脂肪酸是植物细胞的重要组成成分,它形成甘油三酯为生命活动提供能量[1];作为细胞膜的关键成分维持细胞膜的稳态[2];在生物胁迫中参与激素调节与信号转导[3];在响应低温和干旱等逆境胁迫中发挥重要作用[4-5];在果实成熟过程中通过LOX途径、α-氧化途径或β-氧化途径参与合成香气物质[6]。植物中以不饱和脂肪酸为主,饱和脂肪酸转化成不饱和脂肪酸的过程称为去饱和,是由一系列脂肪酸去饱和酶(fatty acid desaturases,FADs)来完成的[7]。FADs 根据溶解度可分为可溶性去饱和酶和膜结合去饱和酶。硬脂酰ACP去饱和酶(FAB2/SAD)是质体基质中唯一已知的可溶性FAD,它在Δ-9位置(从羧基端开始的第9号碳与10号碳之间)引入双键,催化硬脂酸转化为油酸,剩余的FAD 均是膜结合的[8]。根据功能不同,膜结合FADs 又进一步分为FAD2/FAD6(ω-6/Δ-12)、FAD3/FAD7/FAD8(ω-3/Δ-15)、FAD4(Δ-3)、FAD5/ADS(Δ-7)和DES/SLD五大亚族。其中,FAD2 和FAD6 是ω-6/Δ-12 去饱和酶,即FAD2在内质网、FAD6在质体中于油酸的Δ-12位置,也就是ω-6 位置(甲基端开始的第6 号碳与7 号碳之间)插入双键生成亚油酸[9];FAD3 在内质网、FAD7 和FAD8 在质体中于亚油酸的Δ-15 位置(ω-3位置)插入双键生成亚麻酸[10];FAD4 和FAD5/ADS都在质体中分别作用于磷脂酰甘油和单半乳糖二酰基甘油中的棕榈酸合成棕榈油酸[11];DES 和SLD 都是在内质网中参与脂肪酸衍生物鞘脂代谢的酶,SLD催化t18∶0鞘脂长链碱基(LCB)C8位去饱和生成t18∶1 鞘脂[12],DES 催化二氢鞘氨醇(d18:0)生成鞘氨醇-1-磷酸(d18:1),DES通常不会单独起作用,而是和SLD共同参与鞘脂代谢[13]。
1992 年有学者先后从拟南芥突变体中克隆得到FAD3 和FAD2[14-15],1993 年和1994 年从拟南芥中先后克隆出FAD7 和FAD8[16],1993 年从蓝藻中克隆出FAD6[17],SLD 最先于1998 年从向日葵中分离得到[12]。目前研究比较多的是ω-6和ω-3型FAD基因,已经在香蕉[18]、茄子[19]、番茄[20]等中分离出其家族成员。当前对FAD 基因研究热点主要集中在抗逆功能,尤其是低温冷害胁迫,大量研究证实了脂肪酸去饱和酶能够调整膜脂不饱和脂肪酸的比例和成分,从而维持细胞膜流动性,达到增强对低温抵抗力的目的[21]。然而,关于FAD 基因在果实成熟中的功能鲜有报道。
果实中的挥发性芳香物质主要通过脂肪酸代谢途径、萜类代谢途径和氨基酸代谢途径合成。其中,直链脂肪族醇、醛、酮和酯类物质主要来源于脂肪酸的代谢,因此脂肪酸是形成果实香气物质的主要前体物质[22]。脂肪酸代谢途径中对香气物质合成贡献最大的是不饱和脂肪酸的代谢,这是通过LOX酶催化的第一步和随后的反应,通常被称为脂氧合酶途径。脂肪酸去饱和酶FAD 在脂肪酸碳氢链上引入双键,产生不饱和脂肪酸,在将油酸(18∶1)转化为亚油酸(18∶2)和亚麻酸(18∶3)过程中发挥着关键的作用[23]。
猕猴桃果实中因富含有机物质以及人体所必需的多种维生素而备受消费者青睐。猕猴桃在发育阶段(未成熟阶段)主要产生C6 醇和醛等“青香型”芳香物质,但在采收后熟过程中酯类等“果香型”芳香物质增多,因此常会散发出成熟阶段特有的果香味,也影响着消费者的偏好[24]。目前关于猕猴桃采后成熟过程中的香气研究较多,主要集中在贮藏处理方式、成熟度和品种等因素对其香气物质化学成分种类和含量比例的影响方面[25-26],香气脂肪酸代谢合成途径的相关酶基因(如LOX、AAT、ADH等)也已被分离鉴定出来[27-29]。然而对其合成前体物质不饱和脂肪酸的研究鲜有报道,目前发现低温贮藏延缓了猕猴桃果实中亚油酸和亚麻酸的分解,抑制了脂肪酸LOX代谢途径中相关酶活性及其基因的表达,从而造成酯类香气物质的种类减少和相对含量降低,保持了较多醛酮类物质种类和较高的相对含量,维持了果实的特征风味[30],其他相关的研究更多集中在猕猴桃籽和果肉中脂肪酸组分方面,发现其富含多种不饱和脂肪酸[31-32],猕猴桃FAD 基因家族也未进行分离鉴定。红阳猕猴桃属于中华猕猴桃,因其果心具放射状红色条纹而得名,味甜,维生素C含量极高,香气浓郁[33]。因此,笔者以红阳猕猴桃为研究试材,采用生物信息学的方法对猕猴桃FAD基因家族成员进行分离和鉴定,然后再基于转录组数据对各成员在果实采后成熟过程中的表达模式进行分析,最后通过实时荧光定量PCR 验证FADs 基因的表达特性并筛选出差异表达的候选基因,为解析FAD 基因在猕猴桃果实采后成熟过程中不饱和脂肪酸代谢和香气物质形成的生物学功能提供一定的理论基础。
1 材料和方法
1.1 植物材料
