猕猴桃(Actinidia spp.)是猕猴桃科(Actinidiaceae)猕猴桃属(Actinidia Lindl.)植物,是20 世纪初开始人工驯化栽培的特色经济果树,由于果实风味独特、营养丰富、维生素C含量高等优点,被誉为水果之王且深受广大消费者青睐[1-2]。2024年联合国粮农组织FAO(https://www.fao.org/home/zh/)统计数据显示,截至2022 年世界猕猴桃采收面积28.61 万hm2,产量429.15万t,是全球性重要的水果产业。其中中国猕猴桃产量约占世界猕猴桃总产量的2/3,已成为中国重要的特色水果产业之一。猕猴桃为功能性雌雄异株植物,起源和分布中心均在中国,是广大山区常见的一种水果,生长在路旁、林中、水沟边、灌丛中,自然状态下存在着广泛的种间和种内杂交现象,造成了猕猴桃属植物复杂的形态结构变异。在猕猴桃属植物中,多倍化现象普遍存在,例如已知的主栽品种中华猕猴桃红阳、软枣猕猴桃魁绿、美味猕猴桃贵长分别为二倍体、四倍体、六倍体。此外,猕猴桃种内染色体倍性变异也较为常见,不同倍性材料在生态适应[3]、抗逆[4]及果实品质[5]方面存在显著差异。猕猴桃多倍化是适应环境变化保护自身种群发展的进化,不仅使猕猴桃的染色体数目加倍,还影响其基因组的结构和功能,从而丰富猕猴桃遗传多样性[6]。
随着基因组学时代的到来和发展,测序成本不断降低,高通量测序已被广泛应用于植物基因组测序中。在猕猴桃属植物中,中华猕猴桃(Actinidia chinensis)[7]、毛花猕猴桃(Actinidia eriantha)[8]、阔叶猕猴桃(Actinidia latifolia)[9]、山梨猕猴桃(Actinidia rufa)[10]、软枣猕猴桃(Actinidia arguta)[11]、长叶猕猴桃(Actinidia hemsleyana)[12]、葛枣猕猴桃(Actinidia polygama)[10]等基因组已有报道,为其他猕猴桃属植物的全基因组测序、重要性状解析和遗传改良等工作奠定了基础[13]。然而中华猕猴桃、美味猕猴桃、软枣猕猴桃等主要栽培利用的物种普遍存在多倍化的现象,尽管不同倍性种质的基因组信息有共性之处,但多倍体猕猴桃的全基因组信息仍有待解析[14-15]。此外,对萼猕猴桃作为新型猕猴桃砧木,具备较强的抗涝、抗寒、抗病能力,在产区中也已经得到较大规模的推广[16],但缺乏其基因组信息,阻碍了对其重要抗逆性状的解析。因此,考察中华猕猴桃、软枣猕猴桃、对萼猕猴桃的倍性及基因组信息对后续指导多倍体基因组的组装和辅助其他相关研究具有十分重要的意义。
笔者在本研究中选取野生种质中华猕猴桃AcD2301(Actinidia chinensis)、软枣猕猴桃AcD2302(Actinidia arguta)、对萼猕猴桃AcD2303(Actinidia valvata)进行多倍体猕猴桃基因组Survey 分析及系统进化研究,通过流式细胞术、K-mer分析和系统进化树构建,进行染色体倍性、物种杂合率、基因组重复序列比例和基因组大小的评估及系统进化关系研究,以期为多倍体猕猴桃全基因组组装提供参考,也可为深入研究猕猴桃多倍化和系统进化提供理论支持。
1 材料和方法
1.1 试验材料
试验材料中华猕猴桃AcD2301、软枣猕猴桃AcD2302、对萼猕猴桃AcD2303 均为野生资源(表1),保存于贵州省农业科学院果树科学研究所百宜落叶果树试验基地。试验样品采集,剪取顶端幼嫩叶片,液氮速冻后置于-80 ℃超低温冰箱保存备用。
表1 样品采集信息
Table 1 Sample collection information

样品名称Sample name中华猕猴桃AcD2301 Actinidia chinensis AcD2301软枣猕猴桃AcD2302 Actinidia arguta AcD2302对萼猕猴桃AcD2303 Actinidia valvata AcD2303性别Sexuality雌Female雌Female两性花Bisexual flower采样时间Sampling time 2023年5月May,2023 2023年5月May,2023 2023年5月May,2023采样地点Sampling site贵州省贵阳市Guiyang,Guizhou贵州省贵阳市Guiyang,Guizhou贵州省贵阳市Guiyang,Guizhou
1.2 试验方法
1.2.1 染色体倍性检测 以二倍体红阳猕猴桃(Actinidia chinensis‘Hongyang’,2n=58)为内参样本,采用流式细胞术进行染色体倍性检测[17]。分别称取AcD2301、AcD2302和AcD2303新鲜顶端叶片0.2 g,置于培养皿中,用CyStain UV Precise P 试剂盒进行细胞核裂解,提取完成后用50 μm Celltrics滤网过滤至样品管中,加入DAPI荧光染液避光染色2 min后在CyFlow Space 流式细胞仪上进行流式细胞术测试,用FloMax软件分析核悬浮液。
1.2.2 DNA 提取及测序 采用CTAB 法提取猕猴桃基因组总DNA,并通过0.8%琼脂糖凝胶电泳检测DNA 提取质量,同时采用紫外分光光度计对DNA进行定量。利用第二代测序技术Illumina NovaSeq测序平台对样本文库进行双末端测序。采用fastp[18]等软件查看碱基质量分布、Reads平均错误率分布、Reads 测序碱基含量分布,原始数据过滤接头和低质量reads 获得高质量序列,并与核酸库进行比对,排除外源物种污染。
1.2.3 基因组Survey 分析 高质量测序数据基于jellyfish(version 2.3.0)软件设置K-mer为19生成Kmer 频数表和频率直方图,统计总K-mer数、唯一Kmer 数等,并运用GenomeScope 2 工具进行基因组大小、杂合度和重复序列比例的估计[19-20]。
1.2.4 基于SNP的系统进化树构建 基于自测数据(AcD2301、AcD2302、AcD2303)和公共数据库(NGDC、NCBI)下载部分已公布的猕猴桃二代测序数据(表2),在贵州省农业科学院果树科学研究所生物信息学分析平台进行系统进化分析,与参考基因组红阳v4.0[21]进行比对,利用GATK软件[22]进行SNP calling(仅保留二等位基因),用fastTree软件中的Maximum likelihood算法构建系统进化树,并将树文件进行可视化处理。
表2 猕猴桃属植物种名
Table 2 Kiwifruit plant species name and Latin name
注:NGDC.国家生物信息中心(中国);NCBI.美国国家生物技术信息中心(美国)。
Note: NGDC.National Center for Biotechnology Information (China);NCBI.National Center for Biotechnology Information(USA).
