葡萄(Vitis vinifera)是有较高营养价值和经济价值的一种园艺作物[1]。据粮农组织(FAO)统计,2021 年,中国葡萄产量为1 126.99 万t,位居世界第一;种植面积为58.272 8 万hm2,位居世界第四。但近年来中国葡萄产区病毒性病原日益流行,葡萄已知的病毒性病原种类超过100 种[2-4],成为感染病毒种类最多的果树。目前,葡萄感染病毒后一般会出现发芽延迟、节间缩短、叶片畸形、浆果坏死、浆果变酸、花叶、斑驳、脉明、环斑以及木质部凹陷等症状[5-6],严重影响葡萄产业的发展。
灰比诺葡萄病毒(grapevine pinot gris virus,GPGV)是乙型线形病毒科(Betafexiviridae)纤毛病毒属(Trichovirus)的代表成员,基因组为正义单链RNA 分子,编码3 个重叠的开放阅读框(open reading frame,ORF),ORF1 编码RNA 依赖的RNA 聚合酶(RNA-dependent RNA polymerase,RdRp)、ORF2编码运动蛋白(movement protein,MP)、ORF3 编码外壳蛋白(coat protein,CP)。GPGV 所引起的葡萄病害最早始于意大利特伦蒂诺地区葡萄园种植的灰比诺上[1]。但该病毒是在9 a(年)后才被发现,随后世界上大多数主要的葡萄种植区都检测到了GPGV,例如欧洲的法国、德国、俄罗斯、捷克、希腊、斯洛伐克、斯洛文尼亚、土耳其、西班牙和葡萄牙;亚洲的中国、韩国和巴基斯坦;北美洲的美国、加拿大;南美洲的乌拉圭;大洋洲的澳大利亚[7-17]。目前,GPGV在中国葡萄主要种植区较为流行,可造成葡萄叶片斑驳和变形病(grapevine leaf mottling and deformation,GLMD),并且浆果酸度增加,严重影响葡萄以及葡萄酒产业的发展。但是一些几乎无症状的葡萄样本也检测到了GPGV,因此这些症状相关的原因尚不清楚[18]。此外,白花蝇子草(Silene latifolia)和藜(Chenopodium album)是GPGV 的草本寄主[19],葡萄缺节瘿螨(Colomerus newkirk)是GPGV传播的昆虫介体[20]。
近年来,GPGV 的基因组特征、危害症状、地理分布和起源中心受到了广泛关注,较多研究表明GPGV 种群的遗传多样性处于中等水平,且中国也是起源中心的主要指向之一[17]。然而,关于GPGV中国分离物的全基因组序列报道较少。因此,为了深入了解GPGV 的遗传多样性和系统发育关系,利用RT-PCR 和RACE 技术获得了2 条GPGV 的内蒙古分离物(20IM-ViVi1 和20IM-ViVi2),并将其与NCBIGenBank 数据库所下载的其他GPGV 分离物进行了序列一致性分析、系统发育分析、重组分析以及遗传多样性分析,以期为中国GPGV 的防治提供理论基础。
1 材料和方法
1.1 材料
试验材料为前期研究中检测为GPGV阳性的葡萄叶片样品,分别于2020 年6 月8 日和2020 年7 月15 日在内蒙古呼和浩特市周边不同的设施葡萄园采集,并经液氮速冻后存放于-80 ℃冰箱备用。
1.2 总RNA提取
取上述感染GPGV 的葡萄样品100 mg,按照植物总RNA 提取试剂盒(Spectrum™Plant Total RNA Kit)说明书进行总RNA 的提取,通过1%的琼脂糖凝胶电泳和微量分光光度计分别对所提取RNA 的质量和浓度进行检测,并于-80 ℃冰箱保存备用。
1.3 引物设计
利用Vector NTI软件对NCBIGenbank 数据库已报道的所有GPGV 全长基因组序列进行序列比对,在序列保守区设计了3个引物对(GPGV-1F/GPGV-1R、GPGV-2F/GPGV-2R、GPGV-3F/GPGV-3R)用于GPGV 全基因组序列扩增,相邻扩增片段间重叠片段长度均大于200 bp,随后结合上述扩增结果设计了用于扩增GPGV 末端序列的引物(GPGV3、GPGV1)(表1)。引物均由生工生物工程(上海)股份有限公司合成。
表1 本研究所使用的引物信息
Table 1 Primer information used in this study

注:扩增产物所对应的位点以GPGV 国际标准序列(GenBank 登录号:NC_015782)为参考。
Note:The amplified product sites are referenced to the International Standard Sequence of GPGV(GenBank Accession No.:NC_015782).
