杧果(Mangifera indica Linn.)为漆树科杧果属植物,素有“热带果王”之称,具有一定的营养价值与经济价值[1]。杧果作为多年生木本果树,基因组杂合度高,且大部分育种亲本自交亲和性、杂交亲和性不够清晰,人工授粉结实率低,给杂交育种亲本的选配带来了一定难度。
分子标记是继传统形态标记、细胞标记、生化标记之后兴起的新技术。相对于RFLP、PAPD、STS、AFLP等其他分子标记技术,简单重复序列标记(simple sequence repeats,SSR)具有位点丰富、重复性好、鉴别能力强、多态性高、呈共显性[2]等优点,被广泛应用于花生[3-5]、小豆[6]、板栗[7]、石榴[8-9]、水稻[10]、茶树[11]和苹果[12]等多种作物杂交后代的真假鉴定以及遗传多样性分析等。
目前,在杧果的研究中,SSR主要应用于遗传多样性分析、亲缘关系分析以及指纹图谱的构建[13],姚全胜等[14]利用SSR技术对184株杂交实生苗进行鉴定,证实该技术可以对杧果杂种的真实性进行快速验证,但是多亲本、大样本实生后代杂种鉴定研究未见报道。笔者在本研究中以大果、红色、高可溶性固形物含量、丰产稳产为育种目标,选择13 个品种为亲本材料,采用SSR技术对其1001个F1后代株进行真假杂种检测及遗传特性分析,为明确杧果育种亲本杂交亲和性和育种亲本选配提供理论指导。
1 材料和方法
1.1 试验材料
材料种植于中国热带农业科学院品种资源研究所儋州实验基地五队的杧果育种圃,13个杧果品种贵妃、金煌、台农1号、白象牙、红玉、R2E2、椰香、汤米、凯特、Villard、南逗迈、Juile、A61 混合种植,进行自然授粉。从挂果母本树上收获成熟果实,母本品种包括贵妃、台农1号、金煌、Juile、南逗迈、椰香、Villard、R2E2、汤米、红玉、A61、凯特等12个品种,播种后获得实生后代,共1001 份材料,并从13个品种中鉴定出实生后代的可能父本,并分析其遗传特性。
1.2 DNA提取
用打孔器在叶片上打孔,取新鲜组织100 mg,采用磁珠法基因组DNA 提取试剂盒提取杧果的基因组DNA,用紫外分光光度计和1%琼脂糖凝胶电泳检测DNA浓度和纯度,-20 ℃保存备用。
1.3 引物筛选
在前期获得杧果的转录组数据中选取200对引物中进行筛选,由天一辉远生物科技有限公司合成,通过预试验,对每对引物都进行多次重复扩增。以各亲本作为DNA 的模板对合成的200 对引物进行筛选,将筛选得到的引物对样品进行SSR分析。
1.4 SSR-PCR体系
PCR反应体系如表1,扩增反应为:95 ℃预变性5 min,95 ℃变性30 s,52~62 ℃退火30 s,72 ℃延伸30 s,10 个循环,每个循环降1 ℃;95 ℃变性30 s,52 ℃退火30 s,72 ℃延伸30 s,25个循环;72 ℃末端延伸20 min,4 ℃保存。
表1 PCR 反应体系(15 μL)
Table 1 PCR reaction system(15 μL)

反应体系Reaction system 2×Taq PCR Mix ddH2O上游引物Forward primer/(10 mmol·L-1)下游引物Reverse primer/(10 mmol·L-1)DNA用量Dosage/μL 7.5 4.5 1.0 1.0 1.0
1.5 PCR产物检测
采用1%琼脂糖凝胶电泳检测与毛细管荧光电泳检测。
1.6 数据统计与分析
利用Excel 统计扩增的等位基因位点信息。利用Cervus 软件计算等位基因数(Na)、有效基因数(Ne)、观测杂合度(Ho)、期望杂合度(He)、固定指数(F)、Shannon’s 信息指数(I)等遗传参数;利用GenALEx6.0 软件计算多态信息指数(PIC),获得遗传分化系数;利用R语言绘制热点图;利用Structure分析群体遗传结构;采用非加权组平均法(UPGMA),利用筛选出的SSR核心引物对杧果实生后代进行群体间的聚类分析。
2 结果与分析
2.1 SSR引物筛选
通过对每个亲本进行多次重复性检测,筛选出13对稳定扩增、条带清晰的引物对(表2),确定13个多态性和稳定性较好的位点(图1)。
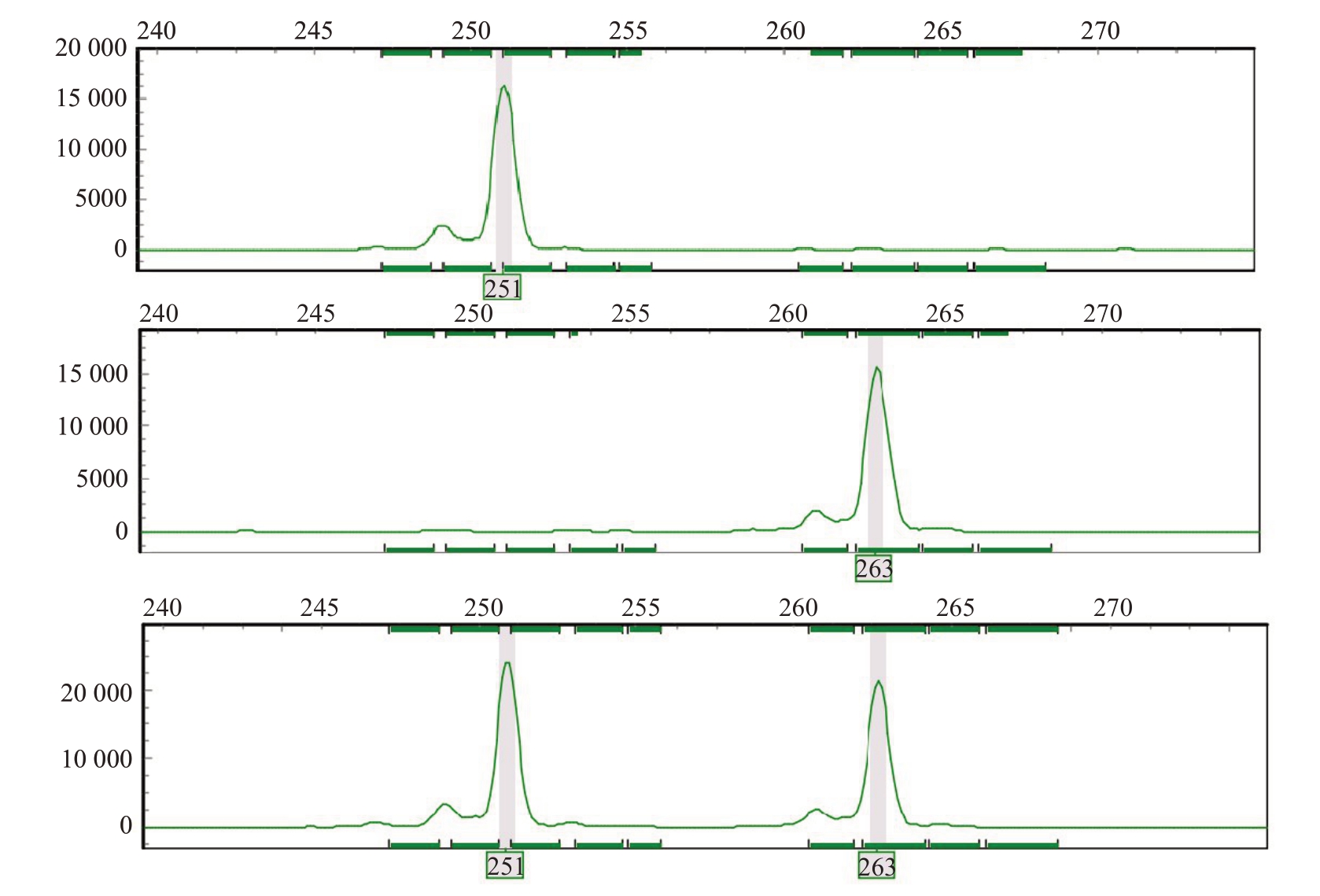
图1 SSR 引物MG-030 在部分杧果样品中的扩增情况
Fig.1 Amplification of SSR primer MG-030 in some mango samples
表2 13 对SSR 引物序列
Table 2 13 pairs of SSR primer sequences

