猕猴桃是猕猴桃科(Actinidiaceae)猕猴桃属(Actinidia Lindl.)落叶藤本果树,该属有54 个种和21 个变种,共75 个分类单元[1]。中国是猕猴桃属植物的原产地,野生资源丰富,地域分布广泛。自20世纪被人工驯化以来,该产业在世界范围内的栽培面积不断扩大[2],因其果实风味独特、营养丰富而深受人们喜爱。随着产业的发展,不同区域间引种频率增加,生产中的细菌性溃疡病也越发严重,对产业造成了严重危害[3]。自1984年日本首次出现该病以来[4],随后短短几年内,迅速成为威胁世界猕猴桃产业的毁灭性病害[5]。中国、新西兰、意大利、韩国等世界各大猕猴桃产区深受其害[6-9]。中国作为猕猴桃生产大国,在四川、安徽、陕西等主产省份也早已出现,并逐渐向其他栽培地区蔓延[10]。因此,开展溃疡病相关研究,了解其致病机制和作用机制、探索病害防控技术,选育抗病新品种,对助力产业高质量发展具有重要意义。随着现代测序技术的发展,分子生物学技术为猕猴桃溃疡病研究提供了一种有效途径。本文旨在综述猕猴桃溃疡病抗性研究方面最新进展,为该产业抗性育种和高效防控技术研发提供理论基础。
1 溃疡病的危害、传播媒介及致病菌
猕猴桃细菌性溃疡病在一年内有冬末春初和秋季两个发病高峰[11]。主要危害叶片、花、果实和枝蔓,病菌侵染多从伤口、皮孔、落叶痕、枝条分叉等部位开始[12]。枝蔓发病初期呈水渍状,后病斑扩大、颜色加深;早期病斑部位流白色黏液,不久转为铁锈红色,剥开病斑皮层后,可见韧皮部腐烂,木质部变黑[13-14]。叶片染病后会形成不规则的黑色斑点,并带有黄色晕圈。溃疡病具有隐蔽性、传染性、暴发性和毁灭性等特点,一旦发生,轻则减产、重则毁园。2010年新西兰首次发现溃疡病之后,感病果园数量迅速增加,到2012 年占全部果园的37%[15],对该国猕猴桃产业造成严重危害。研究发现,不同栽培品种对溃疡病的抗性不一,一些商品性很好的品种抗病能力却很弱,如新西兰的黄肉品种Hort16A 以及中国的红心品种红阳[16]。
猕猴桃溃疡病病菌主要借助风、雨等在果园内和果园间迅速散播,低温潮湿的环境可极大地促进病原菌繁殖,一些不规范的农艺操作以及低温、冷害、冻害等极端气候现象也有助于病菌的进一步传播和流行[17]。苗木、接穗、工具、人员等都可能是携带病菌的重要载体,此外,昆虫[18]、花粉[19]、非猕猴桃属植物[20]等也可以作为中间媒介或中间寄主来实现病菌的传播和侵染。
丁香假单胞杆菌猕猴桃致病变种(Pseudomonas syringae pv. actinidiae,Psa)是引起猕猴桃细菌性溃疡病的致病菌[4],其关键致病因子是存在T3SS(type Ⅲsecretion system)的蛋白分泌系统[10]。该分泌系统能分泌多种有毒效应因子(如HopZ5、Avr-Rpm1等)来破坏植物的免疫防御反应,使病原体快速适应宿主环境[21-22]。Psa 的整合共轭元件(integrative and conjugative elements,ICEs)是可移动元件,赋予其新的表型,且常被认为是细菌病原体产生耐药性进化的机制[22-23]。研究人员通过对目前收集到的Psa 株系进行基因组比较、系统进化和起源分析等,可将目前已发现的Psa 株系分为6 类[24]:第一类是在日本和意大利海沃德品种上采集到的病原菌Psa1[25];第二类是在韩国发现的病原菌Psa2[26];第三类是于2008年首次在意大利发现,并对世界各国产业造成毁灭性伤害的Psa3[27];第四类是在新西兰发现的病原菌Psa4,该类病菌致病能力较弱,仅引起叶斑[9],与前三类存在明显不同,虽然Psa4是从猕猴桃属植物上分离出来的,但由于其表型、遗传和系统发育不同,后将其更名为Pseudomonas syringae pv.actinidifoliorum(Pfm)[28];第五类和第六类是在日本发现的Psa5[29]和Psa6[30]。在已有的研究报道中,并没有发现对溃疡病有效的治愈方法,生产上频繁使用的药物防治使得Psa菌株对铜和链霉素产生了抗药性[31]。因此,选育抗性强的品种来增强自身的抗病性,是解决溃疡病危害最直接的方法。
2 不同种质资源抗病性差异及成因
不同种类和品种的猕猴桃对Psa的抗病性有所不同,研究人员分别从形态结构、生理和分子水平等方面对其抗病能力进行了研究分析。对24 个不同种类或品种的猕猴桃进行抗性评价,不同种类抗性由强到弱的顺序依次为:毛花猕猴桃(A.eriantha)、美味猕猴桃(A.chinensis var.delicious)、中华猕猴桃(A.chinensis)[32]。利用29个猕猴桃种或品种(系)进行抗性评价,在离体枝条接种Psa 病原菌6 d 后,中华猕猴桃红阳、6-65、2-72就开始发病并溢出少量白色黏液,而接种21 d 后软枣猕猴桃(A.arguta)和毛花猕猴桃才开始出现病斑且病斑直径明显小于中华猕猴桃,体现出抗性相对较强[33]。通过采用离体枝条和叶片进行人工接种Psa病原菌的方法对51份软枣猕猴桃进行抗性鉴定,结果显示51份资源中高抗33 份、中抗18 份,无高感、中感和感病种质,体现了软枣猕猴桃具有较好的抗性[34]。采用室内和田间接种方法分别对12个和23个品种进行抗性鉴定,总体抗性趋势为软枣猕猴桃和毛花猕猴桃强于美味猕猴桃,而中华猕猴桃表现最差[35]。近年来,研究人员选育出了多个抗病新品种,如先沃五号[36]、华金3号[37]、金塘一号[38]等。
猕猴桃的组织结构及活性成分与抗病性有一定关系。通过比较不同抗性资源的叶片和枝条结构发现,叶片气孔密度和长度、枝条皮孔密度和长度与病情指数呈显著正相关[39-40]。接种病原菌后,抗病品种金魁的枝条、叶片中可溶性糖和木质素含量也显著高于感病品种金丰[41]。在不同品种中,抗病性越强,叶片中可溶性蛋白质和酚类物质含量越高[42]。韧皮部蔗糖代谢的增加可能引起免疫应答相关酶的增加,但不同品种的抗病能力与蔗糖的含量呈负相关[43]。还有研究发现,抗性砧木也能够有效增强接穗的抗病性[44]。
综上所述,不同猕猴桃种类、品种(系)的抗性情况存在差异,但总体抗性趋势为软枣猕猴桃和毛花猕猴桃强于美味猕猴桃,且强于中华猕猴桃。这种抗性差异可能与生理生化特性及遗传背景等因素有关。因此,准确评价和筛选出高抗猕猴桃细菌性溃疡病的品种(系)至关重要。
3 分子标记在抗病性鉴定中的应用