材料为红阳猕猴桃(Actinidia chinensis‘Hongyang’)果实。于2023 年8 月中旬(盛花后130 d 左右)从江西省奉新县果业局猕猴桃种质资源圃采摘试验果。选取树龄相同、长势良好的母树,采摘大小均匀、果形一致、无病虫害和机械损伤的果实,采摘当天立即运回实验室,经人工挑选剔除癍痂、伤果后置于阴凉通风处发汗到次日。经发汗散除田间热的果实用塑料袋密封贮藏于室温(20±1)℃中,于贮藏后0、4、10 d取样,每次随机挑选18个果实,6个果实为1次重复,设置3次生物学重复,经测量果肉硬度等成熟指标后去掉头部和尾部,剥除果皮、中柱和籽,取果肉部分混合均匀经液氮速冻后保存于-80 ℃备用。
1.2 猕猴桃FAD 基因家族成员鉴定及其蛋白特性分析
从猕猴桃基因组网站(https://kiwifruitgenome.org/)下载Hong Yang v3基因组数据,包括基因序列数据文件、蛋白序列数据文件、GFF3 注释文件等。从Pfam蛋白家族数据库(http://pfam.xfam.org/)下载FA_去饱和酶(PF00487)、FA_去饱和酶2(PF03405)和TMEM189(PF10520)结构域对应的隐马尔科夫模型(HMM)文件,使用TBtools(v2.102)中的Simple HMM Search,以e值≤1e-5为判据,与猕猴桃蛋白数据进行比对,初步筛选出AcFAD基因。然后使用SMART 数据库(https://smart.embl-heidelberg.de/)对初筛得到的候选蛋白序列进行结构域信息验证,最终确定了26 个猕猴桃FAD 基因家族成员。在猕猴桃基因组网站中获得这26 个AcFADs 编码区长度。利用蛋白分子质量计算-SMS2南京德泰生物镜像网站(https://www.detaibio.com/sms2/protein_mw.html)对26 个猕猴桃FAD 基因家族成员进行蛋白质理化性质分析,获得氨基酸数量、蛋白分子质量(MW)、等电点(pI)等。通过Cell-PLoc 2.0网站(http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/)中的Plant-mPLoc对AcFAD蛋白进行亚细胞定位预测。
1.3 猕猴桃FAD 基因家族多序列比对与系统发育分析
拟南芥和黄瓜的FAD 蛋白质序列来自Cheng等[18]和Dong 等[34]的报道,利用MEGA(version 11.0)软件中的ClustalW对猕猴桃、拟南芥和黄瓜的FAD蛋白序列进行多序列比对。系统发育分析采用邻接法(neighbor-joining,NJ),在p-distance、部分删除50%和1000 次Bootstrap 重复参数下构建系统发育树,并使用Evolview v2(https://evolgenius.info//evolview-v2/)对进化树进行美化。
1.4 猕猴桃FAD 基因家族染色体定位和共线性分析
根据猕猴桃和拟南芥的全基因组序列文件和GFF3注释文件,使用TBtools中的Fasta Stats提取猕猴桃全基因组染色体长度并输出染色体骨架文件,利用One Step MCScanX 获得全基因组共线性文件并据此文件使用Text Block Extract and Filter提取猕猴桃FAD 家族基因共线性文件,使用File Transformat for MicroSynteny Viewer 提取配对基因对染色体信息,最后使用Advanced Circos 绘制猕猴桃FAD染色体定位图。使用One Step MCScanX-Super Fast对猕猴桃和拟南芥FAD 基因的复制事件进行共线性分析,并使用Dual Systeny Plot作可视化图。
1.5 猕猴桃FAD基因结构与蛋白保守基序分析
通过基因结构显示服务器GSDS(http://gsds.gao-lab.org/)分析猕猴桃FAD基因的外显子-内含子基因结构。使用MEME 在线网站(https://memesuite.org/meme/tools/meme)分析AcFAD蛋白的保守基序,最大基序数设置为20,并利用TBtools 中的Gene Structure View绘制蛋白保守基序。
1.6 猕猴桃FAD基因家族启动子顺式作用元件分析
使用TBtools 从猕猴桃基因组数据库中提取每个AcFAD 基因起始密码子(ATG)上游1500 bp 启动子序列,利用PlantCARE(http://bioinformatics.psb.ugent.be/webtools/plantcare-/html/)预测各个基因的启动子顺式作用元件,再利用TBtools对获得的顺式作用元件进行可视化绘图。
1.7 AcFADs在果实采后成熟过程中的基因表达分析
将常温贮藏0、4、10 d猕猴桃果实冷冻样品分装3 管设置3次生物学重复,在上海派森诺生物科技有限公司完成转录组测序。将获得的转录组数据提取上述26个AcFAD基因的FPKM值,将log(2FPKM+1)归一化处理后得到的数据使用TBtools 生成基因表达热图。
