,恬心,六月香,金香玉,红酥脆,满天红,美人酥,早白蜜,T109,红玉,中梨金福,秋月;库尔勒香梨为对照。以上试材均采自中国农业科学院郑州果树研究所梨种质资源圃(国家园艺种质资源库郑州梨分库),于春季采集各品种幼嫩叶片,-80 ℃保存备用。
试剂材料:PrimeSTAR HS DNA Polymerase with GC Buffer 酶试剂盒(R044A)购自宝日医生物技术(北京)有限公司(TaKaRa中国);琼脂糖凝胶纯化回收试剂盒(DH101-01)和DH5α感受态购自北京博迈德基因技术有限公司;CTAB 植物基因组DNA快速提取试剂盒(DN14)购于北京艾德莱生物科技有限公司;核酸 Marker (M018 50 bp Ladder)购自康润生物(GenStar);pEASY-Blunt Zero Cloning Kit(CB501)购自北京全式金生物技术 (TransGen Biotech)有限公司;其他药品购自生工生物工程(上海)股份有限公司。测序委托生工生物工程(上海)股份有限公司完成。
1.2 试验方法
1.2.1 梨叶片基因组DNA 提取 使用CTAB 植物基因组DNA 快速提取试剂盒(DN14)提取80 μL 梨叶片基因组DNA,取2 μL进行电泳后在紫外灯下检测基因组DNA 纯度和完整性,取完整性高的DNA于-20 ℃保存备用。
1.2.2 PCR 扩增S-RNase 基因 本实验共设计3 对引物,第一对采用Ishiimizu等[16]所设引物,即上下游引物分别取S 基因高变区两侧保守区序列中特异性较高的序列,正向引物为FTQQYQ:5'-TTTACGCAGCAATATCAG-3',反向引物为anti-IWPNV:5'-AC(A/G)TTCGGCCAAATAATT-3'。第二对引物利用DANMAN 软件比对红香酥等重测序数据设计,FTQQYQ-B:5'-TTTAC(C/G/T)CAGCAATATCAG-3'和anti-GIIWPN:5'-AC(A/G)TTCGGCCAAAT(A/T)AT-(G/T)(G/T)CC-3'。第三对引物为扩增S39-RNase 特异引物,正向引物为S39-F:5'-TTTACTCAGCAATATCAG-3',反向引物为S39-R:5'-ACGTTCGGCCAAATAATG-3'。
PCR反应体系为:基因组DNA 1 μL;10 μmol·L-1上下游引物各0.5 μL;GC Buffer 12.5 μL;dNTP 2 μL;Primer star 0.25 μL;ddH2O 8.25 μL。反应条件:98 ℃预变性3 min;98 ℃变性15 s,4 ℃退火15 s,72 ℃延伸1 min,变性退火延伸35 个循环;72 ℃稳定75 min后降温到4 ℃,取出后放于4 ℃保存备用。
取5 μL PCR 扩增产物点样于1.8%琼脂糖凝胶中,于120 V 电压下电泳30~45 min。电泳结果采用伯乐Bio-Rad GelDoc Go凝胶成像系统拍摄。