分子标记(Molecular Markers)是以个体间核苷酸序列变异为基础的遗传标记,能在DNA水平上直接反映遗传的多态性[45]。与其他生物遗传标记如生化标记、细胞学标记和形态学标记相比,具有不受个体发育时期和外界环境影响、多态性高、易于检测等显著的优点[46]。分子标记技术的发展主要经历了3个阶段,第1 个阶段是以Southern 杂交为基础的限制性片段长度多态性(Restriction fragment length polymorphism,RFLP)标记;第2 个阶段是以PCR 反应为基础的随机扩增多态性(Random amplified polymorphic DNA,RAPD)、扩增片段长度多态性(Amplified fragment length polymorphism,AFLP)、简单重复序列(Simple sequence repeat,SSR)标记;第3个阶段则是以测序为基础的单核苷酸标记(Single nucleotide polymorphism,SNP)[47]。目前,SNP技术已被广泛应用于物种亲缘关系进化分析、种质资源鉴定保存、分子遗传图谱构建、植物抗病基因定位及遗传育种等方面。
猕猴桃属植物种类繁多,地域分布范围广泛,因此对其种质资源进行准确鉴定是合理利用的基础。利用分子标记技术筛选抗性优异种质资源已广泛应用于大豆[48]、水稻[49]、番茄[50]、黄瓜[51]等植物的抗性育种中,猕猴桃中也有少量应用。通过利用ISSR标记进行遗传多样性分析,发现不同品种对溃疡病的抗病能力与遗传有关,且各品种抗性强弱分组与ISSR聚类组有明显相关性,表明了该技术可用于辅助选育抗溃疡病品种[52]。采用SSR技术结合BSA分析方法对杂交F1代群体及40 份资源进行抗病基因(PR)分子标记筛选,获得了与抗病基因连锁的SSR 分子标记UDK97-428116[53]。利用SCoT分子标记确定了9个中华猕猴桃红肉品种的亲缘关系并筛选出较抗溃疡病品种[16]。对6 个品系进行抗病相关的RAPD 分析,结果发现,抗病品系都有一条1458 bp的DNA片段,而感病品系均无该条带[54]。通过构建表达序列标签(EST)文库,并基于同源序列设计EST引物,成功鉴定出部分参与基础防御途径的基因,为猕猴桃抗性育种提供了基础[55]。
4 遗传图谱及QTL 定位在抗病研究中的应用
遗传连锁图谱是通过遗传重组交换结果进行连锁分析所得到的基因或分子标记在染色体上相对位置的线性排列图[56],是数量性状定位(quantitative trait locus,QTL)、分子标记辅助选择育种等研究的理论依据。因此,构建高密度的遗传连锁图谱是植物进化过程、遗传育种及功能基因组学等研究的重要环节。
利用果树基因组中的多种分子标记技术构建高精度的遗传图谱已逐渐成为该学科研究的重要内容。近年来,已在枣[57]、苹果[58]、葡萄[59]、柑橘[60]、樱桃[61]、梨[62]等果树作物中构建了分子遗传图谱。目前,猕猴桃相关分子遗传图谱的构建也在逐步完善。自2001年Testolin等[63]利用AFLP和SSR标记,分别构建中华猕猴桃和硬齿猕猴桃的遗传图谱以来,目前已报道构建的遗传图谱共有11 组(表1)。这些遗传图谱对猕猴桃相关性状定位研究具有重要意义。在目前已构建的图谱中,应用于溃疡病研究的只有两组,分别是2019年以二倍体中华猕猴桃为亲本和2020 年以四倍体中华猕猴桃为亲本构建。在得到的二倍体中华猕猴桃高密度遗传图谱中,通过QTL 定位技术在Hort16A 的27 号染色体上定位到了一个主效QTL,在P1 中定位到了6 个微效QTLs(3、14、15、22、24 和28 号染色体),并验证27、14、22、28之间的互作效应,结果表明,F1代对溃疡病抗性的增强是Hort16A 和P1 上的QTL 加性效应引起的[64]。随后,在四倍体中华猕猴桃遗传图谱中定位到了4个抗Psa的QTLs(LG1、LG2、LG4和LG7);其中,高抗亲本包含3 个QTLs(LG1、LG4 和LG7),耐病亲本包含1 个QTL(LG2)[65]。主要抗性基因与数量抗性因子的结合,可以保持抗性品种的持久性,QTL的加性效应增强了子代对病原菌的抗性[66-67]。
表1 猕猴桃遗传图谱
Table 1 Genetic linkage map of kiwifruit
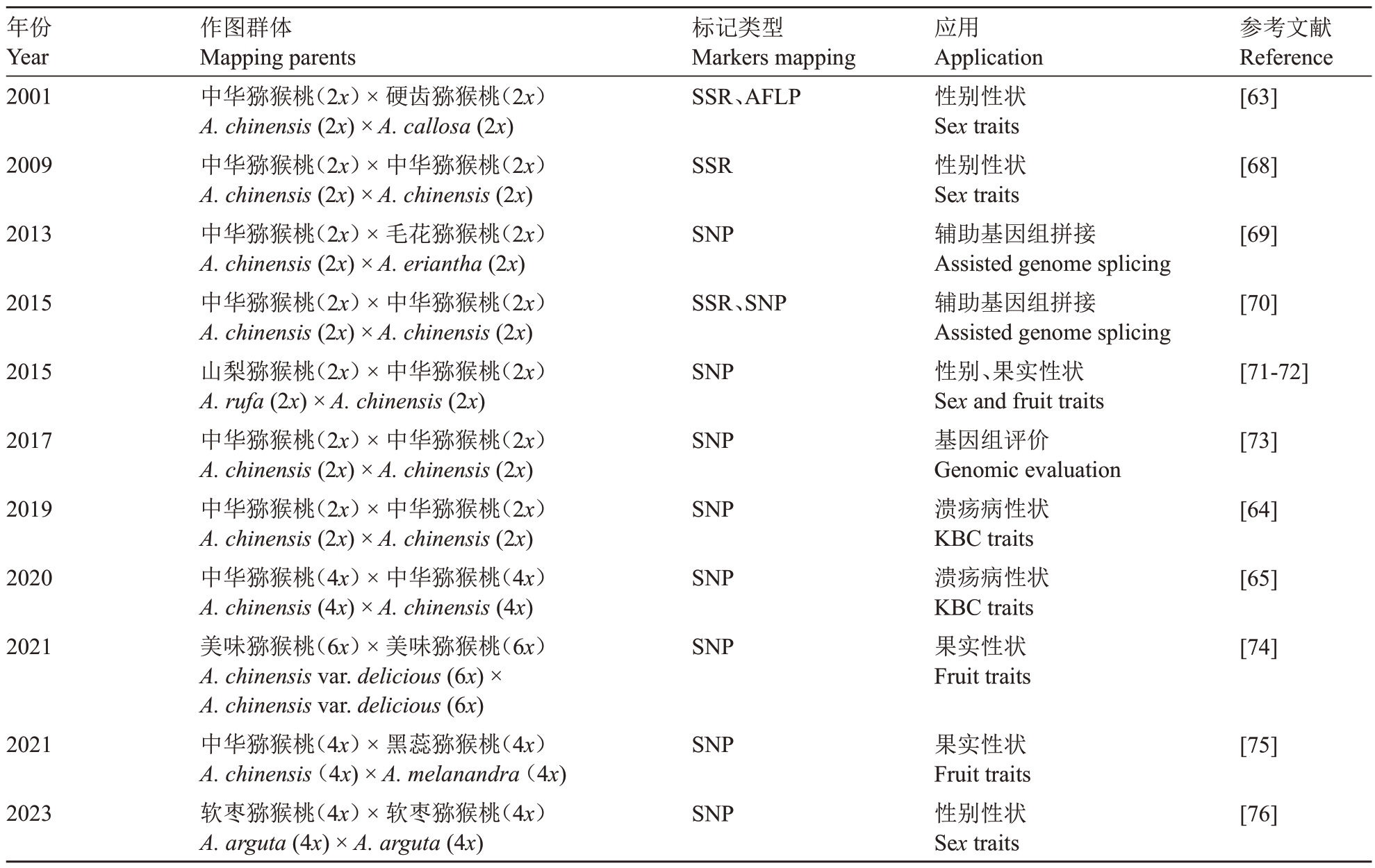
年份Year 2001标记类型Markers mapping SSR、AFLP参考文献Reference[63]2009 SSR [68]2013 SNP [69]2015 SSR、SNP [70]2015 SNP [71-72]2017 SNP [73]2019 SNP [64]2020 SNP [65]2021 SNP应用Application性别性状Sex traits性别性状Sex traits辅助基因组拼接Assisted genome splicing辅助基因组拼接Assisted genome splicing性别、果实性状Sex and fruit traits基因组评价Genomic evaluation溃疡病性状KBC traits溃疡病性状KBC traits果实性状Fruit traits[74]2021 SNP [75]2023作图群体Mapping parents中华猕猴桃(2x)×硬齿猕猴桃(2x)A.chinensis(2x)×A.callosa(2x)中华猕猴桃(2x)×中华猕猴桃(2x)A.chinensis(2x)×A.chinensis(2x)中华猕猴桃(2x)×毛花猕猴桃(2x)A.chinensis(2x)×A.eriantha(2x)中华猕猴桃(2x)×中华猕猴桃(2x)A.chinensis(2x)×A.chinensis(2x)山梨猕猴桃(2x)×中华猕猴桃(2x)A.rufa(2x)×A.chinensis(2x)中华猕猴桃(2x)×中华猕猴桃(2x)A.chinensis(2x)×A.chinensis(2x)中华猕猴桃(2x)×中华猕猴桃(2x)A.chinensis(2x)×A.chinensis(2x)中华猕猴桃(4x)×中华猕猴桃(4x)A.chinensis(4x)×A.chinensis(4x)美味猕猴桃(6x)×美味猕猴桃(6x)A.chinensis var.delicious(6x)×A.chinensis var.delicious(6x)中华猕猴桃(4x)×黑蕊猕猴桃(4x)A.chinensis(4x)×A.melanandra(4x)软枣猕猴桃(4x)×软枣猕猴桃(4x)A.arguta(4x)×A.arguta(4x)SNP果实性状Fruit traits性别性状Sex traits[76]