1.8 果实硬度的测量
使用质构仪(TMS-Touch,美国)测定果实的硬度,每个取样点随机选取18 个果实,重复测量3 次,单位:N[35]。
1.9 果实脂肪酸含量的测定
将猕猴桃果肉冷冻样品粉末2 g 与正己烷∶异丙醇(3∶2,体积比)15 mL 和6.7%Na2SO4 7.5 mL 混合,10 000g 离心10 min(4 ℃),上清液用氮气吹干。加入甲醇∶甲苯∶H2SO(488∶10∶2,体积比)制备脂肪酸甲酯(FAMEs)。冷却后,加入1 mL 庚烷和0.5 g无水Na2SO4进行FAME提取。为了检测脂肪酸含量,使用安捷伦6890 N气相色谱仪,配备火焰电离检测器和DB-Wax 色谱柱(0.25 mm,30 m,0.25 μm;J&W Scientific)。注入器和检测器温度为230 ℃。初始烤箱温度为50 ℃,以25 ℃·min-1速率增加到200 ℃,然后以3 ℃·min-1速率增加到230 ℃。载气为氮气,流速为1 mL·min-1。加入外源十七烷酸(C17∶0)作为内标定量计算脂肪酸含量。
1.10 RNA 提取、cDNA 合成及实时荧光定量PCR分析
使用多糖多酚植物总RNA 提取试剂盒(天根,北京)提取猕猴桃果肉总RNA,每个样品设置3次技术重复,然后通过凝胶电泳和超微量分光光度计(Bio-Rad,美国)测定提取的R N A 的质量和浓度。将提取好的RNA 使用三代逆转录预混液Mon-ScriptTM RTIII All-in-One Mix with dsDNase(莫纳生物,苏州)反转录成第一链cDNA,并用ddH2O稀释3倍,最后采用半定量PCR 检测cDNA 的质量。通过WoLF PSORT 网站(https://www.genscript.com/wolfpsort.html)设计实时荧光定量引物(表1)。实时荧光定量PCR(qPCR)分析参照之前的研究方法[36]。
表1 实时荧光定量PCR 引物
Table 1 Primers for quantitative real-time PCR analysis
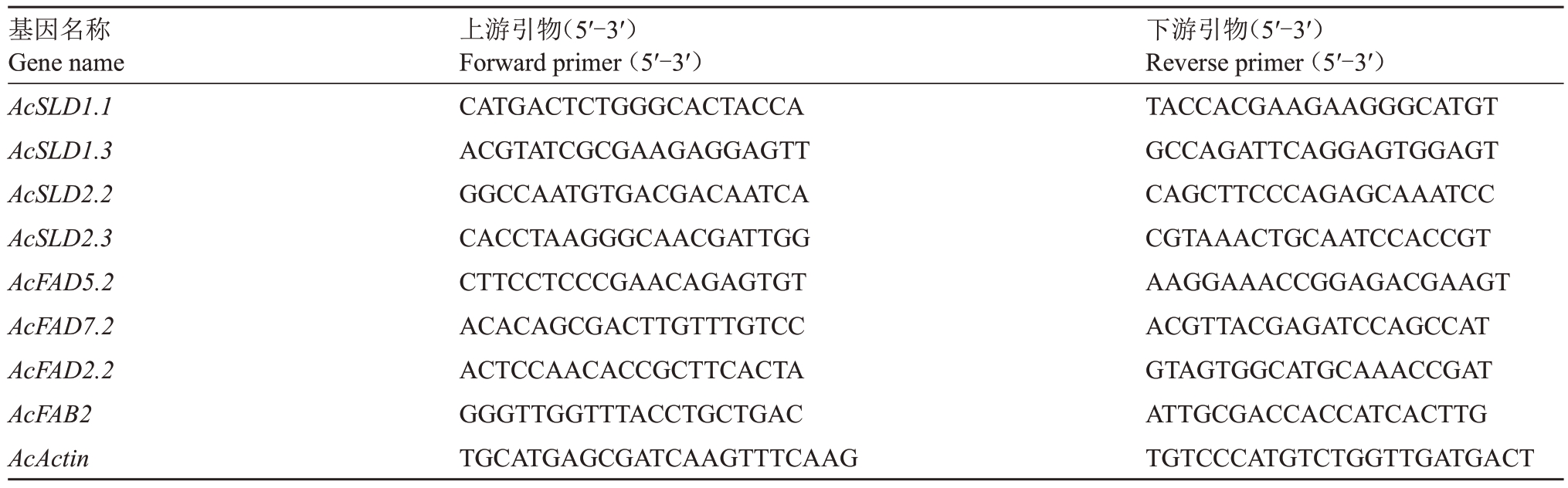
基因名称Gene name AcSLD1.1 AcSLD1.3 AcSLD2.2 AcSLD2.3 AcFAD5.2 AcFAD7.2 AcFAD2.2 AcFAB2 AcActin上游引物(5′-3′)Forward primer(5′-3′)CATGACTCTGGGCACTACCA ACGTATCGCGAAGAGGAGTT GGCCAATGTGACGACAATCA CACCTAAGGGCAACGATTGG CTTCCTCCCGAACAGAGTGT ACACAGCGACTTGTTTGTCC ACTCCAACACCGCTTCACTA GGGTTGGTTTACCTGCTGAC TGCATGAGCGATCAAGTTTCAAG下游引物(5′-3′)Reverse primer(5′-3′)TACCACGAAGAAGGGCATGT GCCAGATTCAGGAGTGGAGT CAGCTTCCCAGAGCAAATCC CGTAAACTGCAATCCACCGT AAGGAAACCGGAGACGAAGT ACGTTACGAGATCCAGCCAT GTAGTGGCATGCAAACCGAT ATTGCGACCACCATCACTTG TGTCCCATGTCTGGTTGATGACT
1.11 数据统计与分析
所有数据结果以均数±标准误差(SD)表示。采用SPSS Statistics 25 软件中的单因素方差检验进行显著性差异分析并使用Duncan 新复极差字母标记法标注显著性,最后使用GraphPad Prism 8.0绘制图表。
2 结果与分析
2.1 猕猴桃FAD 基因家族成员鉴定及其蛋白特性分析
笔者从Hong Yang v3 全基因组中共鉴定出26个FAD 基因家族成员(表2),根据其与拟南芥和黄瓜中FAD 基因亲缘关系的远近命名,包括2 个Ac-FAD2s,2 个AcFAD6s,3 个AcFAD3s,3 个AcFAD7s,1个AcFAB2,1个AcFAD4,3个AcFAD5s,2个AcADSs,7个AcSLDs和2个AcDESs。它们的编码序列(CDS)长度差异较大,范围为321 bp(AcFAD5.1)~4392 bp(AcFAD7.3),编码106~1463个氨基酸。蛋白质理论分子质量(MW)为11.51~161.8 ku,等电点为4.93(AcFAD5.1)~10.09(AcADS1.1),其中只有Ac-FAD7.3、AcFAD5.1 和AcSLD1.4 这3 个蛋白的pI 小于7,说明猕猴桃FAD蛋白大多数为碱性,少数为酸性。亚细胞定位预测结果表明,猕猴桃FAD蛋白定位比较分散,在植物细胞各结构中均有分布。其中AcFAD2蛋白都只定位在内质网中;AcFAD6蛋白只定位在叶绿体中;AcFAD3、AcFAD7 和AcADS 蛋白在叶绿体和内质网中都有定位;AcFAB2 定位在细胞膜和细胞核中;AcFAD4 定位在细胞膜、叶绿体、线粒体和细胞核中;AcFAD5.1 定位在细胞膜、叶绿体、高尔基体和细胞核中,另2 个AcFAD 只定位在内质网中;AcSLD1.1 定位在细胞膜和内质网中,其他AcSLD 只定位在内质网中;AcDES1.2 定位在细胞膜和内质网中,AcDES1.1只定位在内质网中。