1.2.3 PCR 产物胶回收、连接并测序 取50 μL PCR 扩增产物点样于1.8%琼脂糖凝胶中,于120 V电压下电泳30~45 min 后在紫外灯下切胶。切下的胶块参照博迈德琼脂糖凝胶纯化回收试剂盒说明书回收目的DNA片段。
胶回收产物与pEASY-Blunt Zero Cloning Kit Vector 按照4∶1 的比例,于25 ℃连接15 min。连接产物转化到大肠杆菌DH5α 感受态中,涂布于Kana培养板上,37 ℃倒置培养过夜。挑取5 个培养过夜的菌斑进行菌落PCR,筛选出的阳性克隆加入培养液中于37 ℃扩大培养,取部分送生工生物工程(上海)股份有限公司测序。样品的测序结果经比对拼接后,与NCBI中已收录的S-RNase序列进行BLAST比对,最终确定样品的S基因型。
2 结果与分析
2.1 S基因的特异扩增
采用针对梨S-RNase 基因的通用引物FTQQYQ和anti-IWPNV 对23个梨新品种和1个对照(库尔勒香梨)的梨品种基因组DNA 进行PCR 扩增,获得琼脂糖凝胶电泳检测结果(图1)。24 个品种均可以扩增出S 基因特异片段,条带范围在300~600 bp。其中,库尔勒香梨、中梨291、阳光蜜露、红酥脆、满天红、美人酥、早白蜜7 个品种均扩增得到两条S 基因特异条带,而红香酥、中梨1 号、红酥宝、丹霞红、红酥蜜、早红玉、红玛瑙、中梨碧玉、中梨秋香、中梨蜜脆、中梨金福、秋月、恬心、六月香、金香玉、T109 和红玉17 个品种只扩增出一条S 基因特异条带,条带范围在300~500 bp。此外,红酥脆、满天红、美人酥3 个品种的2 条S 基因特异条带的位置十分接近,均是一条在300~400 bp范围内,另一条在500~600 bp范围内。
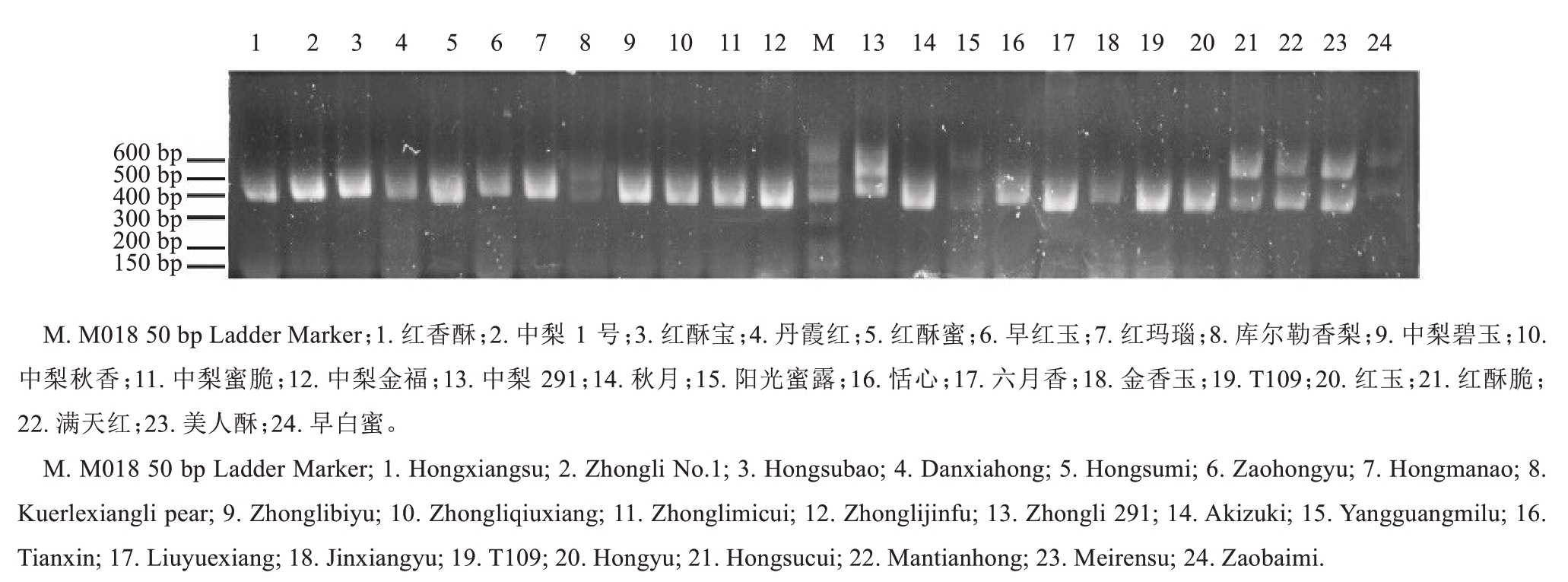
图1 24 个梨品种的PCR 扩增结果
Fig. 1 The PCR amplification of 24 pear cultivars
由于上述17 个品种只扩增出1 条S 基因特异条带,推测原因可能为条带大小非常接近,琼脂糖电泳无法分离,或者引物只能扩增出部分特异的S 基因型。基于以上猜测,对红香酥、中梨1 号、红酥宝、丹霞红、红酥蜜、中梨碧玉、中梨蜜脆、中梨291 等品种的重测序数据进行分析,发现经典的FTQQYQ 和anti-IIWPNV 引物不能扩增出部分梨品种的S 基因。基于此,利用DANMAN 软件比对核苷酸序列,重新设计了新的多态性引物FTQQYQ-B 和anti-GIIWPN(图2)。与之前引物相比,FTQQYQ-B 和anti-GIIWPN分别增加了1个(C/G/T)和3个(A/T)(G/T)(G/T)多态性位点,使得新引物能够扩增出更为丰富的特异性S单倍型序列。
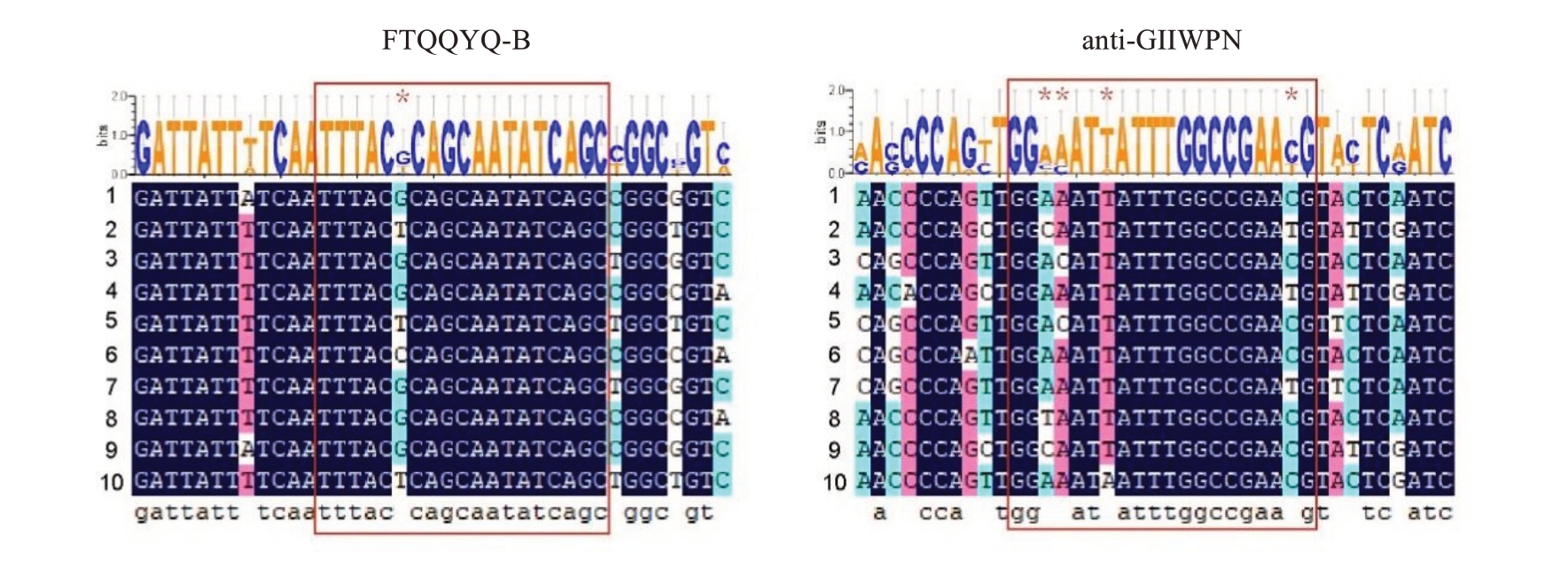