引物编号Primer code MG-023 MG-030 MG-039 MG-046 MG-055 MG-061 MG-063 MG-079 MG-114 MG-146 MG-150 MG-177 MG-187正向引物(5′-3′)Forward primer(5′-3′)CGGAGGCCTACAGGTGAATA GAGACAGGAAGCGAAAGCAG CCGAAATTAATTCCCGTGGT CAATGACCCACATCATCCAA TACCTCGTAATCCAGTCGGC CTCTGCCTGTAAACCACCGT GATTGAGGTTTGGCCAGCTA AGAATCATGGGCAGGCAATA CACAGGCCCAACTTATTGCT ATCTTCTGGTACAGGCGGTG AGCGCCTTAAGATAGCCACA CATTGTCTCTGCACCAAACG CCACGGGAATGTACCTGCTA反向引物(5′-3′)Reverse primer(5′-3′)TGCATTGAACCAAACCTTGA AGTGCTTGGGCGTTAGAGAA TGAAGCTGGACAGTTGAGCA AAGGCCACGTATGTACCAGC CGCACACGCACTCTTTAGAA CATGCAGCATTGCAATTACC GTCCATTGTAGGCCCTGAGA TGAAATTAGGACCCAGCCAG GGAGACCAGAGGTTGACGAA TTTCCCACTCTTCCACGTTC AAGTCCATGAAGCTTGGGTG CTTGGCTATGGCTCCATCAT AGAAGATGCCGTACCAATGC
2.2 各位点遗传多样性
13对引物在杧果中共获得80个等位基因,每对引物扩增的等位基因数为3~9个,其中MG-039、MG-046和MG-187检测到9个等位基因。每个位点有效等位基因数平均为2.557。Shannon’s信息指数平均为1.027,其中MG-023位点的Shannon’s信息指数最高。平均期望杂合度略高于平均观测杂合度。一般认为,0.25<PIC≤0.5时,为中度多态性;PIC>0.5时,为高度多态性。13个位点中PIC值在0.314~0.741之间,表明这13个位点多态性较高(表3)。
表3 杧果SSR 标记位点遗传参数
Table 3 Genetic parameters of mango SSR markers

位点Locus MG-023 MG-030 MG-039 MG-046 MG-187 MG-055 MG-063 MG-079 MG-114 MG-146 MG-150 MG-177 MG-061平均Mean有效个体数N 1022 1015 992 1022 1021 929 968 1017 977 1017 1022 1022 1018 1 003.231等位基因数Na 6799973456456 6.154有效等位基因数Ne 4.487 2.545 2.452 2.044 1.489 2.491 2.801 1.577 2.620 2.222 2.715 3.284 2.516 2.557香农信息指数I 1.550 1.207 1.283 0.961 0.742 0.999 1.061 0.655 1.033 1.086 1.063 1.344 1.028 1.078观测杂合度Ho 0.750 0.643 0.686 0.498 0.324 0.595 0.602 0.389 0.438 0.625 0.623 0.711 0.538 0.571期望杂合度He 0.777 0.607 0.592 0.511 0.328 0.599 0.643 0.366 0.618 0.550 0.632 0.696 0.603 0.579固定指数F 0.036-0.060-0.159 0.025 0.013 0.005 0.063-0.065 0.291-0.137 0.013-0.023 0.107 0.008多态信息指数PIC 0.741 0.564 0.565 0.450 0.314 0.529 0.567 0.320 0.538 0.509 0.562 0.651 0.522 0.526
2.3 群体遗传多样性
在有效个体数(N)与等位基因数(Na)方面,贵妃最高,Juile最低,说明在总群体内,贵妃对应位点实际观测到的等位基因类型最多。有效等位基因数值(Ne)越接近等位基因数(Na)的绝对值时,表明等位基因在群体中分布越均匀,即贵妃、椰香、Juile 相对于其他品种而言,等位基因在群体中分布较均匀。贵妃平均Shannon’s 信息指数(I)最高,Juile 最低,说明贵妃群体中遗传多样性较高,种群分化程度最高,反之,Juile 最低。贵妃的观测杂合度(Ho)与期望杂合度(He)均为最高,表明贵妃的多态性高。固定指数(F)能够衡量种群中基因型的实际频率是否偏离Hardy-Weinberg(哈代温伯格)理论比例,当种群中纯合体过量时,F>0;反之,当杂合体过量时,F<0(表4)。
表4 群体遗传多样性
Table 4 Population genetic diversity

注:Mean 代表均值,SE 代表标准误。
Note:Mean represents mean,SE represents standard error.