溃疡病可能是由数量性状引起[64],而数量性状通常会被多个基因共同作用,且易受到环境因素的影响,遗传情况复杂,因而数量性状的研究非常困难。随着分子标记技术的发展,利用遗传标记和QTL间的遗传连锁现象,可以确定QTL在染色体上的位置和效应,从而提高育种效率、加快新品种选育。
5 溃疡病抗性关键基因挖掘
当植物受到病原菌的感染时,会激发植物的天然防御系统,使植物产生抗病反应。植物的天然免疫系统可分为两个水平,第一是通过植物细胞表面的模式识别受体(Pattern Recognition Receptors,PRRs)识别病原微生物保守成分(Pathogen Associated Molecular Patterns,PAMPs),从而激活与病原相关分子模式成分触发的免疫反应(PAMP-Triggered-Immunity,PTI)[77],第二是病原微生物释放的效应因子触发的免疫反应(Effector-Triggered-Immunity,ETI)[78]。植物强大的免疫系统能抵御大多数病原微生物的侵染,但是丁香假单胞杆菌等部分病菌能向寄主植物细胞内注入毒力蛋白,抑制植物免疫力,从而引起毁灭性病害[79]。核苷酸结合富亮氨酸重复受体(nucleotide-binding and leucine-rich repeat receptors,NLRs)是植物最大的免疫受体家族[80],来自拟南芥和本氏烟草的NLR 蛋白ZAR1 能识别HopZ5 并触发细胞死亡[81]。目前,已鉴定出多个识别Psa效应因子的NLR 蛋白,如RPA1[82]、NbPTR1[83]等,这些蛋白在植物抗病反应中具有关键作用。
NHL(NDR1/HIN1-like)基因家族成员在抵御丁香假单胞杆菌的侵染时具有积极作用。如拟南芥NHL 基因家族的成员NHL2 和NHL3 在抵御丁香假单胞杆菌侵染中发挥重要作用[84];在采用不同丁香假单胞杆菌处理拟南芥时,发现NHL3 基因的表达量均显著增加,且过表达该基因增强了植株对Pseudomonas syringae pv.tomato DC3000(Pst)病菌的抗性[85]。NHL基因在其他病原菌防御反应中也发挥了重要作用。如拟南芥NHL10 基因能调控黄瓜对花叶病毒(CMV)的超敏反应[86];在马铃薯中过表达NHL 基因成员StPOTHR1 增强了植株对疫霉病(Phytophthora infestans)的抗性[87]。
利用转录组学的方法对Psa侵染后猕猴桃中的基因表达进行分析,发现免疫系统PTI、ETI、HR 中多个抗性基因的表达受到诱导,它们可能通过调整代谢过程,并改变次级代谢产物的产生以抑制Psa的生长[88]。研究发现,在受到Psa侵染后,高抗品种华特中编码Pti1 和RPS2 的效应受体及参与水杨酸信号通路的NPR1、TGA 和PR-1 基因均显著上调表达[89]。PR-1基因能在溃疡病菌诱导下显著表达,且其过表达能增强烟草对溃疡病的抗性[90]。AcTGA07基因的过表达显著增强了猕猴桃的抗性,且TGA转录因子能特异性地结合PR 基因的启动子区域并与NPR 蛋白相互作用[91]。NPR1 同源基因AeNPR1a在烟草中过表达后,相比与野生型株系其Psa 和Pst感染症状明显减轻,转基因烟草抗性显著增强;与接种Pst 的拟南芥npr1-1 突变体植株相比,NPR1a回补株系抗性显著增强[92]。使用壳聚糖处理可诱导猕猴桃防御相关基因(PR1、PR5)表达,使其产生系统获得性抗性(systemic acquired resistance,SAR)[93]。
6 展 望
自1984 年在日本首次发现猕猴桃细菌性溃疡病以来,世界各产区深受其害。经过多年的研究积累,对该病害的发病规律与传播途径、致病菌Psa的致病机制、不同种类或品种猕猴桃的抗病特性等研究均已取得一定成果,但一直未有有效的防治方法。因此,在今后的生产实践中应选择抗性强的品种、加强苗木或花粉等媒介物的检疫监测、提高果园栽培管理农艺措施,这对早期病害预防和减少种植者的经济损失具有重要作用。
中国猕猴桃种质资源丰富,倍性多样,充分利用这些宝贵资源进行抗病新品种培育具有重要意义,但传统实生选种或杂交育种不仅耗时长而且目标性状聚合困难,后代易出现性状分离。随着现代测序技术的发展,通过QTL定位、转录组等方法,在猕猴桃中已经挖掘出许多与抗溃疡病相关的基因和转录因子等,这不仅为抗病材料的选育提供了重要的基因资源和分子标记,还可在早期进行目标性状的预判和选择,极大地提高育种效率。但是由于猕猴桃遗传背景复杂,分子标记技术还无法很好的应用于育种实践,今后还需加大稳定性较好的分子标记开发力度。
近年来QTL 定位技术在许多果树重要性状鉴定方面的应用取得重大进展并已在猕猴桃树种中实施,猕猴桃定向高效精准育种策略成为可能。但该树种染色体基数大、倍性复杂且杂合度高,因此构建高密度的遗传图谱较为困难,QTL 精细定位实施还很少。今后应针对不同研究目的,将图谱构建与杂交育种工作相结合,构建高层次和高实用价值的遗传连锁图。
随着基因组、转录组、代谢组等各类技术的发展以及猕猴桃多倍体基因组信息的不断完善,可以建立不同倍性组合的远缘杂交新基因导入群体开展QTL 定位及抗性基因发掘工作,从而开发出溃疡病抗性相关分子标记,为猕猴桃抗性新品种培育提供新途径。虽然已报道许多与抗性相关基因,但整体而言对猕猴桃抗病分子机制及调控网络研究存在不足,在抗病关键基因挖掘方面的研究也有待进一步深入。今后应注重抗性基因的功能验证,以及关键基因在调控网络中的作用,结合免疫途径、代谢途径等相关反应,解析其抗病分子机制。
[1] 黄宏文. 中国猕猴桃种质资源[M]. 北京:中国林业出版社,2013:29-37.HUANG Hongwen.Actinidia germplasm resources in China[M].Beijing:China Forestry Publishing House,2013:29-37.