图2 FTQQYQ-B 和anti-GIIWPN 引物的核苷酸序列比对
Fig. 2 Nucleotide sequence alignment of the primers FTQQYQ-B and anti-GIIWPN
随后,用新的特异引物FTQQYQ-B 和anti-GIIWPN 对24 个梨品种基因组DNA 进行PCR 扩增,电泳检测结果见图3。由图3 可以看出,除中梨1号、红酥蜜、中梨碧玉、中梨蜜脆、秋月扩增出1 条300~400 bp 的特异条带外,其他19 个品种均扩增出2 条明显的S 基因特异性条带,目的片段大小集中在300~600 bp 之间。为了准确鉴定品种的S 基因型,本研究对所有品种的目的条带进行了测序分析。
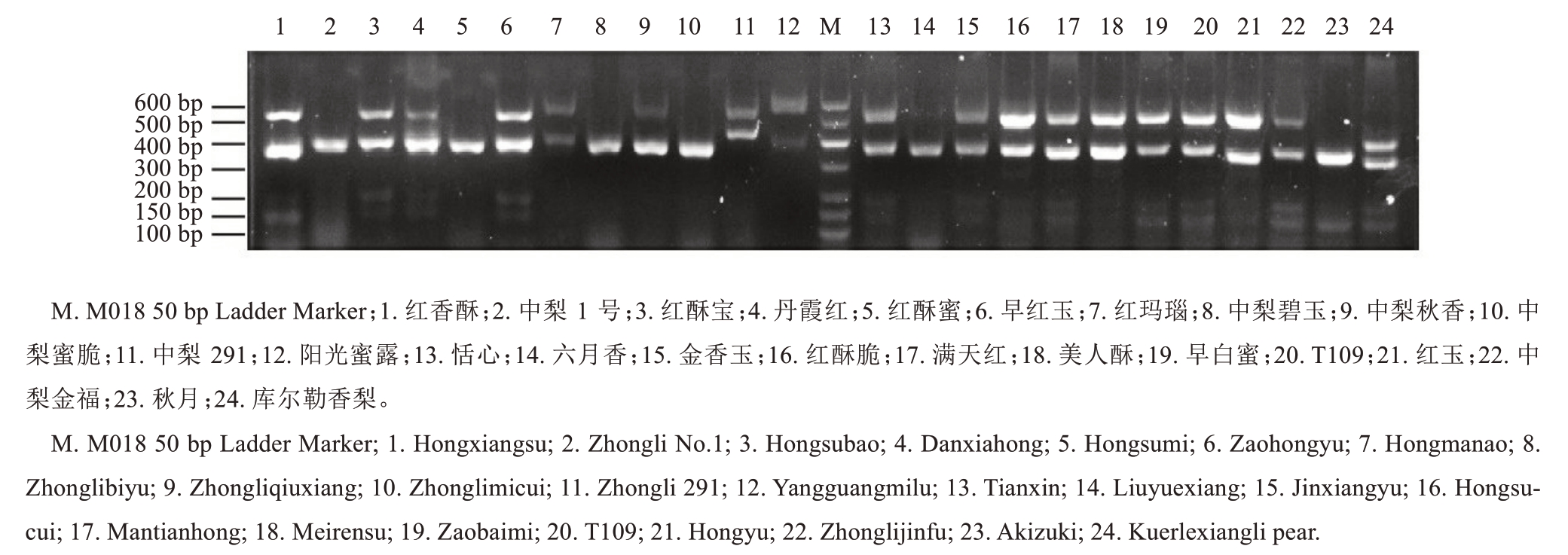
图3 24 个梨品种的S 基因型琼脂糖凝胶电泳
Fig. 3 The agarose electrophoresis of S-RNase gene in 24 pear cultivars
2.2 S基因型的鉴定
将上述24个待测品种的S基因特异条带进行回收、克隆及测序,测序结果通过BLAST 与NCBI 中已公布的梨S-RNase 基因的序列信息进行比较分析,依据S 基因相同时其DNA 序列同源性为100%的原理,确定各品种的S基因型(表1)。
表1 24 个梨品种的PCR 片段长度及S 基因型
Table 1 PCR fragment sizes and S genotypes of 24 pear cultivars

编号No.S基因型S-genotype 2 1 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24品种名Cultivars红香酥 Hongxiangsu中梨1号 Zhongli No.1红酥宝 Hongsubao丹霞红 Danxiahong红酥蜜 Hongsumi早红玉 Zaohongyu红玛瑙 Hongmanao中梨碧玉 Zhonglibiyu中梨秋香 Zhongliqiuxiang中梨蜜脆 Zhonglimicui中梨291 Zhongli 291阳光蜜露 Yangguangmilu恬心 Tianxin六月香 Liuyuexiang金香玉 Jinxiangyu红酥脆 Hongsucui满天红 Mantianhong美人酥 Meirensu早白蜜 Zaobaimi T109红玉 Hongyu中梨金福 Zhonglijinfu秋月 Akizuki库尔勒香梨Kuerlexiangli pear PCR产物大小PCR fragment size/bp 345/528 368/367 375/528 368/528 368/345 375/528 375/528 374/369 368/528 368/374 535/427 535/345 375/528 368/528 