编号Numbering AcD2301种名Species name中华猕猴桃(Actinidia chinensis)AcD2302软枣猕猴桃(Actinidia arguta)AcD2303对萼猕猴桃(Actinidia valvata)CRR635714数据来源data sources自测数据Self-test data自测数据Self-test data自测数据Self-test data NGDC CRR635715 SRR3543582 NGDC NCBI NCBI NCBI NCBI NCBI NCBI NCBI NCBI NCBI NCBI SRR18177732 SRR3723918 SRR3705798 SRR3705797 SRR3723917 SRR3407085 SRR3474219 SRR3474216 SRR3474220中华猕猴桃东红(Actinidia chinensis‘Donghong’)阔叶猕猴桃(Actinidia latifolia)美味猕猴桃(Actinidia chinensis var.deliciosa)湖北猕猴桃(Actinidia hubeiensis)浙江猕猴桃(Actinidia zhejiangensis)黄毛猕猴桃(Actinidia fulvicoma)毛花猕猴桃(Actinidia eriantha)山梨猕猴桃(Actinidia rufa)软枣猕猴桃(Actinidia arguta)葛枣猕猴桃(Actinidia polygama)大籽猕猴桃(Actinidia macrosperma)对萼猕猴桃(Actinidia valvata)
2 结果与分析
2.1 猕猴桃染色体倍性分析
以二倍体红阳猕猴桃(Actinidia chinensis var.‘Hongyang’,2n=58)为内参样本,分析3份猕猴桃样品的倍性,图1 展示为猕猴桃多倍体样品倍性的流式直方图。其流式直方图中横坐标代表荧光强度,纵坐标代表细胞数量,荧光强度与DNA 含量成正比,即峰值的位置反映测试样品的倍性。根据内参物种二倍体红阳猕猴桃(图1-A)的峰值比较,AcD2301(图1-B)和AcD2302(图1-C)的染色体倍性均为四倍体,而AcD2303(图1-D)染色体倍性为六倍体,流式细胞术测得染色体倍性结果与后续全基因组测序结果一致,图中杂峰可能是部分细胞核降解造成的。

图1 猕猴桃多倍体流式直方图
Fig.1 Kiwi polyploid flow cytometry histogram
2.2 猕猴桃基因组测序及质控
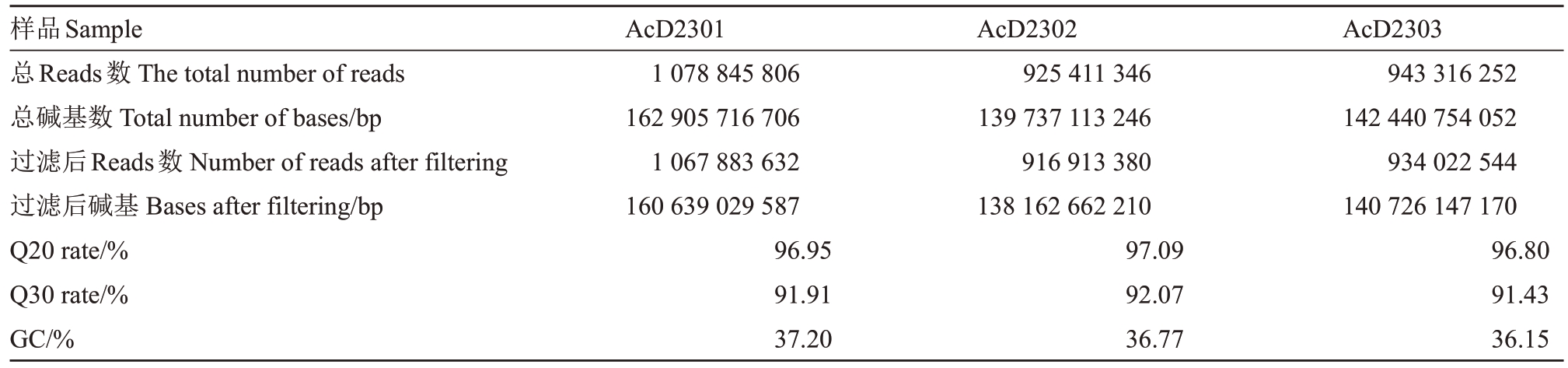