表2 猕猴桃FAD 基因家族信息
Table 2 Information of FAD gene family members in kiwifruit
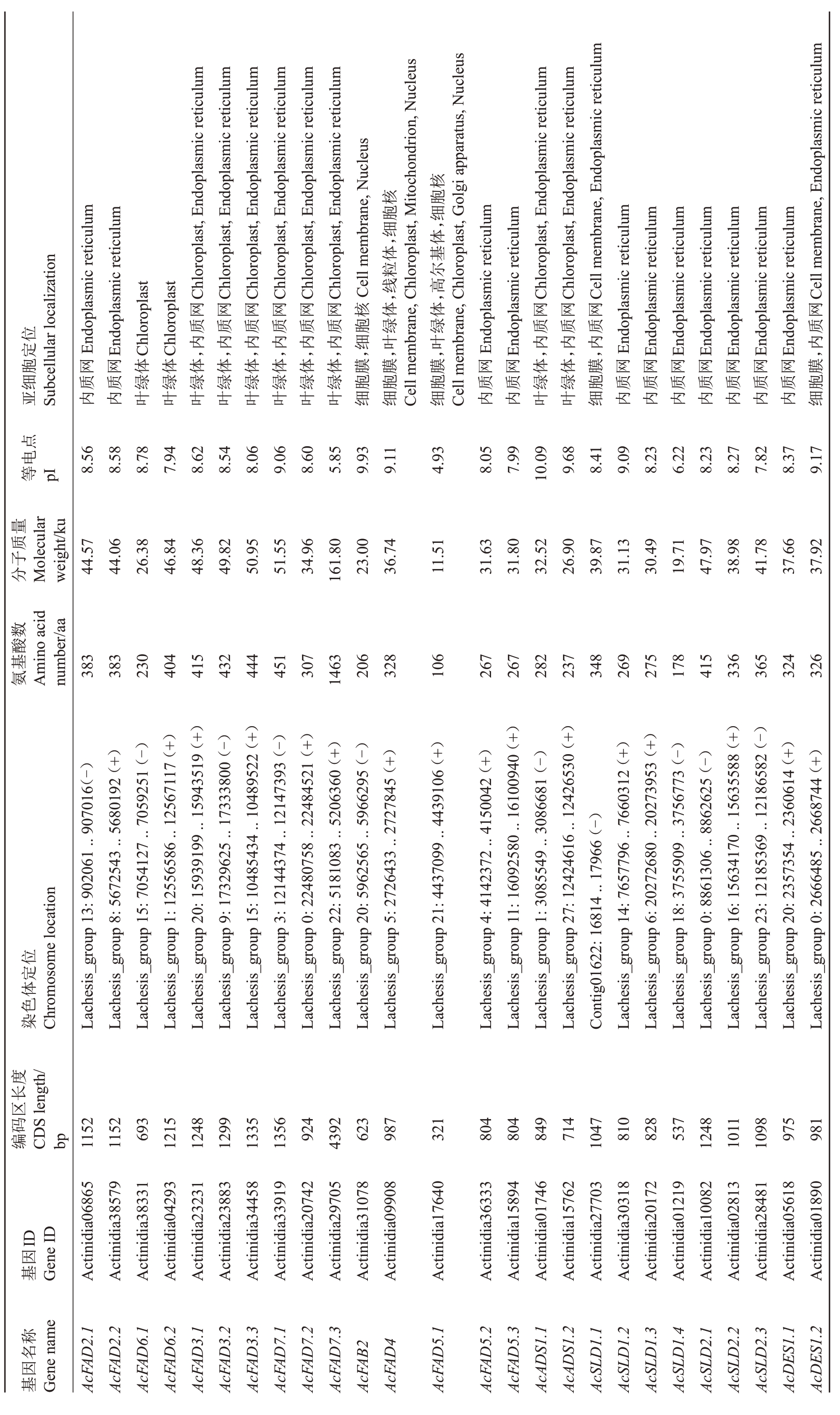
核Endoplasmic reticulum Endoplasmic reticulum 胞Chloroplast,Endoplasmic reticulum Chloroplast,Endoplasmic reticulum,细Chloroplast,Endoplasmic reticulum Chloroplast,Endoplasmic reticulum Chloroplast,Endoplasmic reticulum Chloroplast,Endoplasmic reticulum,细Cellmembrane,Nucleus细Cellmembrane,Chloroplast,Mitochondrion,Nucleus体核体胞亚Subcellular localization 基尔Chloroplast Chloroplast ,线粒,高Chloroplast,Endoplasmic reticulum Chloroplast,Endoplasmic reticulum Cellmembrane,Endoplasmic reticulum Cellmembrane,Endoplasmic reticulum网网网网网网核体体网网网网位质质质质质质胞绿绿质质质质定,内,内,内,内,内,内,细,叶,叶Endoplasmic reticulum Endoplasmic reticulum,内,内,内Endoplasmic reticulum Endoplasmic reticulum Endoplasmic reticulum Endoplasmic reticulum Endoplasmic reticulum Endoplasmic reticulum Endoplasmic reticulum,内胞网网体体体体体体体体膜膜膜网网体体膜网网网网网网网膜细质质绿绿绿绿绿绿绿绿胞胞胞质质绿绿胞质质质质质质质胞内内叶叶叶叶叶叶叶叶细细Cellmembrane,Chloroplast,Golgi apparatus,Nucleus内内叶叶细内内内内内内内细等pI8.56点电8.58 7.94 8.62 8.54 8.06 9.06 8.60 5.85 9.93 9.11 4.93 8.05 7.99 10.09 9.68 8.41 9.09 8.23 6.22 8.23 8.27 7.82 8.37 9.17分Molecular子量质weight/ku 44.57 44.06 26.38 46.84 48.36 49.82 50.95 51.55 34.96 161.80 23.00 36.74 11.51 31.63 31.80 32.52 26.90 39.87 31.13 30.49 19.71 47.97 38.98 41.78 37.66 37.92数8.78酸氨Amino acid基number/aa 383 383 230 404 415 432 444 451 307 1463 206 328 106 267 267 282 237 348 269 275 178 415 336 365 324 326染Chromosomelocation位定体色Lachesis_group13:902061..907016(-)Lachesis_group8:5672543..5680192(+)Lachesis_group15:7054127..7059251(-)Lachesis_group1:12556586..12567117(+)Lachesis_group20:15939199..15943519(+)Lachesis_group9:17329625..17333800(-)Lachesis_group15:10485434..10489522(+)Lachesis_group3:12144374..12147393(-)Lachesis_group0:22480758..22484521(+)Lachesis_group22:5181083..5206360(+)Lachesis_group20:5962565..5966295