375/528 374/535 368/535 368/535 374/535 374/535 374/535 374/528 375/368 345/427 S22S39 S4S1 S3S39 S4S39 S4S22 S3S39 S3S39 S5Sd S4S39 S4S5 S12S28 S12S22 S3S39 S4S39 S3S39 S4S12 S4S12 S4S12 S5S12 S5S12 S5S12 S5S39 S3S4 S22S28
序列经比对分析后,24 个梨品种的S 基因型分别为:红香酥S22S39;中梨1号S1S4;红酥宝、早红玉、红玛瑙、恬心和金香玉S3S39;丹霞红、中梨秋香和六月香S4S39;红酥蜜S4S22;中梨碧玉S5Sd;中梨蜜脆S4S5;中梨291 S12S28;阳光蜜露S12S22;早白蜜、T109 和红玉S5S12;红酥脆、满天红和美人酥S4S12;中梨金福S5S39;秋月S3S4;库尔勒香梨S22S28。其中,作为对照的库尔勒香梨的S基因型与吕文娟等[17]、杨金花等[18]的鉴定结果一致,说明本研究鉴定的梨品种S基因型准确。
2.3 S基因频率的分析
本研究鉴定出的24 个梨品种中包括23 个梨新品种和1 个对照品种库尔勒香梨,共涉及到9 个不同的S 基因,出现频率最高的S 基因为S39和S4,占10/48,其次是S12,占8/48;S3、S5,均占6/48;其余S 基因出现的频率较低,S22占4/48;S28占2/48;S1、Sd均占1/48。因此,不同S 基因在检测的梨品种中出现频率不均衡。
23 个梨新品种中10 个品种含有S39基因型,说明S39基因型在本研究鉴定的新品种中所占比例较大。前文鉴定红香酥的基因型为S22S39,所以根据红香酥的重测序数据设计了扩增S39基因的特异性引物S39-F和S39-R,对24个梨品种DNA进行扩增,检测引物的特异性。从图4 中可以看出,S39-F 和S39-R 引物可以特异扩增出S39基因型,与表1鉴定结果一致,为后续鉴定S39基因型提供了方便快捷的途径。
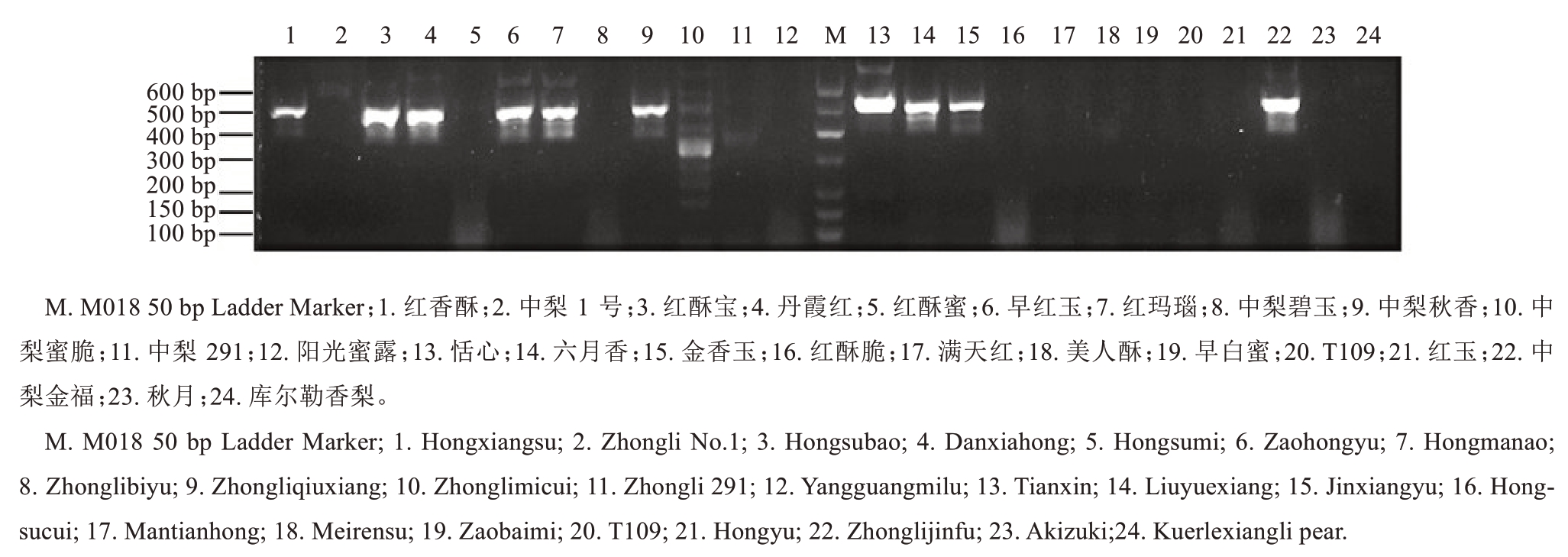
图4 24 个梨品种的S39基因鉴定结果
Fig. 4 Identification results of the S39 gene in 24 pear cultivars
2.4 新品种间授粉亲和性分析
根据鉴定的24 个梨品种的S 基因型,发现红酥宝、早红玉、红玛瑙、恬心和金香玉的S 基因型为S3S39,丹霞红、中梨秋香和六月香的S基因型为S4S39,T109、红玉和早白蜜的S基因型为S5S12,红酥脆、满天红和美人酥的S基因型为S4S12。上述4组品种具有完全相同的S基因型,彼此之间不宜作为授粉品种。