通过二代Illumina NovaSeq 测序平台对AcD2301、AcD2302 和AcD2303 基因DNA 进行测序,分别获得162.91 Gb、139.74 Gb和142.44 Gb原始测序数据,经过过滤后分别获得160.64 Gb、138.16 Gb和140.73 Gb高质量测序数据;测序的质量评估结果显示,AcD2301 的Q20、Q30 值分别为96.95%、91.91%,AcD2302 的Q20、Q30 值分别为97.09%、92.07%,AcD2303 的Q20、Q30 值分别为96.80%、91.43%,表明基因组数据可靠,可用于后续分析。AcD2301、AcD2302和AcD2303基因GC含量分别约为37.20%、36.77%和36.15%(表3)。AcD2301(图2-A、图3-A)、AcD2302(图2-B、图3-B)和AcD2303(图2-C、图3-C)基因组中大部分测序数据质量值均大于35(图2),其碱基错误率均小于0.045(图3),表明其基因组测序的Reads质量较高,测序结果可信度较高。
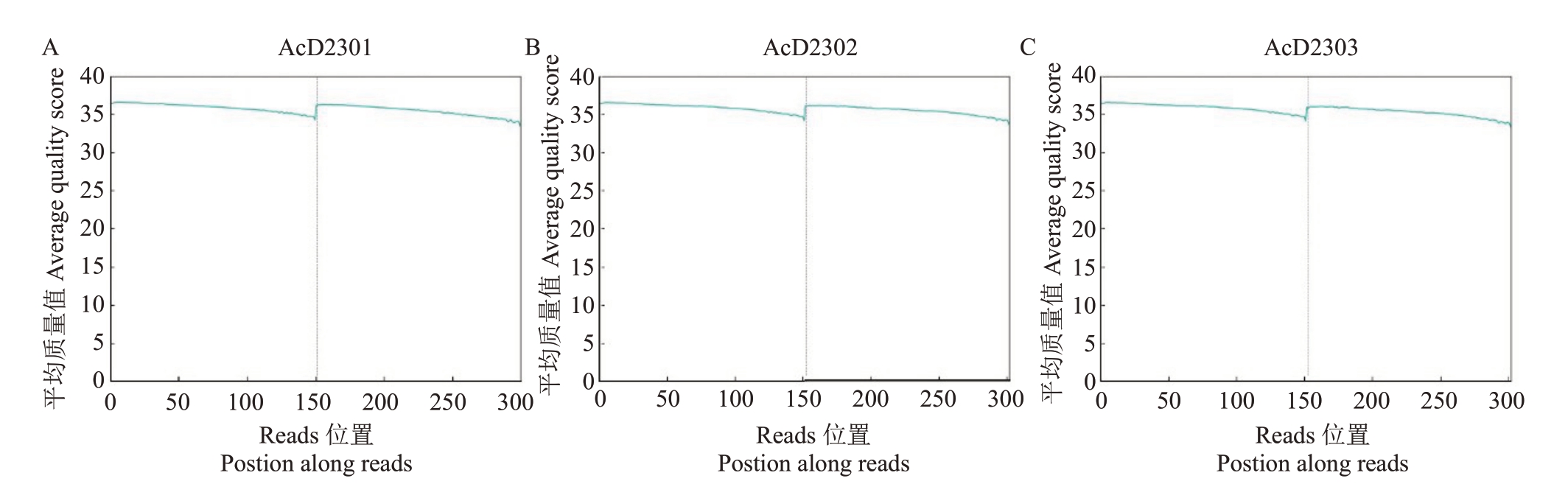
图2 单碱基质量分布
Fig.2 Single base quality distribution
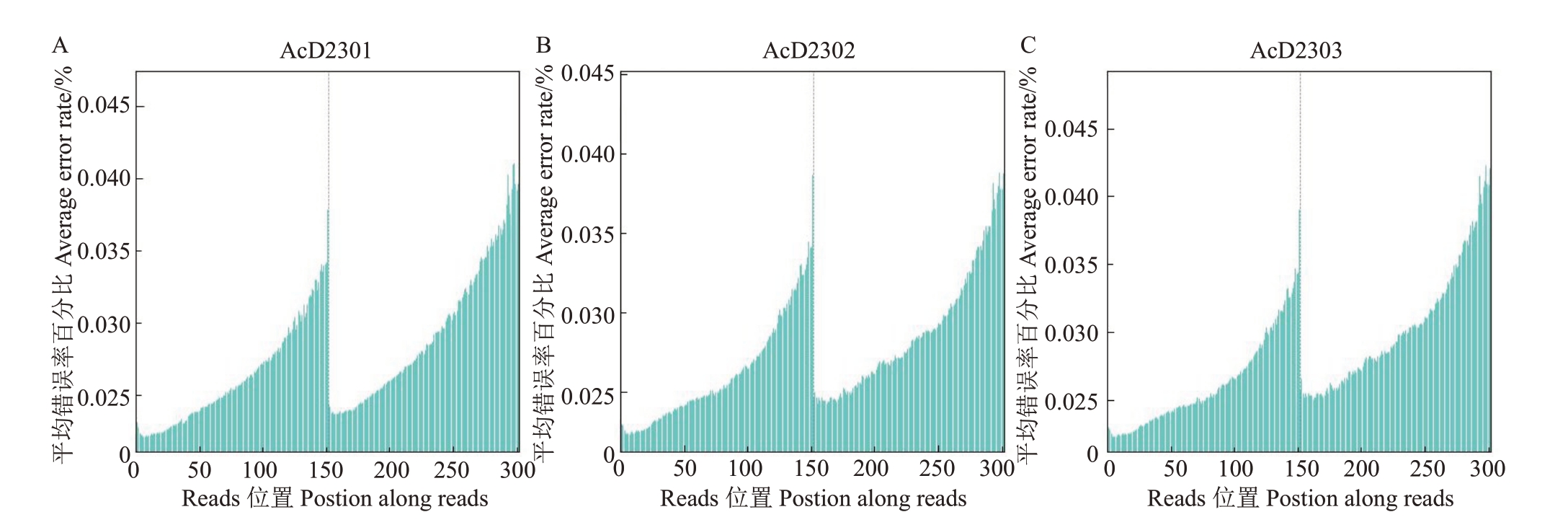
图3 Reads 平均错误率分布
Fig.3 Distribution of average error rate of reads
表3 基因组测序数据统计
Table 3 Statistics of genome sequencing data
样品Sample总Reads数The total number of reads总碱基数Total number of bases/bp过滤后Reads数Number of reads after filtering过滤后碱基Bases after filtering/bp Q20 rate/%Q30 rate/%GC/%AcD2301 1 078 845 806925 411 346943 316 252 162 905 716 706139 737 113 246142 440 754 052 1 067 883 632916 913 380934 022 544 160 639 029 587138 162 662 210140 726 147 170 96.9597.0996.80 91.9192.0791.43 37.2036.7736.15 AcD2302 AcD2303
2.3 猕猴桃基因组测序数据与NT数据库比对
进一步从AcD2301、AcD2302 和AcD2303 基因组测序数据中随机抽取10 000 条Reads 数据使用Blast 软件与核酸库(NT 库)进行比对,挑选最优比对结果按物种统计(表4),结果显示随机选取Reads均能比对上猕猴桃属植物基因组,表明此次测序的基因组数据不存在污染,部分样本与核酸库比对率较低的原因与取样少有关。
表4 NT 比对结果统计
Table 4 NT comparison results statistics
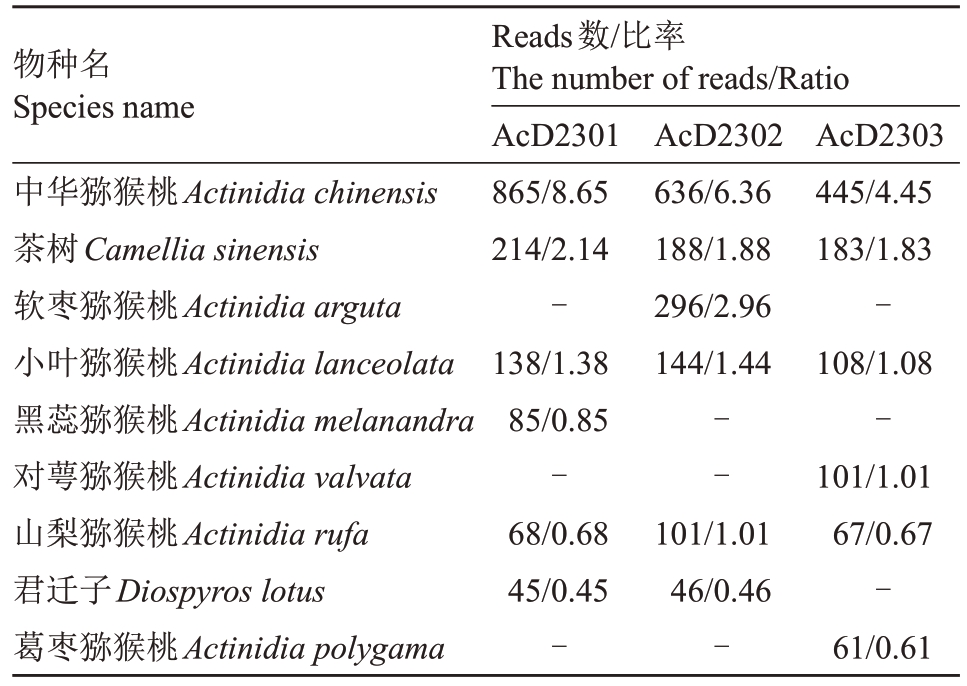