香农信息指数I 0.92 0.07 1.09 0.06 0.78 0.06 1.06 0.07 0.60 0.09 1.06 0.06 0.75 0.07 0.68 0.07 0.99 0.07 1.03 0.06 0.99 0.06 0.88 0.06品种Variety A61贵妃Guifei红玉Hongyu金煌Jinhuang Juile凯特Keitt南逗迈Nan Doc Mai R2E2汤米Tangmi台农1号Tainong No.1 Villard椰香Yexiang固定指数F-0.09 0.06 0.00 0.05-0.14 0.11 0.04 0.05-0.36 0.10 0.10 0.05-0.17 0.06-0.18 0.09 0.01 0.06 0.05 0.06-0.03 0.06 0.02 0.08数据Data Mean SE Mean SE Mean SE Mean SE Mean SE Mean SE Mean SE Mean SE Mean SE Mean SE Mean SE Mean SE有效个体数N 12.91 0.06 173.91 1.30 170.09 3.52 85.00 0.54 1.87 0.07 110.35 1.49 30.04 0.50 127.39 0.58 126.83 0.65 68.43 1.04 50.91 0.70 10.74 0.18等位基因数Na 3.65 0.23 5.43 0.49 4.74 0.40 4.78 0.36 2.09 0.17 5.35 0.41 3.65 0.22 4.52 0.24 4.74 0.33 4.57 0.35 4.30 0.21 3.30 0.19有效等位基因数Ne 2.22 0.14 2.56 0.15 1.93 0.11 2.57 0.18 1.87 0.14 2.51 0.16 1.98 0.20 1.80 0.12 2.38 0.16 2.48 0.15 2.39 0.14 2.23 0.18观测杂合度Ho 0.55 0.05 0.57 0.03 0.55 0.08 0.52 0.03 0.52 0.09 20.50 0.04 0.51 0.06 0.51 0.08 0.52 0.04 0.53 0.04 0.56 0.04 0.48 0.05期望杂合度He 0.51 0.03 0.58 0.03 0.44 0.04 0.56 0.03 0.38 0.05 0.56 0.03 0.41 0.04 0.38 0.04 0.53 0.03 0.56 0.03 0.54 0.03 0.50 0.03无偏期望杂合度uHe 0.53 0.04 0.58 0.03 0.44 0.04 0.57 0.03 0.51 0.07 0.56 0.03 0.42 0.04 0.38 0.04 0.53 0.03 0.57 0.03 0.55 0.03 0.52 0.04
2.4 哈代温伯格平衡检验