红香酥、红酥宝、丹霞红、早红玉、红玛瑙、中梨秋香、恬心、六月香、金香玉和中梨金福均含有S39,中梨1 号、丹霞红、红酥蜜、中梨秋香、中梨蜜脆、六月香、红酥脆、满天红、美人酥和秋月均含有S4,中梨291、阳光蜜露、红酥脆、满天红、美人酥、早白蜜、T109 和红玉均含有S12,中梨碧玉、中梨蜜脆、早白蜜、T109、红玉和中梨金福均含有S5,红酥宝、早红玉、红玛瑙、恬心、金香玉和秋月均含有S3,红香酥、红酥蜜、阳光蜜露和库尔勒香梨均含有S22,中梨291和库尔勒香梨均含有S28。上述7组品种均含有一个相同的S基因,为避免发生结果率低或者结果不实,同一组品种之间不宜相互授粉。
3 讨 论
1999 年Ishimizu 等[16]用PCR-RFLP 体系来鉴定日本梨品种的S 基因型,此方法也适用于中国梨品种,其引物FTQQYQ 和anti-IIWPNV 一直作为扩增中国梨品种S 基因的特异引物。随着分子生物学技术的发展,我国研究者利用这对引物扩增S 等位基因并结合DNA 测序技术进行了大量关于梨S 基因型的鉴定工作[8-14]。本研究中,通过对红香酥、中梨1号、红酥宝、丹霞红、红酥蜜、中梨碧玉、中梨蜜脆、中梨291等梨品种重测序数据的分析,发现经典的S等位基因引物FTQQYQ和anti-IIWPNV并不完全适用于所有梨品种S 基因型的鉴定。由此,根据序列多态性比对结果,重新设计了新的多态性引物FTQQYQ-B 和anti-GIIWPN。与先前的引物相比,FTQQYQ-B 和anti-GIIWPN 引物分别新增了1 个(C/G/T)和3 个(A/T)(G/T)(G/T)多态性位点,这一改进提升了引物的特异性,从而能够扩增出更加丰富且特异性强的S 单倍型序列,扩大了遗传分析的范围,可用于后续新品种S基因型的鉴定工作。
本研究鉴定了24 个梨品种的S 基因型,其中鉴定库尔勒香梨S基因型为S22S28,与吕文娟等[17]和杨金花等[18]鉴定结果一致。本研究表明红酥脆、满天红和美人酥的S 基因型为S4S12,与张琳等[19]鉴定结果一致。此外,S 基因的遗传遵循孟德尔遗传规律,即在杂交后代中,子代的S 基因分别来自其父本和母本。因此,开展果树S基因型鉴定,不仅可以为生产实践中合理配置授粉树和杂交育种提供科学依据,还可以作为鉴定不同品种间亲缘关系的有力证据[20-21]。本研究鉴定的23 个梨新品种的亲本基因型有的已被确定,如火把梨S12S26S44[19],新世纪S3S4[22],早美酥S3Sd[22],早酥S1Sd[23]和幸水S4S5[24]等,故可以通过亲缘关系来进一步验证这23 个梨品种的S 基因型,如早红玉S3S39、红酥宝S3S39、红酥蜜S4S22的亲本均为新世纪S3S4 和红香酥S22S39。丹霞红的S 基因型为S4S39,其亲本为中梨1 号S1S4和红香酥S22S39。红玛瑙为S3S39,亲本为早美酥S3Sd和红香酥S22S39。满天红S4S12、美人酥S4S12和红酥脆S4S12的亲本均为幸水S4S5和火把梨S12S26。中梨秋香S4S39的亲本为满天红S4S12和红香酥S22S39。阳光蜜露S12S22 的亲本为满天红S4S12和库尔勒香梨S22S28。除此之外,也可以利用本研究的结果推测出亲本的S等位基因,如红香酥S基因型为S22S39,亲本为郑州鹅梨和库尔勒香梨S22S28,按照孟德尔遗传规律,红香酥的S22等位基因来自于库尔勒香梨,而另一个等位基因S39来自于郑州鹅梨,故郑州鹅梨一定含有S39等位基因。