(-)Lachesis_group5:2726433..2727845(+)Lachesis_group21:4437099..4439106(+)Lachesis_group4:4142372..4150042(+)Lachesis_group11:16092580..16100940(+)Lachesis_group1:3085549..3086681(-)Lachesis_group27:12424616..12426530(+)Contig01622:16814..17966(-)Lachesis_group14:7657796..7660312(+)Lachesis_group6:20272680..20273953(+)Lachesis_group18:3755909..3756773(-)Lachesis_group0:8861306..8862625(-)Lachesis_group16:15634170..15635588(+)Lachesis_group23:12185369..12186582(-)Lachesis_group20:2357354..2360614(+)Lachesis_group0:2666485..2668744(+)编CDS length/度长区码bp1152 1152 693 1215 1248 1299 1335 1356 924 4392 623 987 321 804 804 849 714 1047 810 828 537 1248 1011 1098 975 981 ID基GeneID因Actinidia06865 Actinidia38579 Actinidia38331 Actinidia04293 Actinidia23231 Actinidia23883 Actinidia34458 Actinidia33919 Actinidia20742 Actinidia29705 Actinidia31078 Actinidia09908 Actinidia17640 Actinidia36333 Actinidia15894 Actinidia01746 Actinidia15762 Actinidia27703 Actinidia30318 Actinidia20172 Actinidia01219 Actinidia10082 Actinidia02813 Actinidia28481 Actinidia05618 Actinidia01890基Genename称名因AcFAD2.1 AcFAD2.2 AcFAD6.1 AcFAD6.2 AcFAD3.1 AcFAD3.2 AcFAD3.3 AcFAD7.1 AcFAD7.2 AcFAD7.3 AcFAB2 AcFAD4 AcFAD5.1 AcFAD5.2 AcFAD5.3 AcADS1.1 AcADS1.2 AcSLD1.1 AcSLD1.2 AcSLD1.3 AcSLD1.4 AcSLD2.1 AcSLD2.2 AcSLD2.3 AcDES1.1 AcDES1.2
2.2 猕猴桃FAD基因的系统进化树分析
为了阐明AcFAD蛋白的功能和进化关系,将26个猕猴桃、19个拟南芥和22个黄瓜的FAD蛋白序列构建了系统发育树。这些FAD 可分为6 个亚族,包括FAD3/FAD7/FAD8(ω-3/Δ-15)、FAD2/FAD6(ω-6/Δ-12)、FAB2(Δ-9)、FAD4(Δ-3)、DES/SLD 和FAD5/ADS(Δ-7),每个亚族分别有6、4、1、1、9 和5 个Ac-FAD成员(图1)。各亚族均有猕猴桃FAD家族成员的分布暗示着AcFAD 蛋白可能具有功能上的多样性。在大多数亚族中猕猴桃与拟南芥亲缘关系较近,与黄瓜的亲缘关系较远。
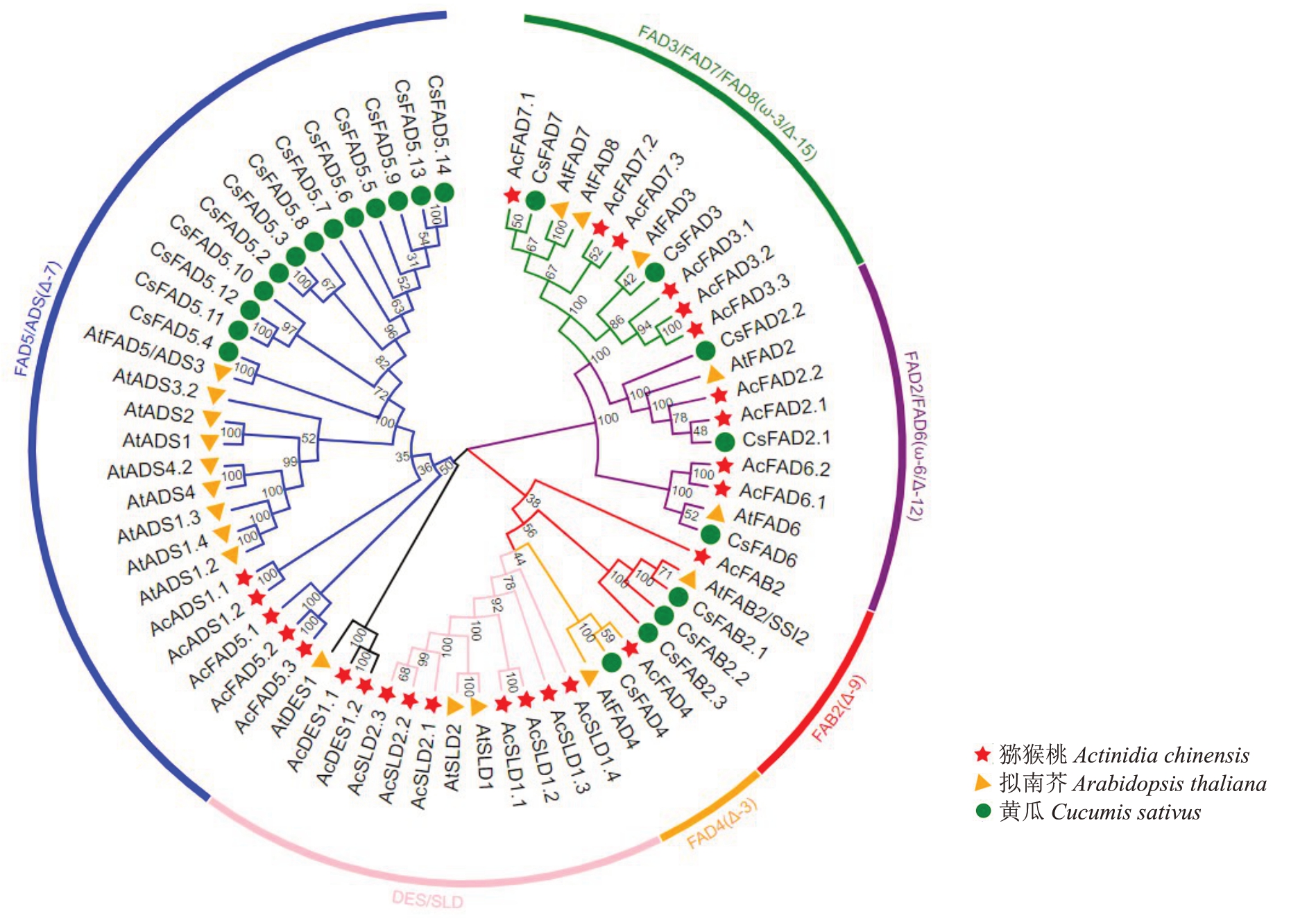
图1 猕猴桃与其他物种FAD 基因家族成员的系统发育树
Fig.1 Phylogenetic tree of the members of the FAD gene family among kiwifruit and other species
2.3 猕猴桃FAD基因的染色体定位和共线性分析
为了研究猕猴桃FAD 基因家族的遗传差异,分析了AcFADs 的染色体定位。结果(图2)显示猕猴桃Hong Yang v3基因组共有29条染色体,25个猕猴桃FAD基因分布在19条不同的染色体(LG)上,1个猕猴桃FAD 基因(AcSLD1.1)分布在Contig(CG)01622重叠群上。大多数FAD基因定位于染色体的前中端。0 和20 号染色体上都分布着3 个FAD 基因,1和15号染色体有2个FAD基因,剩下的15条染色体上(3、4、5、6、8、9、11、13、14、16、18、21、22、23和27 号)都只有1 个FAD 基因。由此可见,猕猴桃FAD基因和染色体存在一定的关联性。