利用Hardy-Weinberg 定律研究12 种不同杧果的基因,其中11 个品种处于平衡状态,其中品种红玉显示显著偏离Hardy-Weinberg(p<0.05)(表5)。
表5 哈代温伯格平衡检验
Table 5 Hardy-Weinberg Equilibrium Test
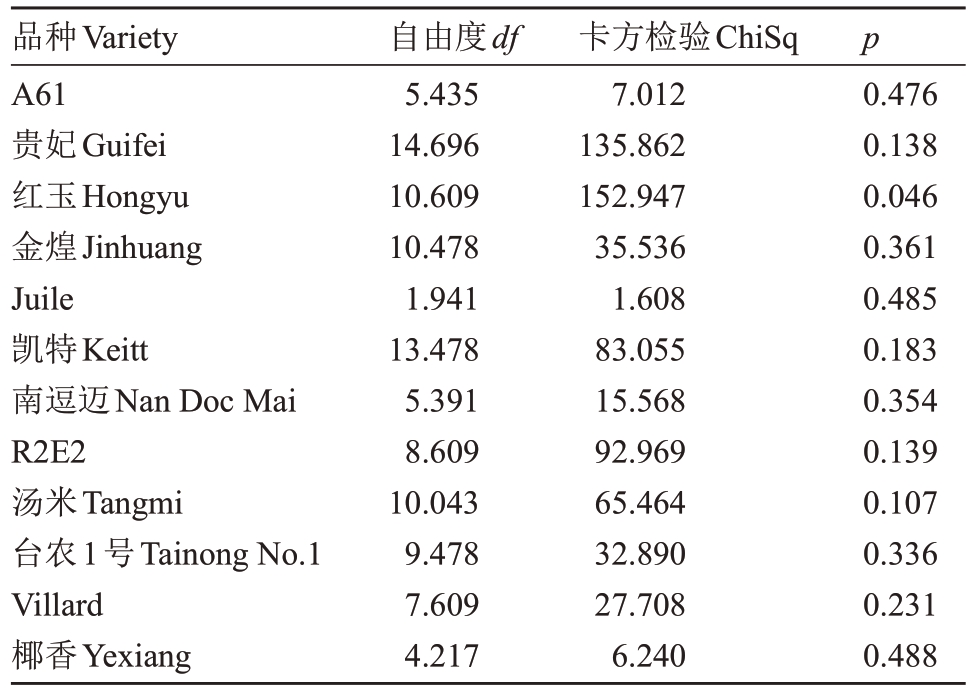
品种Variety A61贵妃Guifei红玉Hongyu金煌Jinhuang Juile凯特Keitt南逗迈Nan Doc Mai R2E2汤米Tangmi台农1号Tainong No.1 Villard椰香Yexiang自由度df 5.435 14.696 10.609 10.478 1.941 13.478 5.391 8.609 10.043 9.478 7.609 4.217卡方检验ChiSq 7.012 135.862 152.947 35.536 1.608 83.055 15.568 92.969 65.464 32.890 27.708 6.240 p 0.476 0.138 0.046 0.361 0.485 0.183 0.354 0.139 0.107 0.336 0.231 0.488
2.5 品种间遗传分化指数Fst
Fst指亲缘关系地方群体间的平均近交系数,当Fst 在0.05~0.15 时,表明群体间存在中等程度的遗传分化,Fst>0.05 时,群体间存在较大程度的遗传分化。通过品种间Fst结果发现,红玉和R2E2、红玉和椰香、红玉和南逗迈、Juile 和R2E2、Juile 和南逗迈、贵妃和南逗迈、贵妃和Juile间遗传分化明显,其余品种间遗传分化不明显(图2)。

图2 品种间遗传分化
Fig.2 Coefficient of genetic differentiation among breeds
2.6 品种间遗传距离与聚类分析
遗传距离作为衡量品种间不同性状综合遗传差异大小的指标,能够直观反映亲本品种间的遗传差异。品种R2E2和红玉、R2E2和Juile间的遗传距离最大,分别为0.737 7和0.622 0,表示该品种间遗传差异较大,其余品种的遗传距离较小,遗传差异不明显(图3)。根据样品间及群体间的遗传距离,运用Phylip 软件采用UPGMA 方法进行群体间聚类分析,凯特和汤米被聚为一类,红玉和Juile 被聚为一类,亲缘关系较近(图4)。

图3 品种间遗传距离
Fig.3 Genetic distance among varieties
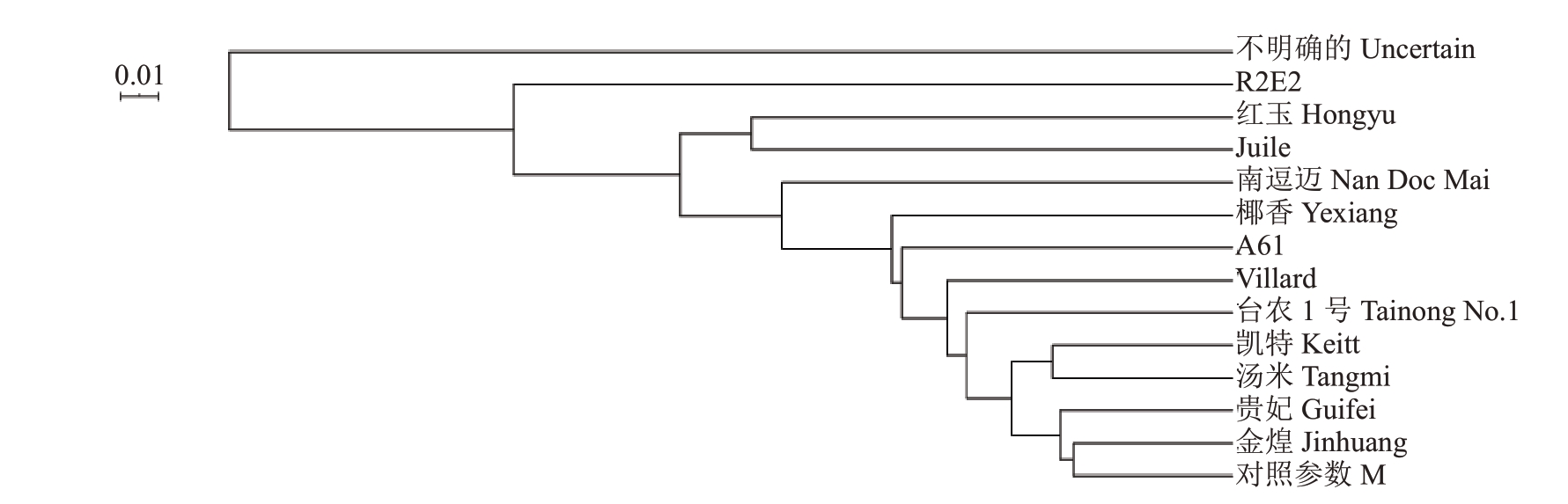
图4 聚类分析
Fig.4 Cluster analysis
2.7 Structure分析