S 基因是可遗传的,亲本S 基因可遗传给子代。本研究也对鉴定的24个梨品种的S基因出现频率进行统计,发现24 个梨品种所包含的9 个S 等位基因的分布频率很不均衡,存在几个明显的高频基因,如S4和S39,这可能是由于人们长期对品种可育性和经济性状进行了持续的选择和优化。在新品种中S39基因出现的频率最高,分析发现含有S39基因的品种亲本之一均为红香酥S22S39,而且发现目前已鉴定的梨品种中较少含有S39基因[5],推测S39可能与某些特殊的性状(如果皮颜色、果心大小和糖酸比等经济性状)相连锁,以致在新品种选育过程中被选择,所以在新品种中出现的频率最高。如果能将S 基因与品种的经济性状的关系进一步研究,研究两者之间是否具有连锁关系,将进一步丰富对自交不亲和领域理论的研究,还将有助于精准筛选出具有优良经济性状的品种,从而显著提高育种效率和品种质量[25]。
本研究所鉴定的新品种,除中梨1 号、红酥脆、满天红和美人酥外,其他品种S 基因型之前未被报道过,而且除秋月外其余均为我国自主培育的新品种,包括中梨1 号、红香酥、丹霞红和早白蜜等代表性优良品种以及具有巨大推广潜力的新秀品种中梨金福和阳光蜜露等。研究鉴定这些新品种的S 基因型,并对其授粉亲和性进行分析,可为新品种商业化栽培配置适宜授粉树提供理论依据。
4 结 论
根据红香酥等重测序数据,设计了新的S 基因型特异引物FTQQYQ-B 和anti-GIIWPN,该引物可扩增出更为丰富的特异性S 单倍型序列。采用PCR技术结合DNA 测序技术,鉴定了23个梨新品种的S基因型,鉴定结果为:红香酥S22S39;中梨1 号S1S4;红酥宝、早红玉、红玛瑙、恬心和金香玉S3S39;丹霞红、中梨秋香和六月香S4S39;红酥蜜S4S22;中梨碧玉S5Sd;中梨蜜脆S4S5;中梨291S12S28;阳光蜜露S12S22;早白蜜、T109和红玉S5S12;红酥脆、满天红和美人酥S4S12;中梨金福S5S39;秋月S3S4;库尔勒香梨S22S28。统计发现新品种中S4和S39基因出现频率最高。
[1] MATTON D P,MAES O,LAUBLIN G,XIKE Q,BERTRAND C,MORSE D,CAPPADOCIA M. Hypervariable domains of self-incompatibility RNases mediate allele-specific pollen recognition[J]. The Plant Cell,1997,9(10):1757-1766.