[2] 齐秀娟,郭丹丹,王然,钟云鹏,方金豹.我国猕猴桃产业发展现状及对策建议[J].果树学报,2020,37(5):754-763.QI Xiujuan,GUO Dandan,WANG Ran,ZHONG Yunpeng,FANG Jinbao. Development status and suggestions on Chinese kiwifruit industry[J]. Journal of Fruit Science,2020,37(5):754-763.
[3] MCCANN H C,LI L,LIU Y F,LI D W,PAN H,ZHONG C H,RIKKERINK E H A,TEMPLETON M D,STRAUB C,COLOMBI E,RAINEY P B,HUANG H W. Origin and evolution of the kiwifruit canker pandemic[J]. Genome Biology and Evolution,2017,9(4):932-944.
[4] CAMERON A,SAROJINI V. Pseudomonas syringae pv. actinidiae:Chemical control,resistance mechanisms and possible alternatives[J].Plant Pathology,2014,63(1):1-11.
[5] CHAPMAN J R,TAYLOR R K,WEIR B S,ROMBERG M K,VANNESTE J L,LUCK J,ALEXANDER B J R. Phylogenetic relationships among global populations of Pseudomonas syringae pv. actinidiae[J]. Phytopathology,2012,102(11):1034-1044.
[6] SCORTICHINI M.Occurrence of Pseudomonas syringae pv.actinidiae on kiwifruit in Italy[J]. Plant Pathology,1994,43(6):1035-1038.
[7] KOH Y J,KIM G H,JUNG J S,LEE Y S,HUR J S. Outbreak of bacterial canker on Hort16A (Actinidia chinensis Planchon)caused by Pseudomonas syringae pv. actinidiae in Korea[J].New Zealand Journal of Crop and Horticultural Science,2010,38(4):275-282.
[8] VANNESTE J L. The scientific,economic,and social impacts of the New Zealand outbreak of bacterial canker of kiwifruit(Pseudomonas syringae pv. actinidiae)[J]. Annual Review of Phytopathology,2017,55:377-399.
[9] BUTLER M I,STOCKWELL P A,BLACK M A,DAY R C,LAMONT I L,POULTER R T M. Pseudomonas syringae pv.actinidiae from recent outbreaks of kiwifruit bacterial canker belong to different clones that originated in China[J]. PLoS One,2013,8(2):e57464.
[10] 王涛,张计育,王刚,贾展慧,潘德林,郭忠仁.猕猴桃细菌性溃疡病研究进展[J].中国农学通报,2020,36(3):123-128.WANG Tao,ZHANG Jiyu,WANG Gang,JIA Zhanhui,PAN Delin,GUO Zhongren. Advances in kiwifruit bacterial canker[J].Chinese Agricultural Science Bulletin,2020,36(3):123-128.
[11] 秦虎强,高小宁,赵志博,朱穗层,李建民,黄丽丽.陕西猕猴桃细菌性溃疡病田间发生动态和规律[J]. 植物保护学报,2013,40(3):225-230.QIN Huqiang,GAO Xiaoning,ZHAO Zhibo,ZHU Suiceng,LI Jianmin,HUANG Lili. The prevalence dynamics and rules of bacterial canker of kiwi fruit in Shaanxi[J].Journal of Plant Protection,2013,40(3):225-230.
[12] 钟彩虹,李黎,潘慧,邓蕾,陈美艳.猕猴桃细菌性溃疡病的发生规律及综合防治技术[J].中国果树,2020(1):9-13.ZHONG Caihong,LI Li,PAN Hui,DENG Lei,CHEN Meiyan.Occurrence rule and comprehensive control of kiwifruit bacterial canker disease[J].China Fruits,2020(1):9-13.
[13] 李黎,钟彩虹,李大卫,张胜菊,黄宏文.猕猴桃细菌性溃疡病的研究进展[J].华中农业大学学报,2013,32(5):124-133.LI Li,ZHONG Caihong,LI Dawei,ZHANG Shengju,HUANG Hongwen.Research progress on bacterial canker disease of kiwifruit[J]. Journal of Huazhong Agricultural University,2013,32(5):124-133.
[14] 李亚巍,王小洁,吴群,李士谣,李双荣,朱立武,刘普.猕猴桃溃疡病菌的田间分布及其传播规律[J].生物学杂志,2019,36(2):46-50.LI Yawei,WANG Xiaojie,WU Qun,LI Shiyao,LI Shuangrong,ZHU Liwu,LIU Pu. The field distribution and prevalence rules of Pseudomonas syringae pv. actinidiae in kiwifruit[J]. Journal of Biology,2019,36(2):46-50.
[15] VANNESTE J L,YU J,CORNISH D A,TANNER D J,WINDNER R,CHAPMAN J R,TAYLOR R K,MACKAY J F,DOWLUT S.Identification,virulence,and distribution of two biovars of Pseudomonas syringae pv.actinidiae in New Zealand[J].Plant Disease,2013,97(6):708-719.