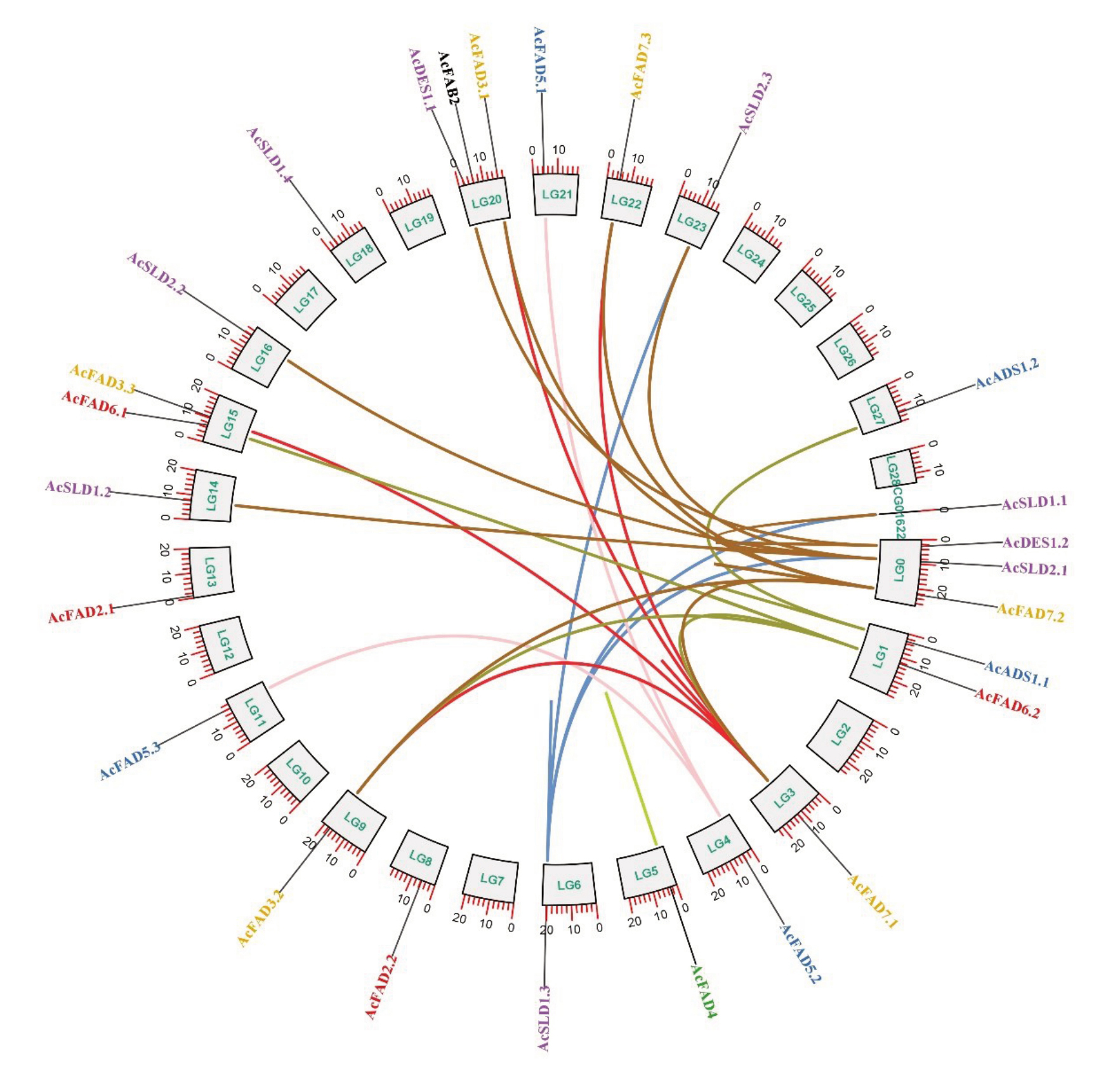
图2 猕猴桃FAD 基因家族染色体定位及种内共线性分析
Fig.2 Chromosomal localization and intraspecific covariance analysis of kiwifruit FAD gene family
不同颜色标注的基因代表猕猴桃FAD 家族6 大亚族内的成员,用不同颜色标记的线条代表基因成员具有共线性(封闭的线条和开放的线条分别表示基因间的片段和串联重复基因对),无线条标记的基因缺乏共线性。LG(Lachesis_group)代表染色体,CG(Contig)代表重叠群,即由一系列重叠的DNA 片段经过拼接后形成的连续序列,代表基因组中的一个特定区域。
Genes labeled in different colors represent members within the six subfamilies of the kiwifruit FAD family, lines labeled in different colors represent gene members with covariance (closed and open lines indicate intergenic segments and tandemly duplicated pairs of genes, respectively), and genes labeled without lines lack covariance.LG(Lachesis_group)stands for chromosome,and CG(Contig)stands for overlapping group,which is a contiguous sequence formed by a series of overlapping DNA segments that have been spliced together to represent a specific region of the genome.
为了进一步了解FAD基因家族的进化关系,进行了猕猴桃物种内以及猕猴桃与拟南芥物种间的共线性分析。种内共线性图谱显示有9对串联重复基因,分别是LG0 中的AcDES1.2、AcFAD7.2 和Ac-SLD2.1;LG1 的AcFAD6.2 和AcADS1.1;LG3 的Ac-FAD7.1;LG4 的AcFAD5.2;LG5 的AcFAD4 和Ac-SLD1.3。此外还鉴定出22 个猕猴桃FAD 基因产生了22 对片段重复基因,其中AcDES1.2(LG0)与AcDES1.1(LG20)存在共线性;AcFAD7.2(LG0)分别与AcFAD7.1(LG3)、AcFAD7.3(LG22)、AcFAD3.1(LG20)、AcFAD3.2(LG9)存在共线性;AcSLD2.1(LG0)与AcSLD2.3(LG23)、AcSLD1.1(CG01622)、AcSLD2.2(LG16)、AcSLD1.2(LG14)存在共线性;AcFAD6.2(LG1)与AcFAD6.1(LG15)、AcFAD7.1(LG3)、AcFAD3.2(LG9)存在共线性;AcADS1.1(LG1)与AcADS1.2(LG27)存在共线性;AcFAD7.1(LG3)与AcFAD3.1(LG20)、AcFAD3.2(LG9)、Ac-FAD7.3(LG22)、AcFAD3.3(LG15)存在共线性;Ac-FAD5.2(LG4)与 AcFAD5.3(LG11)、AcFAD5.1(LG21)存在共线性;AcSLD1.3(LG6)与AcSLD2.1(LG0)、AcSLD2.3(LG23)、AcSLD1.1(CG01622)存在共线性(图2)。由此可见,猕猴桃FAD 基因家族的串联重复事件伴随有片段重复事件,所以基因重复事件(串联重复和片段重复)对AcFADs 的多样性和进化具有促进作用。猕猴桃与拟南芥物种间共线性图谱显示,23 个猕猴桃FAD 基因与17 个拟南芥FAD 基因之间存在32 对共线性关系(图3),说明猕猴桃和拟南芥的FAD同源基因较多。
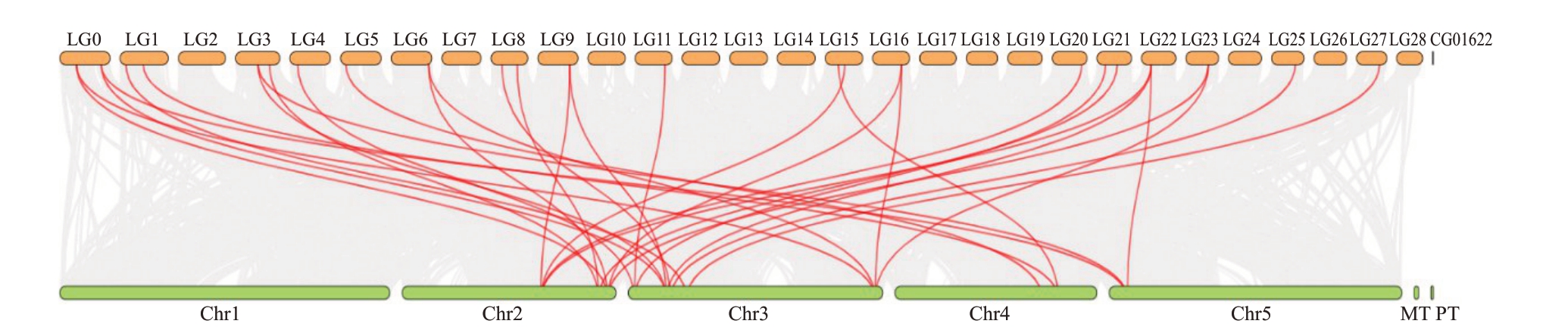
图3 猕猴桃与拟南芥FAD 基因的共线性分析
Fig.3 The collinearity analysis of FAD genes between kiwifruit and Arabidopsis
MT 和PT 分别代表哥伦比亚生态型拟南芥Col-0 线粒体全基因组和叶绿体全基因组。
MT and PT represent the Columbia ecotype Arabidopsis thaliana Col-0 mitochondrial whole genome and chloroplast whole genome,respectively.