物种名Species name中华猕猴桃Actinidia chinensis茶树Camellia sinensis软枣猕猴桃Actinidia arguta小叶猕猴桃Actinidia lanceolata黑蕊猕猴桃Actinidia melanandra对萼猕猴桃Actinidia valvata山梨猕猴桃Actinidia rufa君迁子Diospyros lotus葛枣猕猴桃Actinidia polygama Reads数/比率The number of reads/Ratio AcD2301 865/8.65 214/2.14-138/1.38 85/0.85-68/0.68 45/0.45-AcD2302 636/6.36 188/1.88 296/2.96 144/1.44--101/1.01 46/0.46-AcD2303 445/4.45 183/1.83-108/1.08-101/1.01 67/0.67-61/0.61
2.4 猕猴桃基因组survey分析
高质量数据通过K-mer 分析,预估物种基因组大小,并对物种的杂合度、重复情况进行分析。通过对质控后的猕猴桃基因组数据进行K-mer 分析(图4、表5),可知AcD2301(图4-A)预估单套基因组大小为626 Mb,杂合度为3.00%,重复序列比例为43.70%;AcD2302(图4-B)预估单套基因组大小为668 Mb,杂合度为3.30%,重复序列比例为45.30%;AcD2303(图4-C)预估单套基因组大小为585 Mb,杂合度为8.06%,重复序列比例为40.70%。基于猕猴桃基因组Survey数据分析得出AcD2301(图4-A)同源四倍体支持率为97%,AcD2302(图4-B)同源四倍体支持率为96.7%。
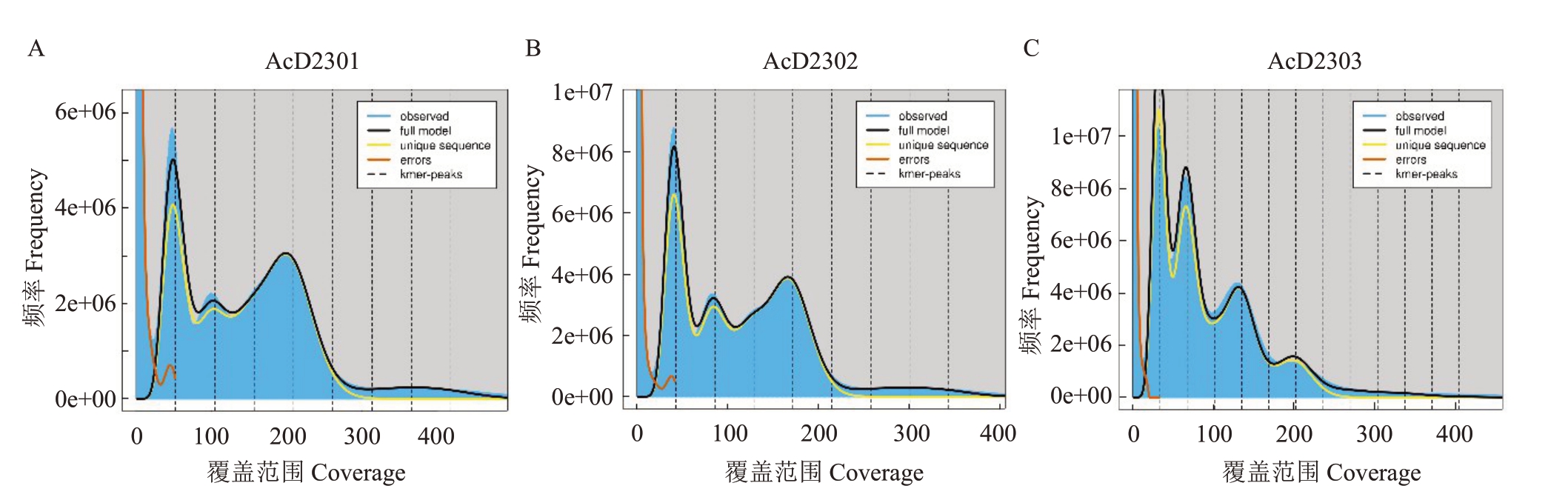
图4 K-mer 分布曲线
Fig.4 K-mer distribution curve
表5 基于19-kmer 基因组信息估计统计
Table 5 Estimated statistics based on 19-kmer genome information
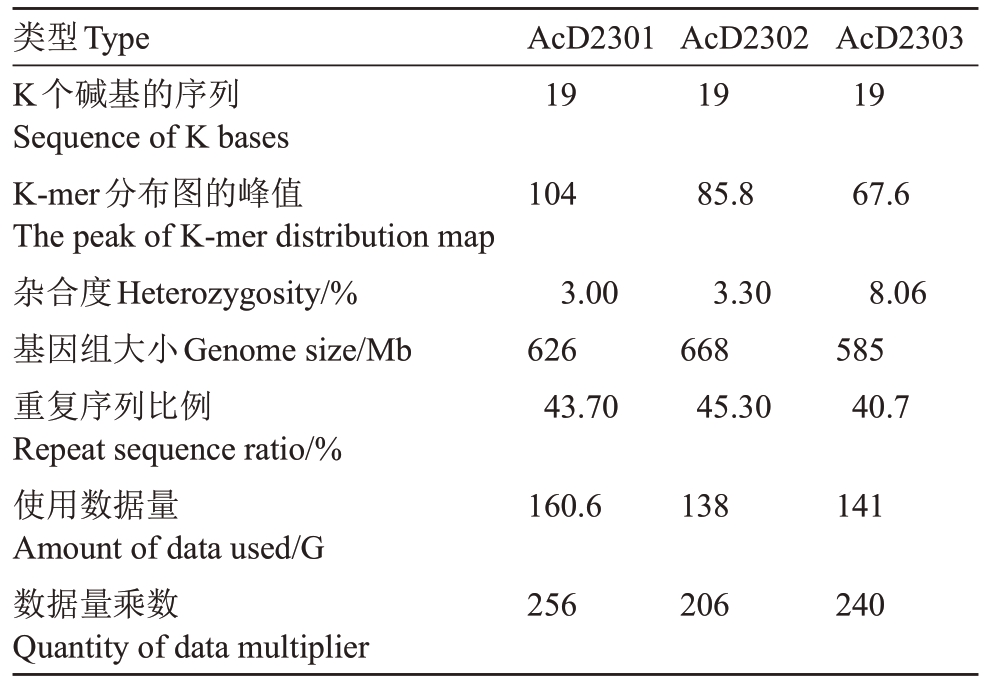
类型Type K个碱基的序列Sequence of K bases K-mer分布图的峰值The peak of K-mer distribution map杂合度Heterozygosity/%基因组大小Genome size/Mb重复序列比例Repeat sequence ratio/%使用数据量Amount of data used/G数据量乘数Quantity of data multiplier AcD2301 19 AcD2302 19 AcD2303 19 104 85.8 67.6 3.00 626 43.70 3.30 668 45.30 8.06 585 40.7 160.6 138 141 256 206 240
2.5 猕猴桃属植物进化树分析