引物名称Primer name GPGV-1F GPGV-1R GPGV-2F GPGV-2R GPGV-3F GPGV-3R GPGV3 GPGV1序列(5'-3')Sequence(5'-3')GATCAATTGATCCCGTGTAGTG RAAGCACCCAACTTTTGGYAG TGCAGGTGTATGAACARGATG CCAAAGCACACATGTCATCAC GGAGGTTTTTGCATTGAATCAG GAAGTTACGYGCTCMTATGAG CCAAATGCCCTATGTGACTG TGACTTTTCGTCAAGTGTGT退火温度Annealing temperature/℃56产物大小Amplified fragment length/bp 2619位点Site/nt 19~2637 56 2892 2339~5230 56 2314 4911~7224 1~937 6892~7260 56 54 937 369
1.4 GPGV全基因组序列扩增
以提取的总RNA 为模板,采用反转录试剂盒(SuperScript™ⅢReverse Transcriptase)合成cDNA,反应条件:50 ℃,1 h,70 ℃,15 min。以cDNA 为模板,在高保真酶(Q5 High-Fidelity 2×Master Mix)的作用下分段扩增GPGV的核苷酸序列,循环参数为:98 ℃变性30 s,56 ℃退火30 s,72 ℃延伸2 min,35个循环。利用SMARTer® RACE 5'/3' Kit 试剂盒扩增GPGV 5'和3'末端序列。PCR产物通过1%琼脂糖凝胶电泳进行检测,琼脂糖凝胶DNA回收试剂盒回收目的片段。纯化后的产物连接至pTOPO-Blunt克隆载体,并转化JM109大肠杆菌感受态细胞,经PCR进行菌液鉴定后,取适量菌液送至华大基因进行测序,剩余菌液用50%甘油保存,置于-80 ℃冰箱备用。
1.5 GPGV基因组序列分析
采用Vector NTI软件将RT-PCR 扩增以及cDNA 末端快速扩增(RACE)得到的GPGV 基因组序列进行组装,获得GPGV 的完整基因组序列。利用NCBIORF finder(https://www.ncbi.nlm.nih.gov/orffinder)进行ORFs预测,获得GPGV分离物的5'端非编码区(5'-UTR)、3'端非编码区(3'-UTR)和ORFs的序列。利用EMBOSS transeq(https://www.ebi.ac.uk/Tools/st/emboss_transeq/)进行ORFs 的翻译,获得氨基酸序列。利用Mega 11 的ClustalW 方法对GPGV在NCBI数据库中所有的完整基因组序列(152个分离物)进行多重序列比对,并以最大似然法(maximum-likelihood method,ML)构建了系统进化树,依MODLES 程序确定了建树参数,自展值设为1000。利用BioEdit 7.2 软件对完整基因组序列以及ORFs的核苷酸与氨基酸序列进行一致性分析。利用RDP4 软件提供的7 种重组检测算法对得到的分离物全基因组序列进行重组分析。利用DnaSP v.6.12.03 对GPGV 分离物进行群体遗传多样性分析[21]。