Structure分析显示,当K=3时,△K=35,值最大(图5),可将所有个体分成3个群体。由群体遗传结构(图6)可知,不同颜色表示不同遗传组成,同一颜色占比最高的能够聚为同一类亚群,即R2E2为一簇,红玉为一簇,A61、贵妃、金煌、Juile、凯特、南逗迈、汤米、台农1号、Villard和椰香聚为一簇,因此12个品种可以划分为三大亚群,遗传背景分为3个类型。
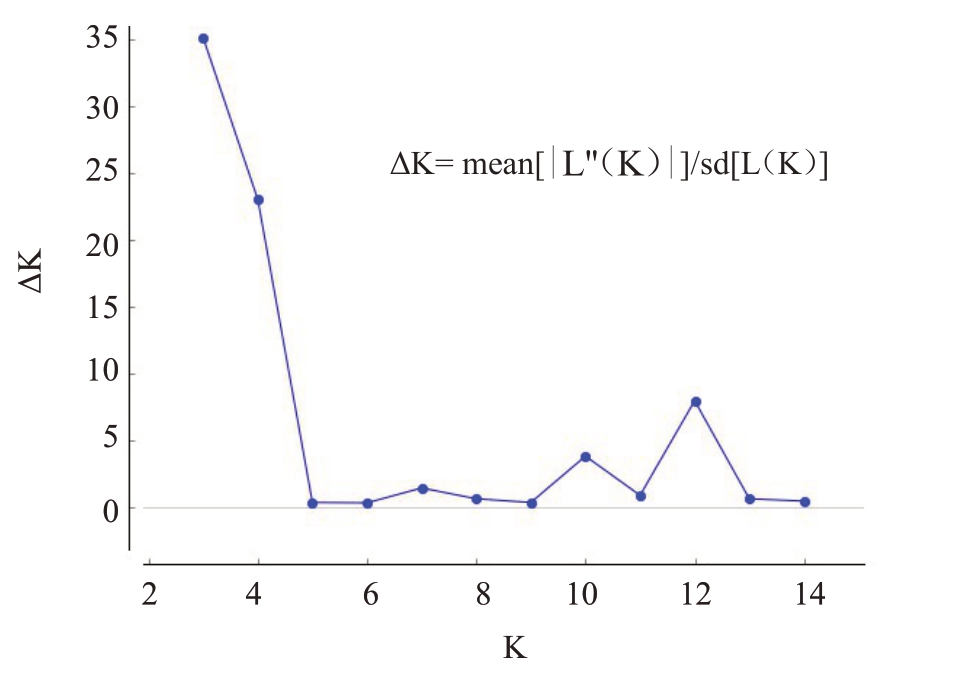
图5 基于ΔK 变化值分析遗传群体分化
Fig.5 Genetic population differentiation based on ΔK variation
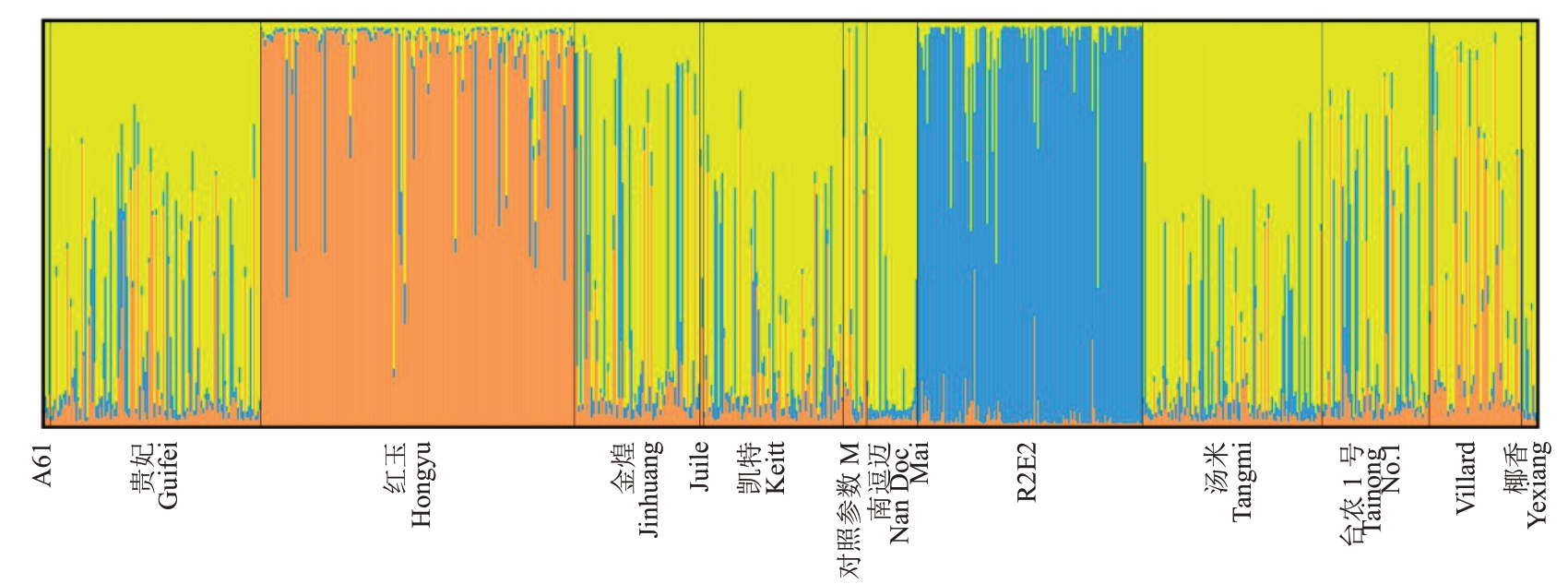
图6 基于Structure 分析的12 个品种的遗传结构
Fig.6 Genetic structure of 12 varieties based on Structure analysis
2.8 亲权鉴定
对双亲本均未知组合进行亲权分析,然后从中挑选出已知母本并不是第一候选亲本将其剔除,其中共剔除191 个样本,剔除比例为19.1%,对809 个子代和候选父本(母本已知)进行亲权关系鉴定,实际鉴定率为87%(表6,表7)。
表6 模拟双亲鉴定率
Table 6 Simulated parental identification rate
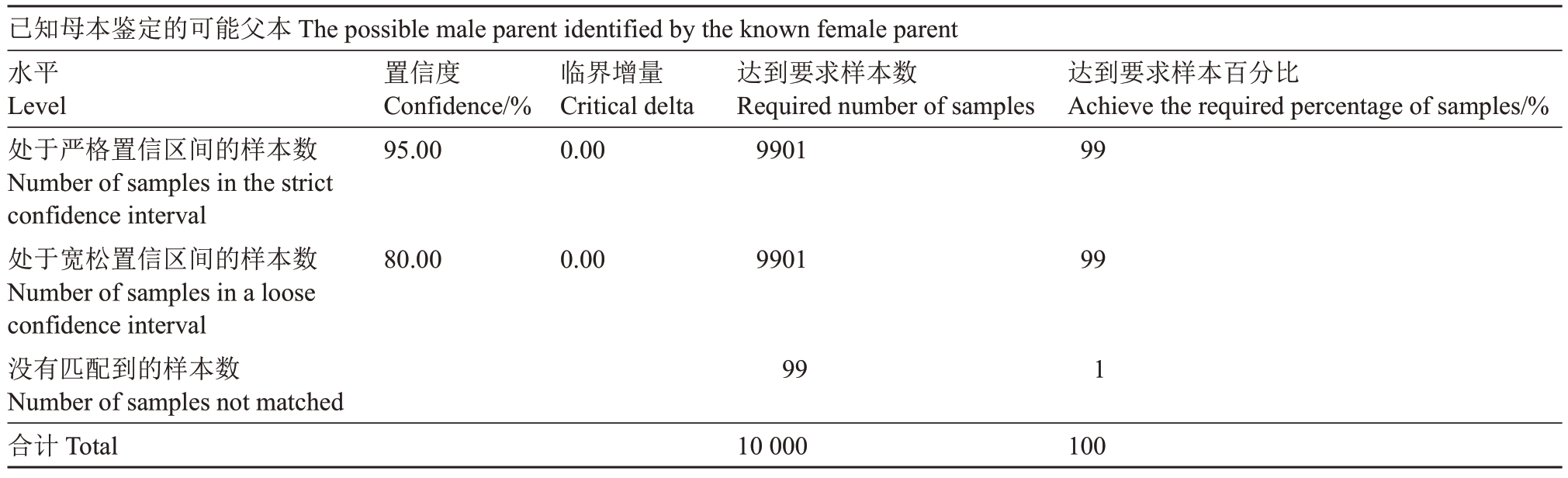
已知母本鉴定的可能父本The possible male parent identified by the known female parent水平Level处于严格置信区间的样本数Number of samples in the strict confidence interval处于宽松置信区间的样本数Number of samples in a loose confidence interval没有匹配到的样本数Number of samples not matched合计Total置信度Confidence/%95.00临界增量Critical delta 0.00达到要求样本数Required number of samples 9901达到要求样本百分比Achieve the required percentage of samples/%99 80.00 0.00 9901 99 99 1 10 000 100
表7 实际双亲鉴定率
Table 7 Actual parental identification rate
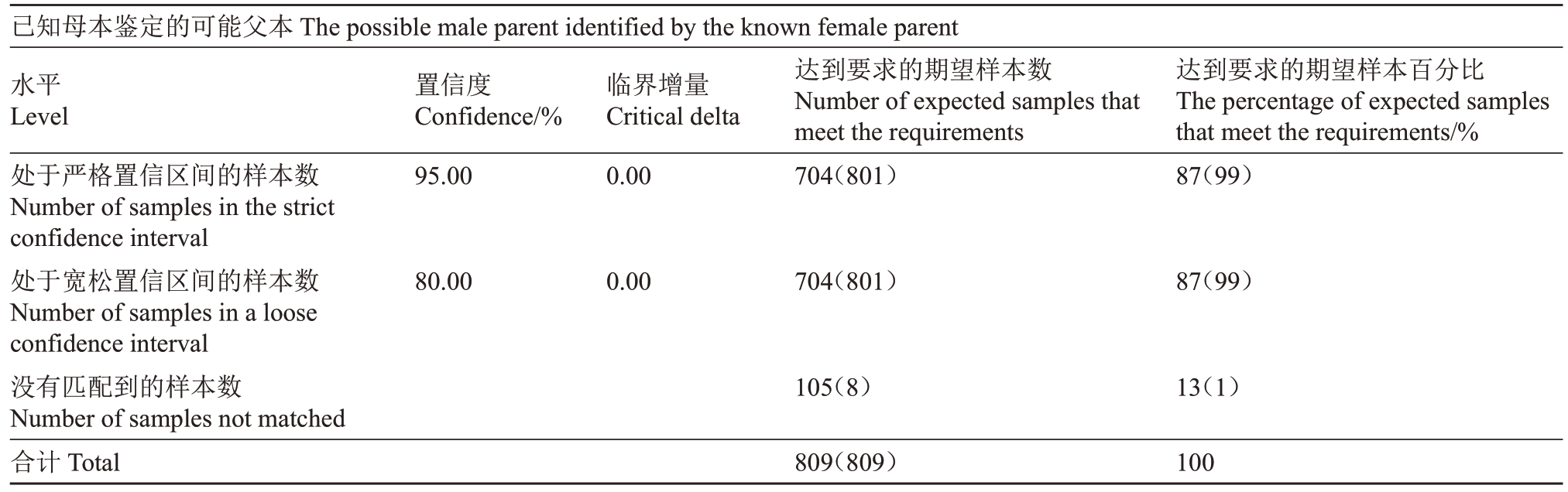
注:括号中的数值代表在置信区间实际样本数量与模拟匹配率。
Note:The values in brackets represent the actual number of samples and the simulation matching rate in the confidence interval.