[16] 王发明,齐贝贝,叶开玉,龚弘娟,莫权辉,蒋桥生,刘平平,李洁维.九个中华类红肉猕猴桃品种的亲缘关系及其溃疡病抗性分析[J].分子植物育种,2021,19(1):193-199.WANG Faming,QI Beibei,YE Kaiyu,GONG Hongjuan,MO Quanhui,JIANG Qiaosheng,LIU Pingping,LI Jiewei. The genetic relationship of nine red-fleshed kiwifruit cultivars(Actinidia chinensis) and their resistance to Pseudomonas syringae pv.actinidiae[J].Molecular Plant Breeding,2021,19(1):193-199.
[17] SCORTICHINI M,MARCELLETTI S,FERRANTE P,PETRICCIONE M,FIRRAO G. Pseudomonas syringae pv. actinidiae:A re-emerging,multi-faceted,pandemic pathogen[J].Molecular Plant Pathology,2012,13(7):631-640.
[18] DONATI I,MAURI S,BURIANI G,CELLINI A,SPINELLI F.Role of Metcalfa pruinosa as a vector for Pseudomonas syringae pv. actinidiae[J]. Plant Pathology Journal,2017,33(6):554-560.
[19] EMILIO S,DAVIDE G. Dissemination of Pseudomonas syringae pv. actinidiae through pollen and its epiphytic life on leaves and fruits[J]. Phytopathologia Mediterranea,2011,50(3):489-496.
[20] LIU P,XUE S Z,HE R,HU J Y,WANG X J,JIA B,GALLIPOLI L,MAZZAGLIA A,BALESTRA G M,ZHU L W. Pseudomonas syringae pv. actinidiae isolated from non-kiwifruit plant species in China[J].European Journal of Plant Pathology,2016,145(4):743-754.
[21] JAYARAMAN J,YOON M,HEMARA L M,BOHNE D,TAHIR J,CHEN R K Y,BRENDOLISE C,RIKKERINK E H A,TEMPLETON M D.Contrasting effector profiles between bacterial colonisers of kiwifruit reveal redundant roles converging on PTI-suppression and RIN4[J]. New Phytologist,2023,238(4):1605-1619.
[22] COLOMBI E,BERTELS F,DOULCIER G,MCCONNELL E,PICHUGINA T,SOHN K H,STRAUB C,MCCANN H C,RAINEY P B. Rapid dissemination of host metabolism-manipulating genes via integrative and conjugative elements[J]. Proceedings of the National Academy of Sciences of the United States of America,2024,121(11):e2309263121.
[23] BOTELHO J,SCHULENBURG H. The role of integrative and conjugative elements in antibiotic resistance evolution[J].Trends in Microbiology,2021,29(1):8-18.
[24] MCCANN H C,RIKKERINK E H A,BERTELS F,FIERS M,LU A,REES-GEORGE J,ANDERSEN M T,GLEAVE A P,HAUBOLD B,WOHLERS M W,GUTTMAN D S,WANG P W,STRAUB C,VANNESTE J L,RAINEY P B,TEMPLETON M D. Genomic analysis of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease[J]. PLoS Pathogens,2013,9(7):e1003503.
[25] SAWADA H,SUZUKI F,MATSUDA I,SAITOU N. Phylogenetic analysis of Pseudomonas syringae pathovars suggests the horizontal gene transfer of argK and the evolutionary stability of hrp gene cluster[J]. Journal of Molecular Evolution,1999,49(5):627-644.
[26] HAN H S,KOH Y J,HUR J S,JUNG J S. Identification and characterization of coronatine-producing Pseudomonas syringae pv. actinidiae[J]. Journal of Microbiology and Biotechnology,2003,13(1):110-118.
[27] VANNESTE J L. Pseudomonas syringae pv. actinidiae (Psa):A threat to the New Zealand and global kiwifruit industry[J]. New Zealand Journal of Crop and Horticultural Science,2012,40(4):265-267.
[28] CUNTY A,POLIAKOFF F,RIVOAL C,CESBRON S,SAUX M F,LEMAIRE C,JACQUES M,MANCEAU C,VANNESTE J. Characterization of Pseudomonas syringae pv. actinidiae(Psa) isolated from France and assignment of Psa biovar 4 to a de novo pathovar:Pseudomonas syringae pv. actinidifoliorum pv.nov[J].Plant Pathology,2015,64:582-596.
[29] FUJIKAWA T,SAWADA H. Genome analysis of the kiwifruit canker pathogen Pseudomonas syringae pv. actinidiae biovar 5[J].Scientific Reports,2016,6:21399.
[30] FUJIKAWA T,SAWADA H. Genome analysis of Pseudomonas syringae pv. actinidiae biovar 6,which produces the phytotoxins,phaseolotoxin and coronatine[J]. Scientific Reports,2019,9(1):3836.
[31] COLOMBI E,STRAUB C,KÜNZEL S,TEMPLETON M D,MCCANN H C,RAINEY P B. Evolution of copper resistance in the kiwifruit pathogen Pseudomonas syringae pv. actinidiae through acquisition of integrative conjugative elements and plasmids[J].Environmental Microbiology,2017,19(2):819-832.
[32] 石志军,张慧琴,肖金平,杨鲁琼,孙志伟,谢鸣,马远.不同猕猴桃品种对溃疡病抗性的评价[J]. 浙江农业学报,2014,26(3):752-759.SHI Zhijun,ZHANG Huiqin,XIAO Jinping,YANG Luqiong,SUN Zhiwei,XIE Ming,MA Yuan.The resistance evaluation of different kiwifruit varieties to canker[J].Acta Agriculturae Zhejiangensis,2014,26(3):752-759.
[33] 宋雅林,林苗苗,钟云鹏,陈锦永,齐秀娟,孙雷明,方金豹.猕猴桃品种(系)溃疡病抗性鉴定及不同评价指标的相关性分析[J].果树学报,2020,37(6):900-908.SONG Yalin,LIN Miaomiao,ZHONG Yunpeng,CHEN Jinyong,QI Xiujuan,SUN Leiming,FANG Jinbao. Evaluation of resistance of kiwifruit varieties (line) against bacterial canker disease and correlation analysis among evaluation indexes[J].Journal of Fruit Science,2020,37(6):900-908.
[34] 温欣. 软枣猕猴桃种质资源溃疡病抗性评价及抗性生理研究[D].北京:中国农业科学院,2020.WEN Xin. Evaluation of resistance to Pseudomonas syringae pv.actinidiae and study on resistance physiology of Actinidia arguta germplasm resources[D].Beijing:Chinese Academy of Agricultural Sciences,2020.
[35] 裴艳刚,马利,岁立云,崔永亮,刘晓敏,龚国淑.不同猕猴桃品种对溃疡病菌的抗性评价及其利用[J]. 果树学报,2021,38(7):1153-1162.PEI Yangang,MA Li,SUI Liyun,CUI Yongliang,LIU Xiaomin,GONG Guoshu.Resistance evaluation and utilization of different kiwifruit cultivars to Pseudomonas syringae pv. actinidiae[J].Journal of Fruit Science,2021,38(7):1153-1162.
[36] 郑丽,夏文娟,陈奇,袁炎良,陈沙,尹海军,柳威.抗溃疡病猕猴桃新品种“先沃五号”的选育[J]. 中国南方果树,2023,52(1):160-162.ZHENG Li,XIA Wenjuan,CHEN Qi,YUAN Yanliang,CHEN Sha,YIN Haijun,LIU Wei. Selecting of canker resistant kiwifruit variety“Xianwo No. 5”[J]. South China Fruits,2023,52(1):160-162.