2 结果与分析
2.1 GPGV 内蒙古分离物全基因组序列扩增与基因组结构特征
将葡萄叶片样品提出的总RNA 反转录为cDNA 后,通过RACE 技术和RT-PCR 技术扩增出3段重叠的基因组序列以及末端序列(图1)。使用Vector NTI软件将各序列片段进行拼接组装,获得了2条GPGV分离物(20IM-ViVi1和20IM-ViVi2)的完整基因组序列(登录号:OR935780、OR935781)。2条全基因组序列长度均为7250 nt,3'和5'非编码区(UTR)长度为95 nt、82 nt,基因组结构均与已报道的GPGV 基因组结构一致,含3 个重叠的ORFs,ORF1(96~5563 nt,1855 aa)编码了病毒甲基转移酶(methyltransferases,MT)、2OG-Fe Ⅱ-Oxy 加氧酶结构域、病毒RNA 解旋酶以及RNA 依赖型的RNA 聚合酶(RdRp),ORF2(5569~6696 nt,375 aa)和ORF3(6581~7168 nt,195 aa)分别编码了运动蛋白(MP)和外壳蛋白(CP)。
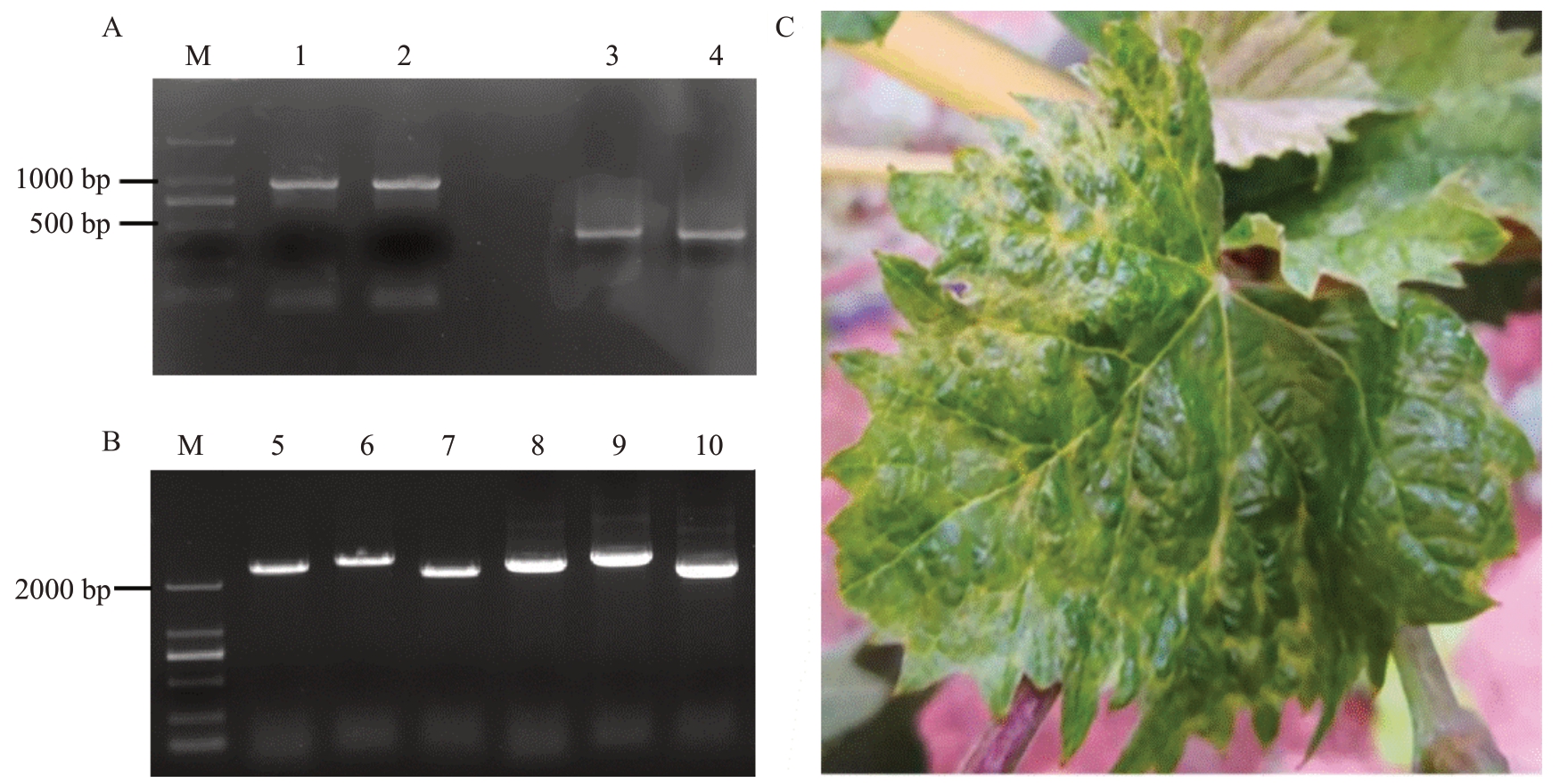
图1 RACE 和RT-PCR 扩增结果及感染GPGV 的葡萄植株田间发病症状
Fig.1 Amplification results of RACE and RT-PCR and field symptomatology of grapevine plants infected with GPGV
A.20IM-ViVi2 的RACE 扩增结果;B.20IM-ViVi2 的RT-PCR 扩增结果;C.感染GPGV 的夏黑葡萄田间发病症状,症状表现为叶片斑驳、皱缩、畸形。M. DL2000marker;1、3、5、6、7 为GPGV 分离物20IM-ViVi1 的RACE 及RT-PCR 扩增结果;2、4、8、9、10 为GPGV 分离物20IM-ViVi2 的RACE 及RT-PCR 扩增结果。
A.The results of the rapid amplification of cDNA ends for 20IM-ViVi1 and 20IM-ViVi2;B.The RT-PCR amplification results for 20IM-ViVi1 and 20IM-ViVi2; C. The field symptoms of Summer Black grapes infected with GPGV, characterized by mottled, wrinkled, and deformed leaves. M.DL2000 marker;Lanes 1,3,5,6,7 show the RACE and RT-PCR amplification results of GPGV isolate 20IM-ViVi1;Lanes 2,4,8,9,10 display the RACE and RT-PCR amplification results for GPGV isolate 20IM-ViVi2.