2.4 猕猴桃FAD基因结构与蛋白保守基序分析
为了深入了解AcFADs结构的异同,提取猕猴桃FAD 基因家族成员的CDS 序列及其基因组序列绘制了外显子-内含子基因结构图(图4),结果显示内含子数量在0~33 个之间,外显子数量在1~34 个之间(一般比对应内含子数目多1 个),其中FAD3/FAD7 家族中除了AcFAD7.3 内含子数量高达33 个外,其他基因成员的内含子数量都稳定在7~8 个;FAD2.1 没有内含子,FAD2.2 有1 个内含子,FAD6 成员各有7个和5个内含子,ADS成员均只有2个内含子,DES 成员均只有1 个内含子,FAD4、SLD、FAD5成员包含1~2 个内含子,FAB2 有4 个内含子。Ac-SLD1.2 和AcSLD2.2 没有5’UTR,AcFAB2 没有3’UTR,AcFAD7.3、AcADS1.2、AcFAD4 以及SLD 其余的5个成员没有5’UTR和3’UTR。由此可见,在同一分支中,大多数成员具有相似的长度和相同数量的结构分布。
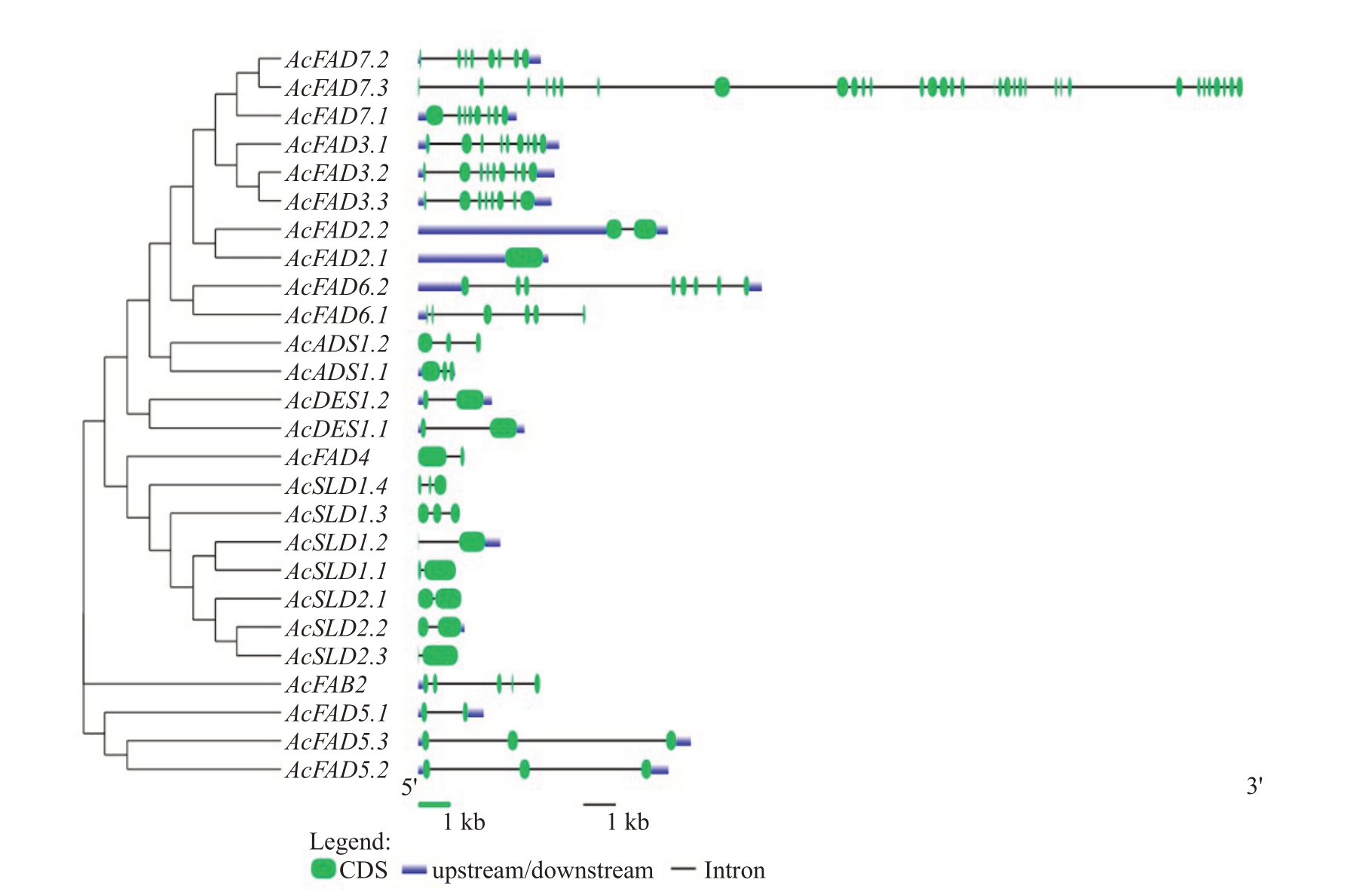
图4 猕猴桃FAD 基因结构
Fig.4 Gene structure of kiwifruit FAD gene
通过在线网站MEME 预测了FAD 蛋白序列中的20 个保守基序(图5),结果发现大多数猕猴桃FAD 蛋白均含有motif 2、motif 4 和motif 14,且motif 2 和motif 4 基本都位于C 端,说明这些基序保守性较强,是典型的FAD结构域,可能行使相似的功能。motif 15是FAD5蛋白特有的基序,motif 18 和motif 19 是DES 蛋白特有的基序,FAD5 与DES蛋白共有motif 17,FAD4和ADS1.2蛋白没有保守基序,FAD3、FAD2和DES家族所有蛋白成员的基序种类、数量和排列方式各自完全一致。同一亚族内的蛋白表现出相似的基序组成,说明同一亚族成员高度保守,具有较近的亲缘关系。但亚族间也存在明显差异,说明一些基因在进化过程中发生了功能分化。
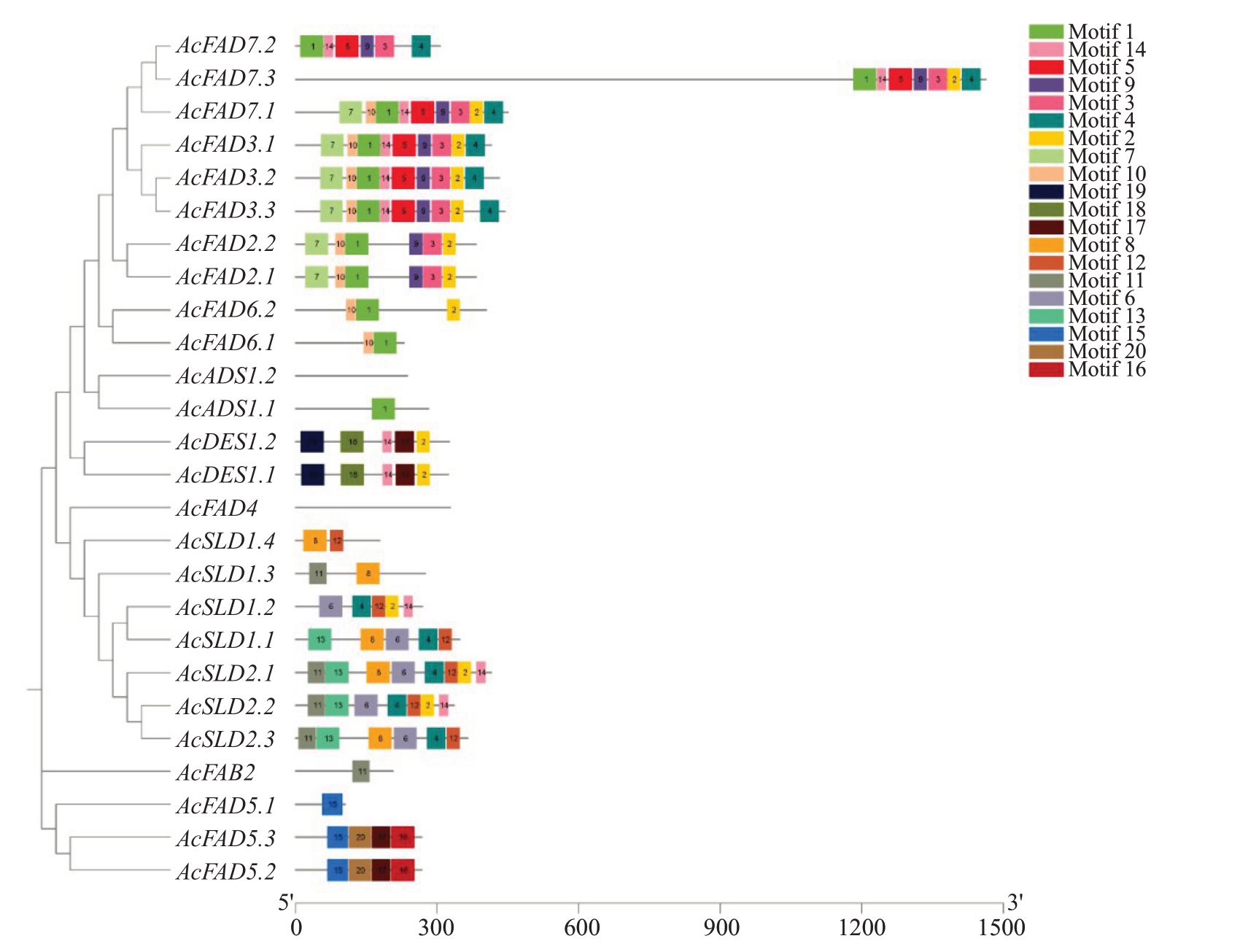
图5 猕猴桃FAD 基因家族基序分析
Fig.5 Motif analysis of kiwifruit FAD gene family
2.5 猕猴桃FAD基因启动子顺式作用元件分析