[37] 廖慧苹,谢玥,周英丹,刘瑶,胥伟秋,李明章.抗溃疡病优质猕猴桃新品种‘华金3 号’[J].园艺学报,2024,51(增刊1):51-52.LIAO Huiping,XIE Yue,ZHOU Yingdan,LIU Yao,XU Weiqiu,LI Mingzhang.A new yellow-fleshed kiwifruit cultivar‘Huajin 3’[J].Acta Horticulturae Sinica,2024,51(Suppl.1):51-52.
[38] 罗双,韩倩容,陈言,戢小梅,刘杰,米绪凯,程运江,邓秀新,蔡礼鸿,张雅娟,曾云流.丰产、抗病加工型猕猴桃新品种‘金塘一号’[J].园艺学报,2023,50(增刊2):23-24.LUO Shuang,HAN Qianrong,CHEN Yan,JI Xiaomei,LIU Jie,MI Xukai,CHENG Yunjiang,DENG Xiuxin,CAI Lihong,ZHANG Yajuan,ZENG Yunliu.Breeding of a new processed kiwifruit cultivar‘Jintang 1’with high yield and high disease resistance[J]. Acta Horticulturae Sinica,2023,50(Suppl. 2):23-24.
[39] 张小桐.猕猴桃对溃疡病抗性评价指标的研究[D].合肥:安徽农业大学,2007.ZHANG Xiaotong. Study on the resistance indexes of kiwifruit to Pseudomonas syringae pv. actinidiae[D]. Hefei:Anhui Agricultural University,2007.
[40] 贺占雪,李欣,朱太富,苏效兰,王连春.野生猕猴桃枝叶组织结构与抗溃疡病的关系分析[J].中国植保导刊,2023,43(10):9-14.HE Zhanxue,LI Xin,ZHU Taifu,SU Xiaolan,WANG Lianchun.Analysis of the relationship between the resistance of wild kiwifruit to canker and the tissue structure of its branches and leaves[J].China Plant Protection,2023,43(10):9-14.
[41] 李淼,檀根甲,李瑶,承河元,周子燕.猕猴桃品种中糖分及木质素含量与抗溃疡病的关系[J].植物保护学报,2005,32(2):138-142.LI Miao,TAN Genjia,LI Yao,CHENG Heyuan,ZHOU Ziyan.Relationship between contents of lignin and soluble sugar in plants of kiwifruit cultivars and their resistance to kiwifruit bacterial canker infected by Pseudomonas syringae pv. actinidiae[J].Journal of Plant Protection,2005,32(2):138-142.
[42] 李聪. 猕猴桃枝叶组织结构及内含物与溃疡病的相关性研究[D].杨凌:西北农林科技大学,2016.LI Cong. Correlation of the relationship between resistance of branch leaves structure and inclusion and kiwifruit canker[D].Yangling:Northwest A&F University,2016.
[43] WANG Y,TAN Z C,ZHEN X,LIANG Y Y,GAO J Y,ZHAO Y H,LIU S B,ZHA M R. Contribution of sucrose metabolism in phloem to kiwifruit bacterial canker resistance[J]. Plants,2023,12(4):918.
[44] 贺占雪,朱太富,李欣,苏效兰,刘惠民,王连春.不同砧穗组合对猕猴桃溃疡病的抗性差异及机制分析[J].河南农业科学,2023,52(1):95-107.HE Zhanxue,ZHU Taifu,LI Xin,SU Xiaolan,LIU Huimin,WANG Lianchun.Analysis of differences and mechanism of resistance to canker among different kiwifruit rootstock- scion combinations[J]. Journal of Henan Agricultural Sciences,2023,52(1):95-107.
[45] 刘明,王继华,王同昌.DNA 分子标记技术[J].东北林业大学学报,2003,31(6):65-67.LIU Ming,WANG Jihua,WANG Tongchang. DNA molecular markers[J]. Journal of Northeast Forestry University,2003,31(6):65-67.
[46] 董晓莉,汤浩茹,甘玲,李明章.DNA 分子标记在猕猴桃上的应用[J].果树学报,2005,22(6):682-686.DONG Xiaoli,TANG Haoru,GAN Ling,LI Mingzhang. Advances in research on application of DNA molecular markers in Actinidia[J].Journal of Fruit Science,2005,22(6):682-686.
[47] 王玉杰,冷春旭,孙中义,王珣,赵曦,李晓娟,赵伟,吴立成.分子标记技术在农作物种子检测中的应用[J].中国种业,2022(3):38-40.WANG Yujie,LENG Chunxu,SUN Zhongyi,WANG Xun,ZHAO Xi,LI Xiaojuan,ZHAO Wei,WU Licheng.Application of molecular marker technology in crop seed detection[J].China Seed Industry,2022(3):38-40.
[48] 刘念析,陈亮,厉志,刘宝泉,刘佳,衣志刚,董志敏,王曙明.大豆抗病分子标记的研究进展[J].作物杂志,2019(4):10-16.LIU Nianxi,CHEN Liang,LI Zhi,LIU Baoquan,LIU Jia,YI Zhigang,DONG Zhimin,WANG Shuming.Advances in molecular markers of soybean disease resistance[J]. Crops,2019(4):10-16.
[49] 邓世峰,王先如,张安存,陈次娥,吴明.分子标记辅助选择在我国水稻抗病育种中的研究进展[J].江西农业,2019(22):40.DENG Shifeng,WANG Xianru,ZHANG Ancun,CHEN Cie,WU Ming. Research progress of molecular marker-assisted selection in rice disease resistance breeding in China[J]. Jiangxi Agriculture,2019(22):40.
[50] 杨再俊,郑家瑞,潘鹏程,潘寅涛,高彬,李云洲.番茄抗病种质资源分子标记筛选[J].山地农业生物学报,2021,40(6):30-36.YANG Zaijun,ZHENG Jiarui,PAN Pengcheng,PAN Yintao,GAO Bin,LI Yunzhou. Molecular marker detection of resistance genes in tomato germplasm[J]. Journal of Mountain Agriculture and Biology,2021,40(6):30-36.
[51] 张桂华,韩毅科,孙小红,李淑菊,魏爱民,杜胜利.与黄瓜抗黑星病基因连锁的分子标记研究[J]. 中国农业科学,2006,39(11):2250-2254.ZHANG Guihua,HAN Yike,SUN Xiaohong,LI Shuju,WEI Aimin,DU Shengli. Molecular marker linked to the resistant gene of cucumber scab[J]. Scientia Agricultura Sinica,2006,39(11):2250-2254.
[52] 刘娟.猕猴桃溃疡病抗性材料评价及其亲缘关系的ISSR 聚类分析[D].雅安:四川农业大学,2015.LIUJuan.Evaluationofresistantvarietiesonkiwifruitbacterialcanker and cluster analysis of genetic relations by ISSR markers[D].Yaan:Sichuan Agricultural University,2015.
[53] 易盼盼,樊红科,雷玉山,王飞.猕猴桃抗溃疡病基因连锁SSR分子标记初步研究[J].西北农林科技大学学报(自然科学版),2015,43(4):91-98.YI Panpan,FAN Hongke,LEI Yushan,WANG Fei. Priliminary study on SSR marker of gene linkage against Pseudomonas syringae pv. actinidiae[J]. Journal of Northwest A& F University(Natural Science Edition),2015,43(4):91-98.