2.2 GPGV分离物序列一致性分析
将所获得的2 个内蒙古GPGV 分离物与其他GPGV 中国分离物进行全基因组序列一致性分析,结果表明,GPGV 中国分离物彼此之间的全基因组序列一致率在82.0%~99.9%之间,其中分离物20IMViVi1与20IM-ViVi2之间具有较高的全基因组序列一致率,为96.4%(图2)。将所获得的2个序列分别与GenBank中其他GPGV全基因组序列进行成对比对,结果表明,分离物20IM-ViVi1 与其他GPGV 完整基因组序列的核苷酸序列一致率在79.7%~96.8%之间,其中与俄罗斯白羽分离物Rk3(GenBank登录号:OL961512)之间的全基因组序列一致率最高,为96.9%;与日本紫葛葡萄(Vitis coignetiae)分离物HJP2(GenBank 登录号:LC601812)之间的全基因组序列一致率最低,为78.8%;分离物20IM-ViVi2与其他GPGV 完整基因组序列的核苷酸序列一致率在79.5%~97.7%之间,其中与中国夏黑分离物SRR2845691GPGV(GenBank 登录号:BK011076)之间的全基因组序列一致率最高,为97.7%;与日本紫葛葡萄分离物H-JP2之间的全基因组序列一致率最低,为79.5%。
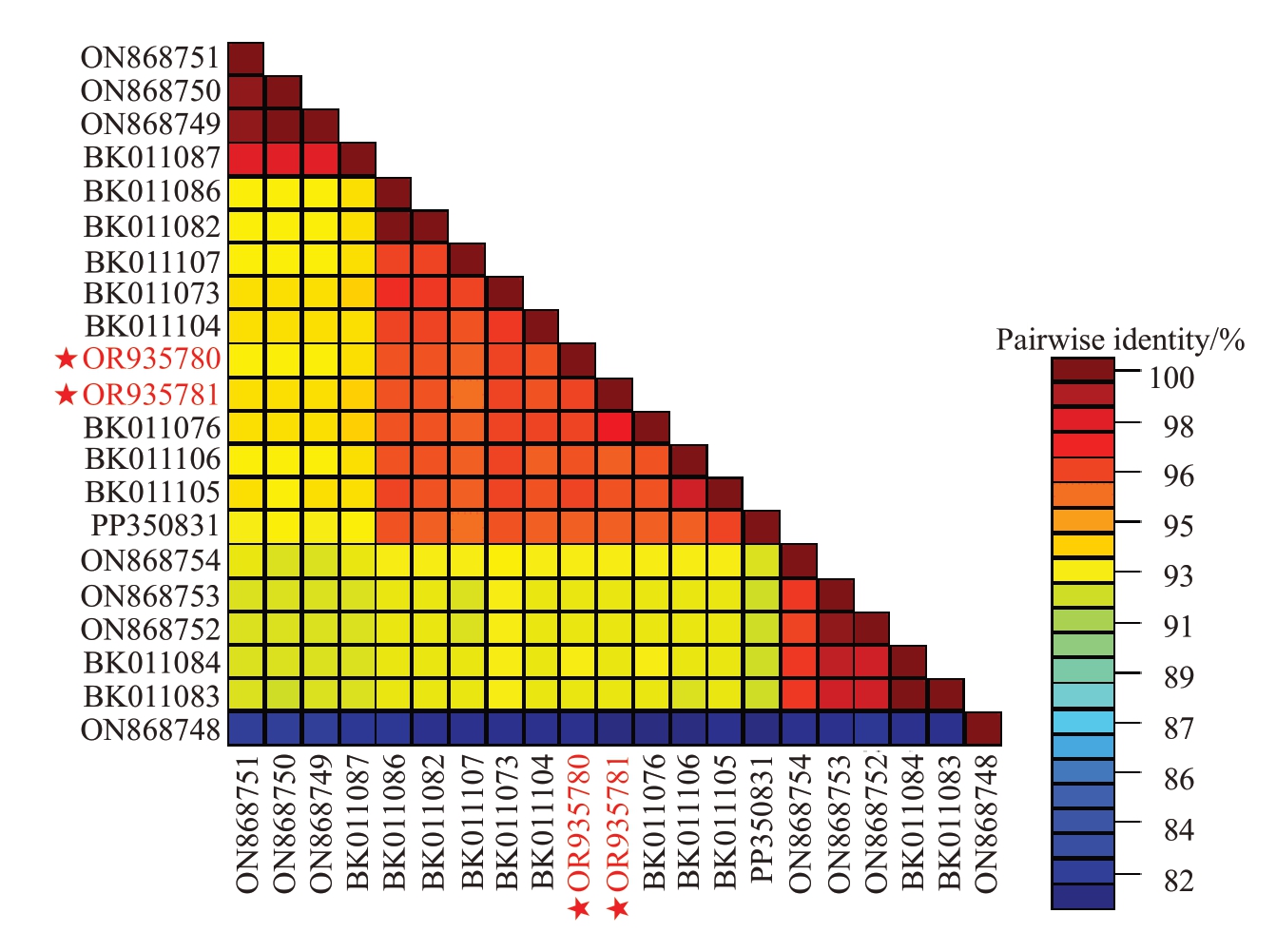
图2 中国不同地区GPGV 分离物的全基因组序列一致性
Fig.2 The complete sequence identities among GPGV isolates from various regions in China
为分析GPGV内蒙古分离物分子多样性的具体区域,将所获得的GPGV 内蒙古分离物的各ORFs与GenBank 中其他GPGV 的ORFs 进行成对比对,结果表明分离物20IM-ViVi1 的RdRp(ORF1)的核苷酸与氨基酸序列一致率的范围分别为78.9%~97.5%、87.6%~98.9%;MP(ORF2)的核苷酸与氨基酸序列一致率的范围分别为80.1%~97.4%、86.6%~98.9%;CP(ORF3)的核苷酸与氨基酸序列一致率的范围分别为84.8%~95.7%、92.3%~100.0%。分离物20IM-ViVi2的RdRp(ORF1)的核苷酸与氨基酸一致率的范围分别为78.6%~97.6%、86.4%~98.7%;MP(ORF2)的核苷酸与氨基酸序列一致率的范围分别为79.9%~98.4%、86.6%~99.2%;CP(ORF3)的核苷酸与氨基酸一致率的范围分别为85.1%~98.1%、92.3%~100.0%。
2.3 GPGV系统进化分析
为明确所获得的GPGV 内蒙古分离物与NCBIGenBank 数据库已报道的GPGV 分离物之间的亲缘关系,利用MEGA11软件以最大似然法构建了系统进化树,MODLES程序判定了最大似然法的最佳进化模型(GTR+G+I)。结果(图3)显示,GPGV现有的152 个完整基因组被分为4 个分支,第Ⅲ分支和第Ⅳ分支又被分为a、b 2个小的分支。其中本研究中所获得的2个分离物20IM-ViVi1与20IM-Vi-Vi2 均聚集在第I分支,并均与中国夏黑分离物SRR2845691-GPGV亲缘关系最近。

图3 基于GPGV 全基因组构建的系统进化树
Fig.3 Phylogenetic tree constructed on the base of the complete genome of GPGV
另外,为明确中国GPGV 分离物的系统发育关系,以最大似然法对22个中国GPGV分离物进行了系统发育树的重建,结果(图4)显示,中国分离物主要分为4 个分支,其中GPGV 新疆分离物Shihezi-1与其他分离物具有较大的遗传距离,亲缘关系较远,被单独分在了第I分支。而本研究中所获得的分离物20IM-ViVi1与20IM-ViVi2均被聚集在第Ⅳ分支,并且均与中国夏黑分离物SRR2845691-GPGV 亲缘关系最近。

图4 基于GPGV 中国分离物所构建的系统发育树
Fig.4 Phylogenetic tree constructed based on the isolates of GPGV from China
2.4 GPGV的遗传多样性分析