为分析猕猴桃属植物的进化关系,筛选了已报道的中华猕猴桃东红、美味猕猴桃和湖北猕猴桃等15种猕猴桃属植物的二代测序数据中的SNP序列,采用Maximum likelihood算法构建系统进化树。该系统进化树显示(图5),15 种猕猴桃属植物分为三大进化分枝,且均得到了较好的支持,其中中华猕猴桃AcD2301为一个独立进化枝,东红猕猴桃为另一个独立进化枝,其余13 种猕猴桃组成一个进化枝。在第三个进化分枝中的美味猕猴桃为一个小进化枝,其余12种猕猴桃组成一个小进化分枝。对于后者湖北猕猴桃单独一组;其余11 种猕猴桃为一组,其中净果组的6种猕猴桃(软枣猕猴桃AcD2302、软枣猕猴桃、葛枣猕猴桃、大籽猕猴桃、对萼猕猴桃AcD2303、对萼猕猴桃)聚为一个小进化分枝,而其余分枝的9种猕猴桃均为斑果组。
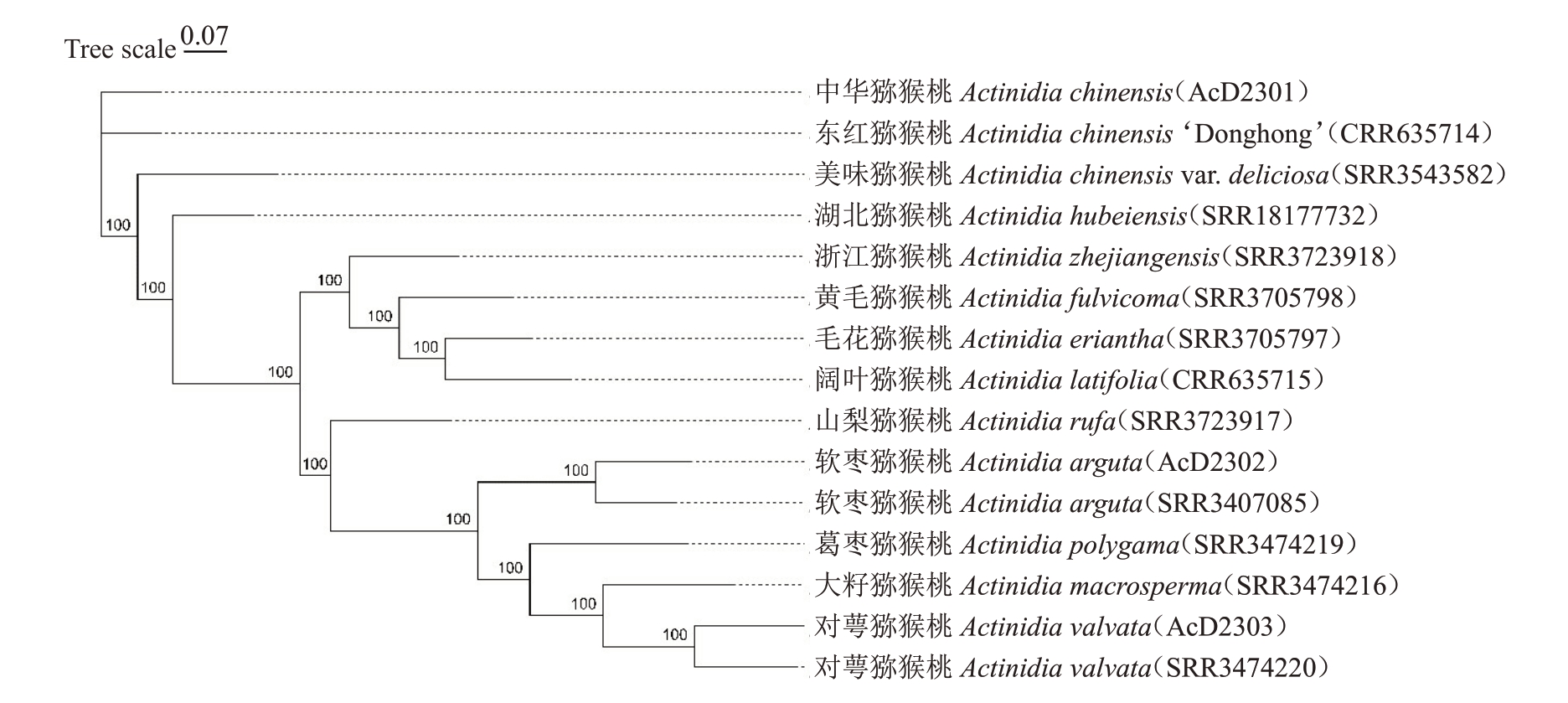
图5 猕猴桃属植物系统进化树
Fig.5 The phylogenetic tree of kiwifruit plants
系统进化树构建中下载并使用已公布的猕猴桃二代测序数据有东红猕猴桃、阔叶猕猴桃(https://ngdc.cncb.ac.cn);美味猕猴桃、湖北猕猴桃、浙江猕猴桃、黄毛猕猴桃、毛花猕猴桃、山梨猕猴桃、软枣猕猴桃、葛枣猕猴桃、大籽猕猴桃、对萼猕猴桃(https://www.ncbi.nlm.nih.gov)。
In the construction of phylogenetic tree,the published next-generation sequencing data of kiwifruit were downloaded and used,including Actinidia chinensis‘Donghong’,Actinidia latifolia (https://ngdc.cncb.ac.cn);Actinidia chinensis var.deliciosa,Actinidia hubeiensis,Actinidia zhejiangensis,Actinidia fulvicoma,Actinidia eriantha,Actinidia rufa,Actinidia arguta,Actinidia polygama,Actinidia macrosperma, and Actinidia valvata (https://www.ncbi.nlm.nih.gov/).
3 讨 论
多倍化是推动植物遗传多样性和适应环境变化的重要机制之一,在植物中广泛存在,其中猕猴桃属植物中多倍化现象非常普遍。猕猴桃多倍化表现为体细胞均增大、果形更加圆润饱满,叶片颜色更深、表皮毛被明显增多、产量高、抗性强等特征[3-5]。尽管以往的研究已经从很大程度上揭示了猕猴桃属物种的基因组信息以及主要倍性,但仍有部分物种尚未明确。本研究中就基于流式细胞术分析AcD2301、AcD2302、AcD2303的染色体倍性分别为四倍体、四倍体和六倍体。而基于猕猴桃基因组Survey 数据分析所得AcD2301 同源四倍体支持率为97%,AcD2302 同源四倍体支持率为96.7%,与上述结果基本一致。以上工作为进一步丰富猕猴桃物种基因组奠定了基础。