[2] EAST E M,MANGELSDORF A J. A new interpretation of the hereditary behavior of self-sterile plants[J]. Proceedings of the National Academy of Sciences of the United States of America,1925,11(2):166-171.
[3] 张绍铃,吴巨友,吴俊,齐永杰,高永彬. 蔷薇科果树自交不亲和性分子机制研究进展[J]. 南京农业大学学报,2012,35(5):53-63.ZHANG Shaoling,WU Juyou,WU Jun,QI Yongjie,GAO Yongbin. Advance in molecular mechanisms of self-incompatibility in Rosaceae fruit trees[J]. Journal of Nanjing Agricultural University,2012,35(5):53-63.
[4] SASSA H,HIRANO H,IKEHASHI H. Identification and characterization of stylar glycoproteins associated with self-incompatibility genes of Japanese pear,Pyrus serotina Rehd.[J]. Molecular & General Genetics,1993,241(1/2):17-25.
[5] 张校立,艾沙江·买买提,徐叶挺,邓莉,王继勋. 梨S 基因与S基因型鉴定的研究进展[J]. 西北农业学报,2018,27(8):1077-1087.ZHANG Xiaoli,Aishajiang·Maimaiti,XU Yeting,DENG Li,WANG Jixun. Present advance of S-gene genotype and S-genotypes in pear[J]. Acta Agriculturae Boreali-occidentalis Sinica,2018,27(8):1077-1087.
[6] ISHIMIZU T,SATO Y,SAITO T,YOSHIMURA Y,NORIOKA S,NAKANISHI T,SAKIYAMA F. Identification and partial amino acid sequences of seven S-RNases associated with self-incompatibility of Japanese pear,Pyrus pyrifolia Nakai[J].Journal of Biochemistry,1996,120(2):326-334.
[7] TAKASAKI T,OKADA K,CASTILLO C,MORIYA Y,SAITO T,SAWAMURA Y,NORIOKA N,NORIOKA S,NAKANISHI T. Sequence of the S9-RNase cDNA and PCR-RFLP system for discriminating S1- to S9-allele in Japanese pear[J]. Euphytica,2004,135(2):157-167.
[8] 乌云塔娜. 中国白梨自交不亲和基因的分离鉴定[D]. 株洲:中南林学院,2003.Wuyun Ta’na. Isolation and identification of self-incompatibility genes of Chinese Pyrus bretschneideri[D]. Zhuzhou:Central South Forestry University,2003.
[9] TAN X F,ZHANG L,Wuyun Ta’na,YUAN D Y,CAO Y F,JIANG A F,LIANG W J,ZENG Y L. Molecular identification of two new self-incompatible alleles (S-alleles) in Chinese pear(Pyrus bretschneideri)[J]. Journal of Plant Physiology and Molecular Biology,2007,33(1):61-70.
[10] 张妤艳,吴俊,衡伟,张绍铃. 京白梨等品种S 基因型鉴定及新基因S28 和S30 的核苷酸序列分析[J]. 园艺学报,2006,33(3):496-500.ZHANG Yuyan,WU Jun,HENG Wei,ZHANG Shaoling. Identification of S-genotypes of pear cultivars and analyses of nucleotide sequences of S28-RNase and S30-RNase[J]. Acta Horticulturae Sinica,2006,33(3):496-500.
[11] 张妤艳,张绍铃,吴俊,张瑞萍,李秀根. 八月酥等14 个梨品种自交不亲和基因(S 基因)型的鉴定[J]. 果树学报,2007,24(2):135-139.ZHANG Yuyan,ZHANG Shaoling,WU Jun,ZHANG Ruiping,LI Xiugen. Identification of S-genotypes in 14 pear cultivars[J].Journal of Fruit Science,2007,24(2):135-139.
[12] 衡伟,张绍铃,张妤艳,吴俊,李秀根. 12 个梨品种S 基因型的鉴定[J]. 园艺学报,2007,34(4):853-858.HENG Wei,ZHANG Shaoling,ZHANG Yuyan,WU Jun,LI Xiugen. Identification of S-genotypes of twelve pear cultivars by analysis of DNA sequence[J]. Acta Horticulturae Sinica,2007,34(4):853-858.
[13] 衡伟,张绍铃,方成泉,吴华清,吴俊. 梨20 个品种S 基因型的鉴定及新S-RNases 基因的克隆[J]. 园艺学报,2008,35(3):313-318.HENG Wei,ZHANG Shaoling,FANG Chengquan,WU Huaqing,WU Jun. Identification of 20 S-genotypes and cloning novel SRNases in Pyrus[J]. Acta Horticulturae Sinica,2008,35(3):313-318.
[14] 陈慧,张树军,张妤艳,衡伟,吴俊,张绍铃. 40 个梨品种S 基因型的鉴定及S 基因频率分析[J]. 南京农业大学学报,2013,36(5):21-26.CHEN Hui,ZHANG Shujun,ZHANG Yuyan,HENG Wei,WU Jun,ZHANG Shaoling. Identification of S-genotypes in forty pear cultivars and analysis of S-RNase genes frequency in Pyrus[J]. Journal of Nanjing Agricultural University,2013,36(5):21-26.