通过DNAsp v.6 软件对152 个GPGV 分离物进行了中性测试以及核苷酸多态性分析,并延ORFs基因组序列走向绘制等比例趋势图。其中中性测试结果(图5)显示,GPGV群体在RdRp、MP以及CP编码区均处的Tajima’s D中性检测值分别为-2.234 92、-2.137 45、-2.059 23,且支撑该数值的p 值均小于0.05,表明GPGV三个ORFs均受到了显著的负向选择。核苷酸多态性分析结果显示,GPGV 群体在RdRp、MP 以及CP 编码区域的核苷酸多态性(π)为0.032 88±0.001 54、0.028 59±0.003 25、0.028 27±0.004 50,表明GPGV具有较高的遗传多样性。此外,亚洲GPGV 分离物的核苷酸多态性(π=0.078 70±0.014 69)显著高于其他大洲GPGV分离物的核苷酸多态性(表2),但亚洲GPGV 分离物的中性检测值并不显著,可能遵循中性进化的原则。

图5 GPGV 群体进化中的中性测试及核苷酸多态性分析
Fig.5 Analysis of neutrality tests and nucleotide polymorphism for the GPGV populations
蓝色线段代表GPGV 群体之间的核苷酸多态性,左侧坐标轴为核苷酸多态性的变化区间;红色线段为GPGV 群体间的Tajima’s D 中性测试,右侧坐标轴为Tajima’s D 值的变化区间。
The blue line segments represent nucleotide polymorphisms among the GPGV populations,with the left y-axis indicating the range of variation in nucleotide polymorphisms;The red line segments depict Tajima’s D neutrality tests between GPGV populations,with the right y-axis indicating the range of variation in Tajima’s D values.
表2 葡萄灰比诺病毒不同编码区选择压力分析参数
Table 2 Analysis parameters of selection pressure on different coding regions of GPGV

注:ENC.有效密码子数;dN、dS 以及dN/dS 比值由DnaSP6 软件估算;通过SLAC 方法计算了葡萄灰比诺病毒群体的dN-dS 值(dN-dS 值除以树的长度总和),这是一个自然选择的指标。Mean(dN/dS)=1,<1 和>1 分别表示每个基因特异性序列数据集为中性进化、负向(净化)选择和正向选择。加粗的字体为FEL、SLAC 以及FUBAR 三种算法共同筛选出的正选择密码子位点。
Note: ENC. Effective number of codons; dN, dS and the dN/dS were estimated by DnaSP6; Normalized dN-dS values (dN-dS divided by the total length of the tree),an indicator of natural selection,was calculated for Shallot yellow stripe virus isolates by using SLAC method.Mean(dN/dS)value=1,<1 and >1 indicate neutral evolution,negative(purifying)selection and positive(diversifying)selection,respectively,for each gene-specific sequence data set.The bolded font indicates the positively selected codon sites identified by the three algorithms(FEL,SLAC and FUBAR).