基于测序技术解析全基因组信息,为植物起源、进化、生殖、发育、抗性和性别调控等研究提供了基础。不同种类的植物基因组大小相差很大,根据目前已经公布的基因组数据中梅溪蕨(Tmesipteris oblanceolata)的基因组大小约160.45 Gb,而旋刺草(Genlisea aurea)的基因组大小仅为约0.063 6 Gb,相差约2500倍[23]。目前主要采用流式细胞术和高通量测序技术等方法评估植物的基因组大小,例如在四数九里香[24]、白及[25]、荆芥[26]等多种植物基因组大小特征评估中都有应用。流式细胞术是通过测量细胞中DNA与荧光染料结合后发出的荧光信号强度,来间接预估基因组大小的相对值,而基因组Survey分析是利用高通量测序技术对植物基因组进行测序和直接获取基因组大小等信息的测序技术,这两种技术结合起来评估基因组大小和特征相对可靠[27]。已报道猕猴桃属植物的基因组大小通常在600 Mb左右,中华猕猴桃为610.1 Mb[7],毛花猕猴桃为619.3 Mb和611.7 Mb[8]等,本研究结果所揭示的单套基因组大小较为相近,AcD2301 为626 Mb、AcD2302 为668 Mb、AcD2303 为585 Mb,均都在600 Mb 左右,但基因组具体大小又取决于不同的种质资源。
基因组学研究还可以揭示物种的遗传多样性、基因组演化历程以及基因功能等,通过构建系统进化树可以直观地展现亲缘关系和进化历程[28]。已有研究通过UPGMA聚类分析得到星毛组的中华猕猴桃与净果组的软枣猕猴桃亲缘关系较远[29],并且与净果组的对萼猕猴桃亲缘关系也较远[30],由此推测同为净果组的软枣猕猴桃和对萼猕猴桃亲缘关系较近,并均与星毛组的中华猕猴桃亲缘关系较远。本研究中构建的猕猴桃属植物系统进化树,证明了软枣猕猴桃AcD2302 与对萼猕猴桃AcD2303 亲缘关系较近,且均与中华猕猴桃AcD2301独立进化而来的结果一致,为阐明物种进化关系及基因组的内在结构奠定了基础。
4 结 论
中华猕猴桃AcD2301、软枣猕猴桃AcD2302、对萼猕猴桃AcD2303 的染色体倍性分别为四倍体、四倍体和六倍体,与全基因组测序预估结果一致;基于全基因组Survey 分析预测基因组大小分别为626 Mb、668 Mb、585 Mb,杂合度为3.00%、3.30%、8.06%,重复序列比例为43.70%、45.30%、40.7%。SNP 系统进化树显示软枣猕猴桃AcD2302 与对萼猕猴桃AcD2303亲缘关系较近,且均与中华猕猴桃AcD2301独立进化而来。
[1] 黄宏文.猕猴桃属分类资源驯化栽培[M].北京:科学出版社,2013.HUANG Hongwen. Actinidia taxonomy germplasm domestication cultivation[M].Beijing:Science Press,2013.