已知母本鉴定的可能父本The possible male parent identified by the known female parent水平Level处于严格置信区间的样本数Number of samples in the strict confidence interval处于宽松置信区间的样本数Number of samples in a loose confidence interval没有匹配到的样本数Number of samples not matched合计Total置信度Confidence/%95.00临界增量Critical delta 0.00达到要求的期望样本数Number of expected samples that meet the requirements 704(801)达到要求的期望样本百分比The percentage of expected samples that meet the requirements/%87(99)80.00 0.00 704(801)87(99)105(8)13(1)100 809(809)
2.9 杂交鉴定结果
从可鉴定的父本中得出,以A61 为母本的3 个杂交后代中,真杂种率为80%;贵妃为母本的116个后代中,自交率为38.46%,真杂种率为61.54%;红玉为母本的167 个后代,其自交率为74.8%,真杂种率为25.2%;金煌为母本的69 个后代中,自交率低,真杂种率为82.61%;Juile为母本的2个杂交后代中,其父本50%来自红玉,50%来自南逗迈;南逗迈为母本的后代中,杂交率低,真杂种率为25.00%,其自交率为75.00%;凯特为母本的77 个后代中,真杂种率为72.73%,自交率为27.27%;母本为R2E2的后代群体中,自交率为82.69%,真杂种率为17.31%;汤米为母本的后代中,自交率为56.57%,真杂种率为43.43%;台农1 号为母本的59 个后代中,自交率为16.95%,真杂种率为83.05%;Villard 为母本的51 个后代中,自交率为66.67%,真杂种率为33.33%;椰香为母本的11 个后代中,自交率为63.64%,真杂种率为36.36%(表8)。结果表明,在12个已知品种中,南逗迈、R2E2、红玉、Villard、椰香自交率高,初步说明其自交亲和性强,极易自交;其余品种的杂交率均高于50%,其品种的杂交亲和性较强。
表8 品种间真杂种率
Table 8 True hybrid rate among varieties%

品种Variety A61贵妃Guifei红玉Hongyu金煌Jinhuang Juile南逗迈Nan Doc Mai凯特Keitt R2E2汤米Tangmi台农1号Tainong No.1 Villard椰香Yexiang样品数Sample 5 117 167 69 2 28 A61贵妃Guifei Juile南逗迈Nan Doc Mai凯特Keitt R2E2汤米Tangmi台农1号Tainong No.1 Villard 椰香Yexiang白象牙Baixiangya 40.00 20.00 3.42 4.19 20.29 38.46 0.60 1.45红玉Hongyu 20.00 17.09 74.85 7.25 50.00金煌Jinhuang 20.00 8.55 3.59 17.39 1.71 2.56 7.69 0.60 11.59 8.55 2.56 2.90 50.00 75.00 15.94 2.50 10.78 5.80 4.19 8.70 6.84 1.20 5.80 2.90 7.14 14.29 3.57 77 124 99 59 5.19 0.81 8.08 1.70 1.30 2.60 3.90 27.27 6.49 3.90 0.81 0.81 1.01 6.78 4.04 15.21 8.87 2.02 1.01 1.01 3.39 5.19 86.29 10.10 15.25 40.26 1.61 56.57 15.25 3.90 0.81 9.10 16.95 7.07 23.73 1.69 51 11 5.88 3.92 1.96 1.96 19.61 9.09 66.67 27.27 63.64
3 讨 论
SSR 分子标记技术作为一种便捷、可靠的方法被广泛应用于不同作物F1代的真伪杂种鉴定及验证[15]、遗传图谱的构建[16]、杂种纯度鉴定[17-20]、遗传多样性分析等。
杂交是品种创新和选育的重要环节,亲本的选择选配是决定杧果杂交育种是否成功的关键。在自然授粉群体中,杧果亲本很难被鉴定,现研究指出目前80%的杧果品种都来自实生选育,仅20%左右的品种通过控制授粉及混合自然授粉所选育[14]。杧果作为定向育种难度大、育种周期长的作物,大部分育种亲本不明确,在国际上常规人工杂交育种坐果率仅为0.3%,所以有必要开展杧果杂交后代的早期鉴定工作,排除假杂种,提高杧果育种效率。
杧果为高度杂合体,在生产过程中后代容易受自花授粉的影响,产生基因变异,且不同品种中极易进行自然杂交。对于遗传变异小的群体,鉴定出的可能父本较多,会导致多个父本剔除不了,笔者在本试验中利用SSR 分子标记技术对其进行亲权鉴定,13 个SSR 位点上,1001 个后代中809 个后代与其父母本基因型基本符合孟德尔遗传定律,初步表明这些位点能够精准鉴定出可能父本,为杧果杂交育种性状遗传提供基础数据,未鉴定出亲本的后代在亲权鉴定中产生位点不匹配的错误,有可能因为无效等位基因使一些杂合子的单条带无法检测出来,Hokanson 等[21]和Kapoor 等[22]的研究指出,模拟结果与实际结果相差较大是无效等位基因的存在造成的。
物种的遗传多样性本质是生物体遗传物质的变异,遗传多样性水平越高,该物种或种群对新环境的适应能力就越强[23]。DNA分子标记能够反映出遗传变异程度,笔者在本研究中通过13 对SSR 标记,对809份杧果样品进行遗传多样性分析,根据群体遗传结构聚类为三大不同亚群。该SSR标记位点显示杧果品种间存在许多等位基因的现象,等位基因数变化范围为3~9,平均等位基因数(Na)为6.154,平均有效等位基因数(Ne)为2.557,Shannon’s信息指数(I)为1.078,观测杂合度(Ho)平均为0.571,期望杂合度(He)平均为0.579,多态性信息含量(PIC)平均为0.526,多态性信息含量在0<PIC≤0.25之间,为低多态性,0.25<PIC≤0.5为中度多态性,PIC>0.5为高度多态性,在本试验的13个标记中有10个为高度多态性,3个为中度多态性。试验中所有SSR位点在样品间表现出好的多态性,各指标之间大多体现较高的一致性,表明引物的多态性较高,能够有效地揭示杧果实生后代的遗传多样性,更好地鉴别现有的主栽品种。
Structure分析是将遗传相似的基因型个体聚在一起,假定每一个被分析的个体在所有的类别中都有共同的祖先,估计各类别的属于该祖先的概率值,是一种分析群体遗传结构的常用方法。本试验中对12 个杧果亲本进行Structure 分析,准确推断出它的品种结构图。结果显示,当K=3 时,ΔK 值最大,说明被检测的品种分为三个簇,R2E2 为一簇,红玉为一簇,A61、贵妃、金煌、Juile、凯特、南逗迈、汤米、台农1号、Villard和椰香聚为一簇,说明这12个亲本的遗传背景分为3 个类型。亲权鉴定结果显示,R2E2品种聚为一簇,红玉品种聚为一簇,其他品种聚为一簇。由此可见,R2E2 和红玉两个品种,将会得到它们的纯合体基因型。