编码区Genetic regions ENC dN dS dN/dS归一化dN-dS值Normalized dN-dS负选择密码子数Number of NS正选择码子位点Codons under PS RDRP 1885 0.007 95 0.122 97 0.064 649 Log(L)-48 777.95 Mean(dN/dS)0.104 0 FEL 1038 SLAC 713 FUBAR 1362 SLAC 585,657,669,1011 FUBAR 189,638,657 MP 459 0.011 23 0.085 04 0.132 055-8 810.42 0.175 0 156 134 212 FEL 470,585,613,622,638,657,1011,1556,1713,1850 300,338,344,346,356,366 300,338,344,346,356,366 32,64 CP 195 0.005 54 0.107 57 0.051 501-4 431.55 0.067 7 99 65 132 32 277,338,344,346,356,366 32
为了评估GPGV进化过程中每个编码区的选择压力变化,使用DNAsp v. 6 计算dN/dS比值(表2)。所有GPGV 编码区的dN 值均小于dS 值(dN/dS 值<1),表明纯化选择限制了群体的变异性。然而,纯化选择压力在整个基因组中并不是均匀分布的(图6)。CP 受到的纯化选择最强,其dN/dS值最低,为0.051 501。MP 受到的纯化选择最弱,其dN/dS值最高,为0.132 055。此外,通过使用HyPhy 软件包中的SLAC 方法进一步表明MP 编码区具有较多的正选择位点(图6),且FEL、FUBAR、SLAC 三种方法均筛选出了强正选择位点(表2),其中共同被筛选的位点为RdRp 编码区的657,MP 编码区的388、344、346、356、366以及CP编码区的32。

图6 基于SLAC 方法对GPGV 分离物进行的选择压力分析
Fig.6 Selection analyses for GPGV populations by SLAC method
A.基于RdRp 编码区进行的选择压力分析;B.基于MP 编码区进行的选择压力分析;C.基于CP 编码区进行的选择压力分析。
A.The selection pressure analysis based on the RdRp coding region;B.The selection pressure analysis based on the MP coding region;C.The selection pressure analysis based on the CP coding region.
3 讨 论
GPGV 是造成葡萄叶片发病、浆果品质变差的主要病原之一[22-23],前人研究表明在大多数情况下GPGV与葡萄发育迟缓以及叶片发生的褪绿、斑驳、畸形等症状有关[1],而在Saldarelli等[18]的调查中发现GPGV 更多的存在于一些几乎无症状的葡萄样品中。Bertazzon等[13]发现有症状的植株的GPGV量高于感染GPGV但无症状的植株。而笔者在本研究中分离得到的GPGV 分离物的寄主症状并不相同,考虑到多种病毒复合侵染情况,2 个GPGV 分离物的致病性有待进一步研究。关于GPGV的草本寄主以及昆虫介体,笔者在本研究中尚未有所收获,然而现有研究表明GPGV 还可感染白花蝇子草、藜等草本植物[19],这些草本植物在世界各地的葡萄园中随处可见。这些草本植物中可能会增强病毒的感染繁殖能力、持久性以及扩大传播范围,进而传播给邻近的葡萄[19]。
近年来,GPGV在世界各地越来越多,但是完整基因组序列的报道仍然有限,以至于其起源问题仍然模糊不清,笔者在本研究中对内蒙古7个地区(呼和浩特、包头、赤峰、通辽、巴彦淖尔、鄂尔多斯、乌海)的69个葡萄样本进行多病毒检测时,其中4个地区(呼和浩特、包头、赤峰、通辽)的16个葡萄样本检测出GPGV,检出率高达23.18%,可见GPGV在内蒙古非常流行,对当地葡萄的产量与品质十分不利。因此,笔者在本研究中对感染GPGV 的阳性样品进行了全基因组扩增、克隆,得到了2个GPGV内蒙古分离物20IM-ViVi1 和20IM-ViVi2 的全基因组序列。内蒙古的2个GPGV分离物基因组结构特征与已报道的一致,含3 个重叠的开放阅读框(ORF),ORF1编码了病毒甲基转移酶(MT)、2OG-Fe Ⅱ-Oxy加氧酶结构域、病毒RNA 解旋酶以及RNA 依赖型的RNA 聚合酶(RdRp),ORF2 和ORF3 分别编码了的运动蛋白(MP)和外壳蛋白(CP)[24-28]。
目前,关于GPGV的起源中心研究较多,大多数研究认为GPGV 或起源于亚洲,其中Pleško 等[12]认为中国极有可能是起源中心。而笔者在本研究中所获得的GPGV 分离物20IM-ViVi1 和20IM-ViVi2 与其他已知的GPGV 全基因组序列一致率分别介于79.7%~96.8%、79.5%~97.7%,其中日本紫葛葡萄分离物H-JP2 与所获得的分离物序列一致率均为最低,序列变异较大[29],序列相似性分析也证实了这一结果。基于GPGV分离物全基因组序列构建的系统进化树显示,笔者在本研究中所获得的夏黑分离物20IM-ViVi2 与南京夏黑分离物SRR2845691-GPGV聚集在同一分支,且亲缘关系最近。在品丽珠、维多利亚、歌海娜以及琼瑶浆等葡萄品种中所克隆到的GPGV 全基因组序列也存在近缘关系。因此,寄主差异在一定程度上影响了GPGV 的遗传进化,基于GPGV中国分离物重建的系统进化树同样证明了这一点。基于GPGV全基因组序列所构建的系统进化树中还显示出GPGV 分离物具有较强的地理特异性,如GPGV 中国分离物大多数聚集在第I分支,GPGV澳大利亚分离物和意大利分离物大多数聚集在第Ⅲ分支,GPGV 法国分离物和加拿大分离物大多数聚集在第Ⅳ分支。基于GPGV中国分离物全基因组序列重建的系统进化树表明,在本研究中所获的分离物与中国东部地区所克隆到的GPGV分离物具有较近的亲缘关系。
物种起源中心通常是其遗传多样性最高的地区[30-31],笔者在本研究中对GPGV进行群体遗传多样性分析,结果表明GPGV 亚洲分离物的遗传多样性(π=0.031 95±0.001 35)高于其他四大洲的GPGV分离物,因此,亚洲应是GPGV的起源中心。但考虑到基于GPGV全基因组序列所构建的ML树第一节点为日本分离物,Hily 等[17]所预测中国是起源地这一观点便不成立。目前,已知的日本分离物有限且变异较大,早期断定的起源时间有待GPGV 序列进一步增多后重新分析[17]。Tajima’s D中性测试结果显示,GPGV 法国/欧洲分离物受到了显著的负向选择,表明近期GPGV在法国/欧洲发生了种群扩张。
4 结 论