[2] 钟彩虹.猕猴桃栽培理论与生产技术[M].北京:科学出版社,2020.ZHONG Caihong.Cultivation theory and production technology of kiwifruit[M].Beijing:Science Press,2020.
[3] 刘彩艳,李大卫,杨石建,潘志立,金若涵,陈芳,郭雯.不同倍性猕猴桃的适生区预测及生态位分化[J].应用生态学报,2021,32(9):3167-3176.LIU Caiyan,LI Dawei,YANG Shijian,PAN Zhili,JIN Ruohan,CHEN Fang,GUO Wen.Potential suitable area and niche shift of different ploidy kiwifruit[J].Chinese Journal of Applied Ecology,2021,32(9):3167-3176.
[4] 金若涵.不同倍性猕猴桃木质部解剖结构和水分传导功能关系的研究[D].昆明:云南大学,2022.JIN Ruohan.Relationship between xylem anatomical structure and hydraulic function of kiwifruit with different ploidy levels[D].Kunming:Yunnan University,2022.
[5] 李志,方金豹,齐秀娟,林苗苗,陈锦永,顾红.不同倍性雄株对软枣猕猴桃坐果及果实性状的影响[J].果树学报,2016,33(6):658-663.LI Zhi,FANG Jinbao,QI Xiujuan,LIN Miaomiao,CHEN Jinyong,GU Hong.Effects of male plants with different ploidy on the fruit set and fruit characteristics in Actinidia arguta kiwifruit[J].Journal of Fruit Science,2016,33(6):658-663.
[6] 李文萃.中华猕猴桃和多倍体美味猕猴桃的抗冷基因表达及进化研究[D].北京:中国环境科学研究院,2024.LI Wencui.Study on the expression and evolution of cold resistance genes in Actinidia chinensis and polyploid Actinidia deliciosa[D].Beijing:Chinese Research Academy of Environmental Sciences,2024.
[7] HUANG S X,DING J,DENG D J,TANG W,SUN H H,LIU D Y,ZHANG L,NIU X L,ZHANG X,MENG M,YU J D,LIU J,HAN Y,SHI W,ZHANG D F,CAO S Q,WEI Z J,CUI Y L,XIA Y H,ZENG H P,BAO K,LIN L,MIN Y,ZHANG H,MIAO M,TANG X F,ZHU Y Y,SUI Y,LI G W,SUN H J,YUE J Y,SUN J Q,LIU F F,ZHOU L Q,LEI L,ZHENG X Q,LIU M,HUANG L,SONG J,XU C H,LI J W,YE K Y,ZHONG S L,LU B R,HE G H,XIAO F M,WANG H L,ZHENG H K,FEI Z J,LIU Y S.Draft genome of the kiwifruit Actinidia chinensis[J].Nature Communications,2013,4:2640.
[8] WANG Y Z,DONG M H,WU Y,ZHANG F,REN W M,LIN Y Z,CHEN Q Y,ZHANG S J,YUE J Y,LIU Y S.Telomere-totelomere and haplotype-resolved genome of the kiwifruit Actinidia eriantha[J].Molecular Horticulture,2023,3(1):4.
[9] HAN X,ZHANG Y L,ZHANG Q,MA N,LIU X Y,TAO W J,LOU Z Y,ZHONG C H,DENG X W,LI D W,HE H.Two haplotype-resolved,gap-free genome assemblies for Actinidia latifolia and Actinidia chinensis shed light on the regulatory mechanisms of vitamin C and sucrose metabolism in kiwifruit[J].Molecular Plant,2023,16(2):452-470.
[10] AKAGI T,VARKONYI-GASIC E,SHIRASAWA K,CATANACH A,HENRY I M,MERTTEN D,DATSON P,MASUDA K,FUJITA N,KUWADA E,USHIJIMA K,BEPPU K,ALLAN A C,CHARLESWORTH D,KATAOKA I.Recurrent neo-sex chromosome evolution in kiwifruit[J].Nature Plants,2023,9(3):393-402.
[11] LU X M,YU X F,LI G Q,QU M H,WANG H,LIU C,MAN Y P,JIANG X H,LI M Z,WANG J,CHEN Q Q,LEI R,ZHAO C C,ZHOU Y Q,JIANG Z W,LI Z Z,ZHENG S,DONG C,WANG B L,SUN Y X,ZHANG H Q,LI J W,MO Q H,ZHANG Y,LOU X,PENG H X,YI Y T,WANG H X,ZHANG X J,WANG Y B,WANG D,LI L,ZHANG Q,WANG W X,LIU Y B,GAO L,WU J H,WANG Y C.Genome assembly of autotetraploid Actinidia arguta highlights adaptive evolution and enables dissection of important economic traits[J].Plant Communications,2024,5(6):100856.
[12] QI X Q,XIE X D,YANG M J,ATAK A,ZHONG C H,LI D W.Characterization of the complete chloroplast genome of Actinidia hemsleyana[J].Mitochondrial DNA.Part B,Resources,2021,6(11):3259-3260.
[13] 孙雷明,方金豹.我国猕猴桃种质资源的保存与研究利用[J].植物遗传资源学报,2020,21(6):1483-1493.SUN Leiming,FANG Jinbao.Conservation,research and utilization of kiwifruit germplasm resources in China[J].Journal of Plant Genetic Resources,2020,21(6):1483-1493.
[14] 滕晓梅,廖川江,刘畅,徐雨生.葡萄叶猕猴桃(猕猴桃科)叶绿体全基因组测序[J].种子科技,2021,39(21):23-25.TENG Xiaomei,LIAO Chuanjiang,LIU Chang,XU Yusheng.Whole genome sequencing of chloroplasts from Actinidia chinensis(Actinidiaceae)[J].Seed Science&Technology,2021,39(21):23-25.
[15] 王连润,陶磅,万红,陈霞,李坤明,沙毓沧,丁仁展.云南4 种野生猕猴桃基因组大小测定与比较[J].中国南方果树,2022,51(4):100-103.WANG Lianrun,TAO Bang,WAN Hong,CHEN Xia,LI Kunming,SHA Yucang,DING Renzhan.Detection and comparison of genome size of four wild kiwifruit varieties from Yunnan[J].South China Fruits,2022,51(4):100-103.