[54] 李淼,檀根甲,李瑶,丁克坚,产祝龙,承河元.不同猕猴桃品种RAPD 分析及其与抗溃疡病的关系[J].植物保护,2009,35(3):41-46.LI Miao,TAN Genjia,LI Yao,DING Kejian,CHAN Zhulong,CHENG Heyuan.Analysis of the relationships between different kiwifruit cultivars and their resistance to Pseudomonas syringae pv.actinidiae by RAPD[J].Plant Protection,2009,35(3):41-46.
[55] FRASER L G,DATSON P M,TSANG G K,MANAKO K I,RIKKERINK E H,MCNEILAGE M A.Characterisation,evolutionary trends and mapping of putative resistance and defence genes in Actinidia (kiwifruit)[J]. Tree Genetics & Genomes,2015,11(2):21.
[56] 王倩,王斌.DNA 分子标记在果树遗传学研究上的应用[J].遗传,2000,22(5):339-344.WANG Qian,WANG Bin. The application of DNA molecular markers in fruit tree genetics[J]. Hereditas(Beijing),2000,22(5):339-344.
[57] 王中堂.枣高密度遗传连锁图谱构建与农艺性状QTL 定位[D].杨凌:西北农林科技大学,2020.WANG Zhongtang. High-density genetic map construction and QTL mapping of agronomic traits of Ziziphus jujuba Mill.[D].Yangling:Northwest A&F University,2020.
[58] 李红莲,梁英海,王珊珊,赵晨辉,张冰冰,宋宏伟.苹果抗病性状QTL 的研究进展[J].分子植物育种,2022,20(17):5741-5746.LI Honglian,LIANG Yinghai,WANG Shanshan,ZHAO Chenhui,ZHANG Bingbing,SONG Hongwei. Research progress on QTL related to apple disease resistance traits[J].Molecular Plant Breeding,2022,20(17):5741-5746.
[59] LI P,TAN X B,LIU R T,RAHMAN F U,JIANG J F,SUN L,FAN X C,LIU J H,LIU C H,ZHANG Y. QTL detection and candidate gene analysis of grape white rot resistance by interspecific grape (Vitis vinifera L. × Vitis davidii Foex.) crossing[J].Horticulture Research,2023,10(5):uhad063.
[60] 杨宏宾.柑橘果面蜡质合成关键基因挖掘及重要采后性状的QTL 定位[D].武汉:华中农业大学,2021.YANG Hongbin. The mining of key genes involved in cuticular wax synthesis and QTL mapping of the important postharvest traits in citrus fruit[D].Wuhan:Huazhong Agricultural University,2021.
[61] QI X L,DONG Y X,LIU C L,SONG L L,CHEN L,LI M.A 5.2-kb insertion in the coding sequence of PavSCPL,a serine carboxypeptidase-like enhances fruit firmness in Prunus avium[J].Plant Biotechnology Journal,2024,22(6):1622-1635.
[62] 王龙,王苏珂,薛华柏,苏艳丽,杨健,李秀根.梨栽培相关性状的QTL 分析[J].果树学报,2018,35(增刊1):61-65.WANG Long,WANG Suke,XUE Huabai,SU Yanli,YANG Jian,LI Xiugen.QTL analysis for cultivated traits in pears[J].Journal of Fruit Science,2018,35(Suppl.1):61-65.
[63] TESTOLIN R,HUANG W G,LAIN O,MESSINA R,VECCHIONE A,CIPRIANI G.A kiwifruit (Actinidia spp.) linkage map based on microsatellites and integrated with AFLP markers[J].Theoretical and Applied Genetics,2001,103(1):30-36.
[64] TAHIR J,HOYTE S,BASSETT H,BRENDOLISE C,CHATTERJEE A,TEMPLETON K,DENG C,CROWHURST R,MONTEFIORI M,MORGAN E,WOTTON A,FUNNELL K,WIEDOW C,KNAEBEL M,HEDDERLEY D,VANNESTE J,MCCALLUM J,HOEATA K,NATH A,CHAGNÉ D,GEA L,GARDINER S E.Multiple quantitative trait loci contribute to resistance to bacterial canker incited by Pseudomonas syringae pv. actinidiae in kiwifruit (Actinidia chinensis)[J]. Horticulture Research,2019,6:101.
[65] TAHIR J,BRENDOLISE C,HOYTE S,LUCAS M,THOMSON S,HOEATA K,MCKENZIE C,WOTTON A,FUNNELL K,MORGAN E,HEDDERLEY D,CHAGNÉ D,BOURKE P M,MCCALLUM J,GARDINER S E,GEA L. QTL mapping for resistance to cankers induced by Pseudomonas syringae pv.actinidiae (Psa) in a tetraploid Actinidia chinensis kiwifruit population[J].Pathogens,2020,9(11):967.
[66] YOUNG N D. QTL mapping and quantitative disease resistance in plants[J]. Annual Review of Phytopathology,1996,34:479-501.
[67] QUENOUILLE J,PAULHIAC E,MOURY B,PALLOIX A.Quantitative trait loci from the host genetic background modulate the durability of a resistance gene:A rational basis for sustainable resistance breeding in plants[J].Heredity,2014,112(6):579-587.
[68] FRASER L G,TSANG G K,DATSON P M,DE SILVA H N,HARVEY C F,GILL G P,CROWHURST R N,MCNEILAGE M A.A gene-rich linkage map in the dioecious species Actinidia chinensis (kiwifruit) reveals putative X/Y sex-determining chromosomes[J].BMC Genomics,2009,10:102.
[69] HUANG S X,DING J,DENG D J,TANG W,SUN H H,LIU D Y,ZHANG L,NIU X L,ZHANG X,MENG M,YU J D,LIU J,HAN Y,SHI W,ZHANG D F,CAO S Q,WEI Z J,CUI Y L,XIA Y H,ZENG H P,BAO K,LIN L,MIN Y,ZHANG H,MIAO M,TANG X F,ZHU Y Y,SUI Y,LI G W,SUN H J,YUE J Y,SUN J Q,LIU F F,ZHOU L Q,LEI L,ZHENG X Q,LIU M,HUANG L,SONG J,XU C H,LI J W,YE K Y,ZHONG S L,LU B R,HE G H,XIAO F M,WANG H L,ZHENG H K,FEI Z J,LIU Y S. Draft genome of the kiwifruit Actinidia chinensis[J].Nature Communications,2013,4:2640.
[70] SCAGLIONE D,FORNASIERO A,PINTO C,CATTONARO F,SPADOTTO A,INFANTE R,MENESES C,MESSINA R,LAIN O,CIPRIANI G,TESTOLIN R. A RAD-based linkage map of kiwifruit (Actinidia chinensis Pl.) as a tool to improve the genome assembly and to scan the genomic region of the gender determinant for the marker-assisted breeding[J].Tree Genetics&Genomes,2015,11(6):115.
[71] ZHANG Q,LIU C Y,LIU Y F,VANBUREN R,YAO X H,ZHONG C H,HUANG H W. High-density interspecific genetic maps of kiwifruit and the identification of sex-specific markers[J].DNA Research,2015,22(5):367-375.