[15] 梁文杰,谭晓风,乌云塔娜. 梨自交不亲和基因克隆及其进化分析[J]. 果树学报,2021,38(10):1621-1637.LIANG Wenjie,TAN Xiaofeng,Wuyun Ta’na. Cloning and phylogenetic analysis of S-RNase genes in genus Pyrus plants[J].Journal of Fruit Science,2021,38(10):1621-1637.
[16] ISHIMIZU T,INOUE K,SHIMONAKA M,SAITO T,TERAI O,NORIOKA S. PCR-based method for identifying the S-genotypes of Japanese pear cultivars[J]. Theoretical and Applied Genetics,1999,98(6):961-967.
[17] 吕文娟,冯建荣,刘小芳,刘海楠,李文慧,钟颖. ‘库尔勒香梨’自交不亲和S-RNase 等位基因全长的克隆与分析[J]. 分子植物育种,2017,15(5):1639-1647.LÜ Wenjuan,FENG Jianrong,LIU Xiaofang,LIU Hainan,LI Wenhui,ZHONG Ying. Cloning and analysis of self-incompatibility S-RNase allelic genes in Korla fragrant pear[J]. Molecular Plant Breeding,2017,15(5):1639-1647.
[18] 杨金花,徐叶挺,田雯,张校立. 14 个新疆与中亚梨品种S 基因型的鉴定[J]. 分子植物育种,2023,21(10):3199-3206.YANG Jinhua,XU Yeting,TIAN Wen,ZHANG Xiaoli. Identification of S-genotypes of fourteen pear cultivars in Xinjiang and central Asia[J]. Molecular Plant Breeding,2023,21(10):3199-3206.
[19] 张琳,谭晓风,胡姣,龙洪旭,袁德义,李秀根. 火把梨及其后代育成品种的S 基因型鉴定[C]//南宁:第二届中国林业学术大会—S9 木本粮油产业化论文集,2009:379-385.ZHANG Lin,TAN Xiaofeng,HU Jiao,LONG Hongxu,YUAN Deyi,LI Xiugen. Determination of S-genotypes of Huobali pear and its progenies[C]//Nanning:Proceedings of the 2nd China Forestry Academic Conference-S9 Woody Oil and Cereal Industrialization,2009:379-385.
[20] 何敏,谷超,吴巨友,张绍铃. 果树自交不亲和机制研究进展[J].园艺学报,2021,48(4):759-777.HE Min,GU Chao,WU Juyou,ZHANG Shaoling. Recent advances on self-incompatibility mechanism in fruit trees[J]. Acta Horticulturae Sinica,2021,48(4):759-777.
[21] 付燕,杨芩,彭舒,蒋瑶,刘伦沛. 枇杷属植物3 个新S 基因鉴定及大渡河枇杷分类地位的新证据[J]. 果树学报,2018,35(11):1316-1323.FU Yan,YANG Qin,PENG Shu,JIANG Yao,LIU Lunpei. Identification of three novel S-RNase alleles in Eriobotrya Lindl.and new evidence for the taxonomic status of E. prinoides var.dadunensis[J]. Journal of Fruit Science,2018,35(11):1316-1323.
[22] 乌云塔娜,谭晓风,李秀根,张林,邓建军. 13 个‘新世纪’梨后代品种S 基因型的鉴定[J]. 林业科学,2007,43(9):116-122.Wuyun Ta’na,TAN Xiaofeng,LI Xiugen,ZHANG Lin,DENG Jianjun. Identification of S-genotypes of 13 progenies of ‘Shinseiki’ pear[J]. Scientia Silvae Sinicae,2007,43(9):116-122.
[23] 许高歌. 桃和‘早酥’梨自交(不)亲和性分子机制的研究[D].南京:南京农业大学,2012.XU Gaoge. Molecular mechanism of self-compatibility in peach and ‘Zaosu’[D]. Nanjing:Nanjing Agricultural University,2012.
[24] 王春雷,高季平,赵宪坤. 利用酶联免疫反应鉴定梨树S 基因型[J]. 江苏农业学报,2015,31(5):1124-1128.WANG Chunlei,GAO Jiping,ZHAO Xiankun. S genotyping in pear by PCR-ELISA[J]. Jiangsu Journal of Agricultural Sciences,2015,31(5):1124-1128.
[25] 姜新,曹晓艳,王大江,冯建荣,刘月霞,樊新民. 南疆杏品种自交不亲和S-RNase 基因型的鉴定[J]. 果树学报,2012,29(4):569-576.JIANG Xin,CAO Xiaoyan,WANG Dajiang,FENG Jianrong,LIU Yuexia,FAN Xinmin. Identification of self-incompatibility S-RNase genotypes for apricot cultivars in South of Xinjiang area[J]. Journal of Fruit Science,2012,29(4):569-576.