获得了2 条GPGV 完整基因组序列,分析结果表明,2 个GPGV 分离物基因组全长为7250 nt,含3个ORFs,与现有GPGV分离物全基因组序列具有较高的一致率(日本分离物H-JP2 除外),一致率范围分别为79.7%~96.8%、79.5%~97.7%。此外基于所有GPGV 全基因组序列组构建的系统进化树可分为4个分支,本研究所获得的分离物均具聚在第I分支。重组分析未发现与本研究所获得分离物相关的重组事件。通过遗传多样性分析发现亚洲GPG 分离物具有较高的遗传多样性,或是起源中心,但种群扩张发生在欧洲。本研究可为GPGV 分子特征、进化关系以及遗传多样性研究奠定基础。
[1] GIAMPETRUZZIA,ROUMIV,ROBERTO R,MALOSSINIU,YOSHIKAWA N,LA NOTTE P,TERLIZZIF,CREDIR,SALDARELLIP.A new grapevine virus discovered by deep sequencing of virus- and viroid-derived small RNAs in cv. Pinot Gris[J].Virus Research,2012,163(1):262-268.
[2] FUCHS M. Grapevine viruses:A multitude of diverse species with simple but overall poorly adopted management solutions in the vineyard[J]. Journal of Plant Pathology,2020,102(3):643-653.
[3] GOMEZ T S,ALONSO R,LUNA F,LANZA V M,BUSCEMA F. Occurrence of nine grapevine viruses in commercial vineyards of Mendoza,Argentina[J].Viruses,2023,15(1):177.
[4] BELKINA D,KARPOVA D,POROTIKOVA E,LIFANOV I,VINOGRADOVA S.Grapevine virome of the don ampelographic collection in Russia has concealed five novel viruses[J].Viruses,2023,15(12):2429.
[5] KAUR K,RINALDO A,LOVELOCK D,RODONIB,CONSTABLE F. The genetic variability of grapevine pinot gris virus(GPGV)in Australia[J].Virology Journal,2023,20(1):211.
[6] TARQUINIG,PAGLIARIL,ERMACORA P,MUSETTIR,FIRRAO G. Trigger and suppression of antiviral defenses by grapevine pinot gris virus (GPGV):Novel insights into virushost interaction[J]. Molecular Plant-Microbe Interactions,2021,34(9):1010-1023.
[7] BEUVE M,CANDRESSE T,TANNIÈRES M,LEMAIRE O.First report of grapevine pinot gris virus(GPGV)in grapevine in France[J].Plant Disease,2015,99(2):293.
[8] REYNARD J S,SCHUMACHER S,MENZEL W,FUCHS J,BOHNERT P,GLASA M,WETZEL T,FUCHS R. First report of grapevine pinot gris virus in german vineyards[J]. Plant Disease,2016,100(12):2545.
[9] GAZEL M,CAGLAYAN K,ELÇIE,ÖZTÜRK L. First report of grapevine pinot gris virus in grapevine in Turkey[J]. Plant Disease,2016,100(3):657.
[10] 夏炎,黄松,武雪莉,刘一琪,王苗苗,宋春晖,白团辉,宋尚伟,庞宏光,焦健,郑先波.基于宏病毒组测序技术的苹果病毒病鉴定与分析[J].园艺学报,2022,49(7):1415-1428.XIA Yan,HUANG Song,WU Xueli,LIU Yiqi,WANG Miaomiao,SONG Chunhui,BAITuanhui,SONG Shangwei,PANG Hongguang,JIAO Jian,ZHENG Xianbo.Identification and analysis of apple viruses diseases based on virome sequencing technology[J].Acta Horticulturae Sinica,2022,49(7):1415-1428.
[11] RASOOL S,NAZ S,ROWHANIA,GOLINO D A,WESTRICK N M,FARRAR K D,AL RWAHNIH M. First report of grapevine pinot gris virus infecting grapevine in Pakistan[J]. Plant Disease,2017,101(11):1958.
[12] PLEŠKO IM,MARN M V,SELJAK G,ŽEŽLINA I. First report of grapevine pinot gris virus infecting grapevine in Slovenia[J].Plant Disease,2014,98(7):1014.
[13] BERTAZZON N,FILIPPIN L,FORTE V,ANGELINIE.Grapevine pinot gris virus seems to have recently been introduced to vineyards in Veneto,Italy[J]. Archives of Virology,2016,161(3):711-714.
[14] JO Y,CHOIH,CHO J K,YOON J Y,CHOIS K,CHO W K.In silico approach to reveal viral populations in grapevine cultivar Tannat using transcriptome data[J]. Scientific Reports,2015,5:15841.