4 结 论
在杧果F1代杂种鉴定中,杧果品种台农1号、凯特、Juile、金煌、红玉、贵妃的杂交亲和性较强;南逗迈、R2E2、红玉、Villard、椰香品种极易自交,且品种间存在许多等位基因的现象,遗传多样性较为丰富。该结果为进一步开展杧果遗传改良与群体构建、亲本选择提供了前期依据。
[1] 李志强,党志国,赵志常,黄建峰,高爱平,陈业渊.芒果杂交F1 代群体的遗传多样性分析及遗传图谱的构建[J].分子植物育种,2016,14(4):953-958.LIZhiqiang,DANG Zhiguo,ZHAO Zhichang,HUANG Jianfeng,GAO Aiping,CHEN Yeyuan. Genetic diversity analyze of mango’s F1 hybrids and construction of genetic map[J].Molecular Plant Breeding,2016,14(4):953-958.
[2] 李文秀,贺军军,张华林,罗萍.SSR 分子标记鉴定橡胶树F1真伪杂种[J].热带作物学报,2021,42(5):1305-1309.LIWenxiu,HE Junjun,ZHANG Hualin,LUO Ping. Identification of F1 hybrids of Hevea brasiliensis by SSR markers[J].Chinese Journal of Tropical Crops,2021,42(5):1305-1309.
[3] 刘洪,徐振江,饶得花,鲁清,李少雄,刘海燕,陈小平,梁炫强,洪彦彬.基于形态学性状和SSR 标记的花生品种遗传多样性分析和特异性鉴定[J].作物学报,2019,45(1):26-36.LIU Hong,XU Zhenjiang,RAO Dehua,LU Qing,LIShaoxiong,LIU Haiyan,CHEN Xiaoping,LIANG Xuanqiang,HONG Yanbin. Genetic diversity analysis and distinctness identification of peanut cultivars based on morphological traits and SSR markers[J].Acta Agronomica Sinica,2019,45(1):26-36.
[4] 王辉,PAWAN K,李双铃,任艳,袁美,庄伟建,VARSHNEY R K,郭宝珠,谢联辉.SSR 分子标记在花生杂种鉴定中的应用[J].福建农林大学学报(自然科学版),2015,44(4):350-354.WANG Hui,PAWAN K,LIShuangling,REN Yan,YUAN Mei,ZHUANG Weijian,VARSHNEY R K,GUO Baozhu,XIE Lianhui.Application of SSR markers in peanut F1 hybrid identification[J]. Journal of Fujian Agriculture and Forestry University(Natural Science Edition),2015,44(4):350-354.
[5] 胡东青,王秀贞,唐月异,高华援,陈殿绪,吴琪,张树伟,王传堂. 利用SSR 标记鉴定花生真杂种[J]. 花生学报,2012,41(4):22-25.HU Dongqing,WANG Xiuzhen,TANG Yueyi,GAO Huayuan,CHEN Dianxu,WU Qi,ZHANG Shuwei,WANG Chuantang.Identification of hybrids in groundnut using SSR markers[J].Journal of Peanut Science,2012,41(4):22-25.
[6] 刘俊睿,谢梦娇,闫龙,李晗,杨凯,孙新展,张运,牛晓,孙东京,李永强,万平.SSR 分子标记鉴定小豆F1 真假杂种[J].北京农学院学报,2014,29(1):1-5.LIU Junrui,XIE Mengjiao,YAN Long,LIHan,YANG Kai,SUN Xinzhan,ZHANG Yun,NIU Xiao,SUN Dongjing,LIYongqiang,WAN Ping. Identification of F1 hybrid in adzuki bean (Vigna angularis) by SSR markers[J]. Journal of Beijing University of Agriculture,2014,29(1):1-5.
[7] 白晓倩,陈于,张仕杰,赵玉强,王武,朱灿灿.基于表型性状和SSR 标记的板栗品种遗传多样性分析及分子身份证构建[J].植物遗传资源学报,2022,23(4):972-984.BAIXiaoqian,CHEN Yu,ZHANG Shijie,ZHAO Yuqiang,WANG Wu,ZHU Cancan.Genetic diversity analysis and fingerprinting of chestnut varieties based on phenotypic traits and SSR markers[J]. Journal of Plant Genetic Resources,2022,23(4):972-984.
[8] 洪文娟.石榴种质资源SSR 分子标记遗传多样性分析及指纹图谱构建[D].北京:北京林业大学,2020.HONG Wenjuan. Genetic diversity analysis and finger prints construction of pomegranate germplasm resources based on SSR markers[D].Beijing:Beijing Forestry University,2020.
[9] 赵丽华.石榴(Punica granatum L.)种质资源遗传多样性及亲缘关系研究[D].重庆:西南大学,2010.ZHAO Lihua. Study on genetic diversity and genetic relationship of pomegranate (Punica granatum L.) germplasm[D].Chongqing:Southwest University,2010.
[10] 刘奇燕,丁显萍,张萍,张雯,韩海英,王亚清,朱昌栋.杂交水稻种子SSR 分子标记纯度鉴定[J]. 安徽农学通报,2021,27(5):9-10.LIU Qiyan,DING Xianping,ZHANG Ping,ZHANG Wen,HAN Haiying,WANG Yaqing,ZHU Changdong.Purity identification of SSR molecular markers in hybrid rice seeds[J].Anhui Agricultural Science Bulletin,2021,27(5):9-10.