[72] 刘春燕. 猕猴桃种间高密度遗传图谱的构建及果实性状QTLs 定位[D]. 武汉:中国科学院研究生院(武汉植物园),2016.LIU Chunyan. Construction of high-density interspecific genetic maps and identification of QTLs for fruits in kiwifruit[D]. Wuhan:Wuhan Botanical Garden,Chinese Academy of Sciences,2016.
[73] LIU C Y,LI D W,ZHOU J H,ZHANG Q,TIAN H,YAO X H.Construction of a SNP-based genetic linkage map for kiwifruit using next-generation restriction-site-associated DNA sequencing(RADseq)[J].Molecular Breeding,2017,37(11):139.
[74] POPOWSKI E,THOMSON S J,KNÄBEL M,TAHIR J,CROWHURST R N,DAVY M,FOSTER T M,SCHAFFER R J,TUSTIN D S,ALLAN A C,MCCALLUM J,CHAGNÉ D.Construction of a high-density genetic map for hexaploid kiwifruit (Actinidia chinensis var. deliciosa) using genotyping by sequencing[J].G3,2021,11(7):jkab142.
[75] MACNEE N,HILARIO E,TAHIR J,CURRIE A,WARREN B,REBSTOCK R,HALLETT I C,CHAGNÉ D,SCHAFFER R J,BULLEY S M. Peridermal fruit skin formation in Actinidia sp.(kiwifruit) is associated with genetic loci controlling russeting and cuticle formation[J].BMC Plant Biology,2021,21(1):334.
[76] WANG R,XING S Y,BOURKE P M,QI X Q,LIN M M,ESSELINK D,ARENS P,VOORRIPS R E,VISSER R G F,SUN L M,ZHONG Y P,GU H,LI Y K,LI S K,MALIEPAARD C,FANG J B. Development of a 135K SNP genotyping array for Actinidia arguta and its applications for genetic mapping and QTL analysis in kiwifruit[J].Plant Biotechnology Journal,2023,21(2):369-380.
[77] JONES J D G,DANGL J L. The plant immune system[J]. Nature,2006,444(7117):323-329.
[78] 汪巧.植物抗病机理研究进展综述[J].安徽农学通报,2015,21(8):24-30.WANG Qiao. Advances on the mechanism of plant disease resistance[J].Anhui Agricultural Science Bulletin,2015,21(8):24-30.
[79] NOMURA K,DEBROY S,LEE Y H,PUMPLIN N,JONES J,HE S Y.A bacterial virulence protein suppresses host innate immunity to cuse plant disease[J].Science,313(5784):220-223.
[80] KOURELIS J,VAN DER HOORN R A L. Defended to the nines:25 years of resistance gene cloning identifies nine mechanisms for R protein function[J]. The Plant Cell,2018,30(2):285-299.
[81] ZHENG X J,ZHOU Z Y,GONG Z,HU M J,AHN Y J,ZHANG X J,ZHAO Y,GONG G S,ZHANG J,ZUO J R,HAN G Z,HOON S K,ZHOU J M. Two plant NLR proteins confer strain-specific resistance conditioned by an effector from Pseudomonas syringae pv. actinidiae[J]. Journal of Genetics and Genomics,2022,49(8):823-832.
[82] YOON M,RIKKERINK E H A. Rpa1 mediates an immune response to avrRpm1Psa and confers resistance against Pseudomonas syringae pv. actinidiae[J]. The Plant Journal,2020,102(4):688-702.
[83] YEH S M,YOON M,SCOTT S,CHATTERJEE A,HEMARA L M,CHEN R K Y,WANG T C,TEMPLETON K,RIKKERINK E H A,JAYARAMAN J,BRENDOLISE C. NbPTR1 confers resistance against Pseudomonas syringae pv. actinidiae in kiwifruit[J]. Plant,Cell & Environment,2024,47(11):4101-4115.
[84] DÖRMANN P,GOPALAN S,HE S Y,BENNING C. A gene family in Arabidopsis thaliana with sequence similarity to NDR1 and HIN1[J]. Plant Physiology and Biochemistry,2000,38(10):789-796.
[85] VARET A,PARKER J,TORNERO P,NASS N,NÜRNBERGER T,DANGL J L,SCHEEL D,LEE J. NHL25 and NHL3 two NDR1/HIN1-1ike genes in Arabidopsis thaliana with potential role(s) in plant defense[J]. Molecular Plant-Microbe Interactions,2002,15(6):608-616.
[86] ZHENG M S,TAKAHASHI H,MIYAZAKI A,HAMAMOTO H,SHAH J,YAMAGUCHI I,KUSANO T. Up-regulation of Arabidopsis thaliana NHL10 in the hypersensitive response to Cucumber mosaic virus infection and in senescing leaves is controlled by signalling pathways that differ in salicylate involvement[J].Planta,2004,218(5):740-750.
[87] CHEN Q S,TIAN Z D,JIANG R,ZHENG X A,XIE C H,LIU J. StPOTHR1,a NDR1/HIN1-like gene in Solanum tuberosum,enhances resistance against Phytophthora infestans[J].Biochemical and Biophysical Research Communications,2018,496(4):1155-1161.
[88] WANG T,WANG G,JIA Z H,PAN D L,ZHANG J Y,GUO Z R.Transcriptome analysis of kiwifruit in response to Pseudomonas syringae pv. actinidiae infection[J]. International Journal of Molecular Sciences,2018,19(2):373.
[89] SONG Y L,SUN L M,LIN M M,CHEN J Y,QI X J,HU C G,FANG J B. Comparative transcriptome analysis of resistant and susceptible kiwifruits in response to Pseudomonas syringae pv.actinidiae during early infection[J]. PLoS One,2019,14(2):e0211913.
[90] 张敏,宋雅林,林苗苗,王然,李玉阔,孙艳香,方金豹,苏彦苹,孙雷明,齐秀娟.猕猴桃病程相关蛋白PR-1 基因的克隆和功能分析[J].果树学报,2024,41(8):1524-1533.ZHANG Min,SONG Yalin,LIN Miaomiao,WANG Ran,LI Yukuo,SUN Yanxiang,FANG Jinbao,SU Yanping,SUN Leiming,QI Xiujuan.Cloning and function analysis of PR-1 gene in Actinidia[J].Journal of Fruit Science,2024,41(8):1524-1533.
[91] LIU W,ZHAO C,LIU L,HUANG D,MA C,LI R,HUANG L L. Genome-wide identification of the TGA gene family in kiwifruit(Actinidia chinensis spp.)and revealing its roles in response to Pseudomonas syringae pv.actinidiae(Psa)infection[J]. International Journal of Biological Macromolecules,2022,222:101-113.
[92] SUN L M,FANG J B,ZHANG M,QI X J,LIN M M,CHEN J Y.Molecular cloning and functional analysis of the NPR1 homolog in kiwifruit (Actinidia eriantha)[J]. Frontiers in Plant Science,2020,11:551201.
[93] BEATRICE C,LINTHORST J M H,CINZIA F,LUCA R. Enhancement of PR1 and PR5 gene expressions by chitosan treatment in kiwifruit plants inoculated with Pseudomonas syringae pv. actinidiae[J]. European Journal of Plant Pathology,2017,148(1):163-179.