[15] GLASA M,PREDAJŇA L,KOMÍNEK P,NAGYOVÁ A,CANDRESSE T,OLMOS A. Molecular characterization of divergent grapevine pinot gris virus isolates and their detection in Slovak and Czech grapevines[J]. Archives of Virology,2014,159(8):2103-2107.
[16] WU Q,HABILIN. The recent importation of grapevine pinot gris virus into Australia[J].Virus Genes,2017,53(6):935-938.
[17] HILY J M,POULICARD N,CANDRESSE T,VIGNE E,BEUVE M,RENAULT L,VELT A,SPILMONT A S,LEMAIRE O. Datamining,genetic diversity analyses,and phylogeographic reconstructions redefine the worldwide evolutionary history of grapevine pinot gris virus and grapevine berry inner necrosis virus[J].Phytobiomes Journal,2020,4(2):165-177.
[18] SALDARELLIP,GIAMPETRUZZIA,MORELLIM,MALOSSINIU,PIROLO C,BIANCHEDIP,GUALANDRIV. Genetic variability of grapevine pinot gris virus and its association with grapevine leaf mottling and deformation[J]. Phytopathology,2015,105(4):555-563.
[19] GUALANDRIV,ASQUINIE,BIANCHEDIP,COVELLIL,BRILLIM,MALOSSINIU,BRAGAGNA P,SALDARELLIP,SI-AMMOUR A. Identification of herbaceous hosts of the grapevine pinot gris virus(GPGV)[J].European Journal of Plant Pathology,2017,147(1):21-25.
[20] MALAGNINIV,DE LILLO E,SALDARELLIP,BEBER R,DUSO C,RAIOLA A,ZANOTELLIL,VALENZANO D,GI-AMPETRUZZIA,MORELLIM,RATTIC,CAUSIN R,GUALANDRIV.Transmission of grapevine pinot gris virus by Colomerus vitis(Acari:Eriophyidae)to grapevine[J].Archives of Virology,2016,161(9):2595-2599.
[21] LIBRADO P,ROZAS J. DnaSP v5:a software for comprehensive analysis of DNA polymorphism data[J]. Bioinformatics,2009,25(11):1451-1452.
[22] REYNARD J S,BRODARD J,ZUFFEREY V,RIENTH M,GUGERLIP,SCHUMPP O,BLOUIN A G.Nuances of responses to two sources of grapevine leafroll disease on pinot noir grown in the field for 17 years[J].Viruses,2022,14(6):1333.
[23] MILJANIĆ V,JAKŠE J,KUNEJ U,RUSJAN D,ŠKVARČ A,ŠTAJNER N. Virome status of preclonal candidates of grapevine varieties (Vitis vinifera L.) from the Slovenian wine-growing region primorska as determined by high-throughput sequencing[J].Frontiers in Microbiology,2022,13:830866.
[24] BERTAZZON N,FORTE V,FILIPPIN L,CAUSIN R,MAIXNER M,ANGELINIE.Association between genetic variability and titre of Grapevine Pinot Gris virus with disease symptoms[J].Plant Pathology,2017,66(6):949-959.
[25] 范旭东,张尊平,任芳,胡国君,李正男,董雅凤.我国灰比诺葡萄病毒分离物检测及基因序列分析[J].植物病理学报,2018,48(4):466-473.FAN Xudong,ZHANG Zunping,REN Fang,HU Guojun,LIZhengnan,DONG Yafeng. Detection and sequence analyses of grapevine pinot gris virus isolates from China[J]. Acta Phytopathologica Sinica,2018,48(4):466-473.
[26] MURRAY G G R,WANG F,HARRISON E M,PATERSON G K,MATHER A E,HARRIS S R,HOLMES M A,RAMBAUT A,WELCH J J.The effect of genetic structure on molecular dating and tests for temporal signal[J]. Methods in Ecology and Evolution,2016,7(1):80-89.
[27] TARQUINIG,DE AMICIS F,MARTINIM,ERMACORA P,LOIN,MUSETTIR,BIANCHIG L,FIRRAO G. Analysis of new grapevine pinot gris virus (GPGV) isolates from NortheastItaly provides clues to track the evolution of a newly emerging clade[J].Archives of Virology,2019,164(6):1655-1660.
[28] XIAO H,SHABANIAN M,MCFADDEN-SMITH W,MENG B. First report of grapevine pinot gris virus in commercial grapes in Canada[J].Plant Disease,2016,100(5):1030.
[29] ABE J,NABESHIMA T. First report of grapevine pinot gris virus in wild grapevines (Vitis coignetiae) in Japan[J]. Journal of Plant Pathology,2021,103(2):725.
[30] VALOUZIH,SHAHMOHAMMADIN,GOLNARAGHIA,MOOSAVIM R,OHSHIMA K.Genetic diversity and evolutionary analyses of potyviruses infecting narcissus in Iran[J]. Journal of Plant Pathology,2022,104(1):237-250.
[31] JONES D R. Plant viruses transmitted by Thrips[J]. European Journal of Plant Pathology,2005,113(2):119-157.