[11] 雷雨,段继华,黄飞毅,康彦凯,罗意,陈宇宏,丁玎,姚利娜,董丽娟,李赛君.茶树杂交F1真假杂种的SSR 鉴定及遗传多样性分析[J].植物遗传资源学报,2021,22(3):748-757.LEIYu,DUAN Jihua,HUANG Feiyi,KANG Yankai,LUO Yi,CHEN Yuhong,DING Ding,YAO Lina,DONG Lijuan,LISaijun. Identification and genetic diversity of tea F1 hybrids based on SSR markers[J].Journal of Plant Genetic Resources,2021,22(3):748-757.
[12] 高源,王大江,王昆,丛佩华,李连文,朴继成.基于荧光SSR 分析中国原产苹果属植物17 个种的遗传多样性和遗传结构[J].果树学报,2020,37(11):1611-1622.GAO Yuan,WANG Dajiang,WANG Kun,CONG Peihua,LILianwen,PIAO Jicheng. Genetic diversity and population structure of 17 species of Malus Mill. native to China based on fluorescent SSR analysis[J]. Journal of Fruit Science,2020,37(11):1611-1622.
[13] 陈道勤,邱永生,王勤南,黄锦福,吉家乐.芒果人工杂交授粉结实率影响因素研究[J].现代农业科技,2020(18):64.CHEN Daoqin,QIU Yongsheng,WANG Qinnan,HUANG Jinfu,JIJiale. Study on the influencing factors of mango artificial hybridization pollination seed setting rate[J].Modern Agricultural Science and Technology,2020(18):64.
[14] 姚全胜,雷新涛,黄忠兴,朱敏,王松标,黄丽芳,马小卫,詹儒林,武红霞.杧果炭疽病抗性杂交群体的构建及SSR 分子标记鉴定[J].果树学报,2010,27(2):265-269.YAO Quansheng,LEIXintao,HUANG Zhongxing,ZHU Min,WANG Songbiao,HUANG Lifang,MA Xiaowei,ZHAN Rulin,WU Hongxia. Construction of the resistance to anthracnose for crossed mango population and identification by SSR molecular markers[J].Journal of Fruit Science,2010,27(2):265-269.
[15] 薛华柏,王芳芳,王磊,杨健,王龙,王苏珂,苏艳丽,乔玉山,李秀根.‘满天红’ב红香酥’杂种鉴定及遗传变异Genic-SSR分析[J].果树学报,2017,34(8):925-934.XUE Huabai,WANG Fangfang,WANG Lei,YANG Jian,WANG Long,WANG Suke,SU Yanli,QIAO Yushan,LIXiugen. Identification of the hybrids and analysis of genetic variation of a pear progeny derived from crossing between‘Mantianhong’and‘Hongxiangsu’by Genic-SSR[J]. Journal of Fruit Science,2017,34(8):925-934.
[16] 魏秀清,许玲,章希娟,许家辉.龙眼优良杂交株系的SSR 鉴定[J].东南园艺,2017,5(5):6-9.WEIXiuqing,XU Ling,ZHANG Xijuan,XU Jiahui. Identification of individual hybrid progeny in longan (Dimocarpus longan)by SSR[J].Southeast Horticulture,2017,5(5):6-9.
[17] 韩国辉.基于EST-SSR、Genomic-SSR 和SCoT 标记的柑橘连锁图谱构建及杂种和多倍体遗传分析[D]. 重庆:西南大学,2012.HAN Guohui.Construction of molecular linkage map and genetic analysis of hybrids and polyploidy of citrus based on ESTSSR,Genomic-SSR and SCoT markers[D]. Chongqing:Southwest University,2012.
[18] 刘伟,罗心平,张惠云,蒋侬辉,肖志丹,袁沛元,邱燕萍,凡超,杨晓燕,高贤玉,左艳秀,向旭.荔枝新种质‘燎原’的分子标记鉴定[J].分子植物育种,2016,14(1):177-185.LIU Wei,LUO Xinping,ZHANG Huiyun,JIANG Nonghui,XIAO Zhidan,YUAN Peiyuan,QIU Yanping,FAN Chao,YANG Xiaoyan,GAO Xianyu,ZUO Yanxiu,XIANG Xu. Identification of a novel litchi germplasm‘Liaoyuan’by molecular markers[J].Molecular Plant Breeding,2016,14(1):177-185.
[19] GONAIT,MANABE T,INOUE E,HAYASHIM,YAMAMOTO T,HAYASHIT,SAKUMA F,KASUMIM.Overcoming hybrid lethality in a cross between Japanese pear and apple using gamma irradiation and confirmation of hybrid status using flow cytometry and SSR markers[J].Scientia Horticulturae,2006,109(1):43-47.
[20] 马凯,赵钰,张恒,韩立群,赵国庆,王继勋.基于SSR 标记的中亚生态区核桃(Juglans regia L.)遗传多样性与种群结构分析[J].果树学报,2021,38(11):1854-1867.MA Kai,ZHAO Yu,ZHANG Heng,HAN Liqun,ZHAO Guoqing,WANG Jixun.Analysis of genetic diversity and population structure of walnut (Juglans regia L.) in Central Asia ecological region based on SSR markers[J].Journal of Fruit Science,2021,38(11):1854-1867.
[21] HOKANSON S C,SZEWC-MCFADDEN A K,LAMBOY W F,MCFERSON J R. Microsatellite (SSR) markers reveal genetic identities,genetic diversity and relationships in a Malus × domestica Borkh. core subset collection[J]. Theoretical and Applied Genetics,1998,97(5):671-683.
[22] KAPOOR M,MAWAL P,SHARMA V,GUPTA R C.Analysis of genetic diversity and population structure in Asparagus species using SSR markers[J]. Journal,Genetic Engineering & Biotechnology,2020,18(1):50.
[23] 姚国琼,杨帆,严苓方,孙正海,李伟.基于转录组SSR 的三角梅遗传多样性分析[J/OL].分子植物育种,2014:1-19[2022-03-15].https://kns.cnki.net/kcms/detail/46.1068.S.20220228.2219.017.html.YAO Guoqiong,YANG Fan,YAN Lingfang,SUN Zhenghai,LIWei. Analysis of Bougainvillea glabra Choisy genetic diversity based on SSR of transcriptome[J/OL]. Molecular Plant Breeding,2024:1-19[2022-03-15]. https://kns.cnki.net/kcms/detail/46.1068.S.20220228.2219.017.html.