[16] 魏远新,郭兴利,张振营,孙继中,李金良,李书林.对萼猕猴桃新型砧木及其绿枝嫁接关键技术[J].北方果树,2023(6):35-36.WEI Yuanxin,GUO Xingli,ZHANG Zhenying,SUN Jizhong,LI Jinliang,LI Shulin.New rootstock of Actinidia chinensis and key technology of green branch grafting[J].Northern Fruits,2023(6):35-36.
[17] 吕雅雅,白事麟,韩宜洁,邱娟,师小军.基于流式细胞术对棱叶韭染色体的倍性鉴定和基因组大小分析[J/OL].分子植物育种,2024(2024-03-19)[2024-09-11].https://link.cnki.net/urlid/46.1068.S.20240318.1332.006.LÜ Yaya,BAI Shilin,HAN Yijie,QIU Juan,SHI Xiaojun.Estimation ploidy and genome size of Allium caeruleum Pall.by flow cytometry[J/OL].Molecular Plant Breeding,2024(2024-03-19)[2024-09-11].https://link.cnki.net/urlid/46.1068.S.20240-318.1332.006.
[18] CHEN S F,ZHOU Y Q,CHEN Y R,GU J.Fastp:An ultra-fast all-in-one FASTQ preprocessor[J].Bioinformatics,2018,34(17):i884-i890.
[19] MARÇAIS G,KINGSFORD C.A fast,lock-free approach for efficient parallel counting of occurrences of k-mers[J].Bioinformatics,2011,27(6):764-770.
[20] VURTURE G W,SEDLAZECK F J,NATTESTAD M,UNDERWOOD C J,FANG H,GURTOWSKI J,SCHATZ M C.GenomeScope:Fast reference-free genome profiling from short reads[J].Bioinformatics,2017,33(14):2202-2204.
[21] YUE J Y,CHEN Q Y,WANG Y Z,ZHANG L,YE C,WANG X,CAO S,LIN Y Z,HUANG W,XIAN H,QIN H Y,WANG Y L,ZHANG S J,WU Y,WANG S H,YUE Y,LIU Y S.Telomere-to-telomere and gap-free reference genome assembly of the kiwifruit Actinidia chinensis[J].Horticulture Research,2023,10(2):uhac264.
[22] ZHOU Y,KATHIRESAN N,YU Z C,RIVERA L F,YANG Y J,THIMMA M,MANICKAM K,CHEBOTAROV D,MAULEON R,CHOUGULE K,WEI S,GAO T T,GREEN C D,ZUCCOLO A,XIE W B,WARE D,ZHANG J W,MCNALLY K L,WING R A.A high-performance computational workflow to accelerate GATK SNP detection across a 25-genome dataset[J].BMC Biology,2024,22(1):13.
[23] FERNÁNDEZ P,AMICE R,BRUY D,CHRISTENHUSZ M J M,LEITCH I J,LEITCH A L,POKORNY L,HIDALGO O,PELLICER J.A 160 Gbp fork fern genome shatters size record for eukaryotes[J].iScience,2024,27(6):109889.
[24] 邢琴琴,周韬,卜家豪,沈植国,韩文军.四数九里香全基因组Survey 分析[J/OL].分子植物育种,2023(2023-12-08)[2024-09-11].https://link.cnki.net/urlid/46.1068.S.20231207.1357.012.XING Qinqin,ZHOUTao,BU Jiahao,SHEN Zhiguo,HANWenjun.Genome Survey Analysis in Murraya tetramera Huang[J/OL].Molecular Plant Breeding,2023(2023-12-08)[2024-09-11].https://link.cnki.net/urlid/46.1068.S.20231207.1357.012.
[25] 杨渊,黄明进,王大昌,阮宝丽,杨秋悦,杨洋,罗影子,覃玉强.基于流式细胞术与基因组Survey 分析白及基因组大小及特征[J].中成药,2023,45(11):3677-3682.YANG Yuan,HUANG Mingjin,WANG Dachang,RUAN Baoli,YANG Qiuyue,YANG Yang,LUO Yingzi,QIN Yuqiang.Analysis of genome size and characteristics of Bletilla striata based on flow cytometry and genomic survey[J].Chinese Traditional Patent Medicine,2023,45(11):3677-3682.
[26] 姜涛,刘灵娣,田伟,刘铭,温春秀.基于流式细胞术和K-mer分析的荆芥基因组大小评估[J].特产研究,2023,45(6):1-5.JIANG Tao,LIU Lingdi,TIAN Wei,LIU Ming,WEN Chunxiu.Genome-determination of Nepeta cataria based on flow cytometry and K-mer analysis[J].Special Wild Economic Animal and Plant Research,2023,45(6):1-5.
[27] 饶静云,刘义飞,黄宏文.中华猕猴桃不同倍性间杂交后代倍性分离和遗传变异分析[J].园艺学报,2012,39(8):1447-1456.RAO Jingyun,LIU Yifei,HUANG Hongwen.Analysis of ploidy segregation and genetic variation of progenies of different interploidy crosses in Actinidia chinensis[J].Acta Horticulturae Sinica,2012,39(8):1447-1456.
[28] LIU Y B,ZHOU Y,CHENG F,ZHOU R C,YANG Y Q,WANG Y C,ZHANG X T,SOLTIS D E,XIAO N W,QUAN Z J,LI J S.Chromosome-level genome of putative autohexaploid Actinidia deliciosa provides insights into polyploidisation and evolution[J].The Plant Journal,2024,118(1):73-89.
[29] 刘娟,廖明安,谢玥,周良强,李明章.猕猴桃属16 个雄性材料遗传多样性的ISSR 分析[J].植物遗传资源学报,2015,16(3):618-623.LIU Juan,LIAO Ming’an,XIE Yue,ZHOU Liangqiang,LI Mingzhang.Genetic diversity of 16 male Actinidia cultivars based on ISSR[J].Journal of Plant Genetic Resources,2015,16(3):618-623.
[30] 张慧,张世鑫,吴绍华,田维敏,彭小列,刘世彪.猕猴桃属33份种质资源的AFLP 遗传多样性分析[J].生物学杂志,2018,35(2):29-33.ZHANG Hui,ZHANG Shixin,WU Shaohua,TIAN Weimin,PENG Xiaolie,LIU Shibiao.Genetic diversity of 33 kiwifruit germplasms based on AFLP markers[J].Journal of Biology,2018,35(2):29-33.