为探索猕猴桃FAD基因家族顺式作用元件的功能,对该家族上游1500 bp 的启动子序列进行了顺式作用元件预测(图6)。结果预测出47 种顺式作用元件,其中光响应元件数量最多,共计298 个(ACE 2个,AE-box 8 个,AT1-motif 4 个,ATC-motif 5 个,ATCT-motif 7个,Box 4 68个,Box Ⅱ1个,CAG-motif 1 个,chs-CMA1a 5 个,chs-CMA2a 4 个,GA-motif 4个,GATA-motif 20 个,G-Box 62 个,GT1-motif 28 个,I-box 9个,LAMP-element 4个,MRE 22个,Sp1 12个,TCCC-motif 10 个,TCT-motif 21 个,TGA-element 1个);其次是植物激素响应元件(196 个),包括茉莉酸甲酯响应元件72 个(CGTCA-motif 36 个,TGACGmotif 36 个)、脱落酸响应元件59 个(ABRE)、生长素响应元件24 个(AAAC-motif 1 个,AuxRE 1 个,AuxRR-core 4个,TGA-box 2个,TGA-element 16个)、水杨酸响应元件21个(TCA-element)和赤霉素响应元件20 个(GARE-motif 8 个,P-box 7 个,TATC-box 5个);再者是逆境胁迫响应元件(110 个),包括厌氧诱导所必需的元件69 个(ARE 60 个,GC-motif 9个)、低温响应元件23个(LTR)、参与防御和应激反应元件11 个(TC-rich repeats)和参与干旱诱导的MYB结合位点元件7个(MBS);最后是植物生长发育相关元件(54 个),包括MYBHv1 结合位点元件13个(CCAAT-box)、与分生组织表达有关的元件12个(CAT-box)、参与玉米蛋白代谢调控的元件8 个(O2-site)、参与胚乳表达的元件7个(AACA_motif 1个,GCN4_motif 6 个)、参富含AT 的DNA 结合蛋白(ATBP-1)的结合位点元件6个(AT-rich element)、与昼夜节律控制有关的元件3个(circadian)、参与细胞周期调控的元件2 个(MSA-like)、参与叶绿体中胚层细胞分化的元件2个(HD-Zip 1)和参与黄酮类生物合成基因调控的MYB 结合位点元件1 个(MBSI)。此外,还有3个最大诱导剂介导的激活元件(2个)(AT-rich sequence)。除了AcADS1.1外,其他Ac-FADs启动子序列上均有激素响应元件,因此这些基因极有可能参与激素调控的生理过程;所有猕猴桃FAD 基因家族成员都含有逆境胁迫响应元件,所以猕猴桃FAD 基因在逆境胁迫应答过程中发挥着重要的作用;AcFAD6.2、AcFAD3.2、AcADS1.1 和AcADS1.2没有调控生长发育的顺式作用元件。
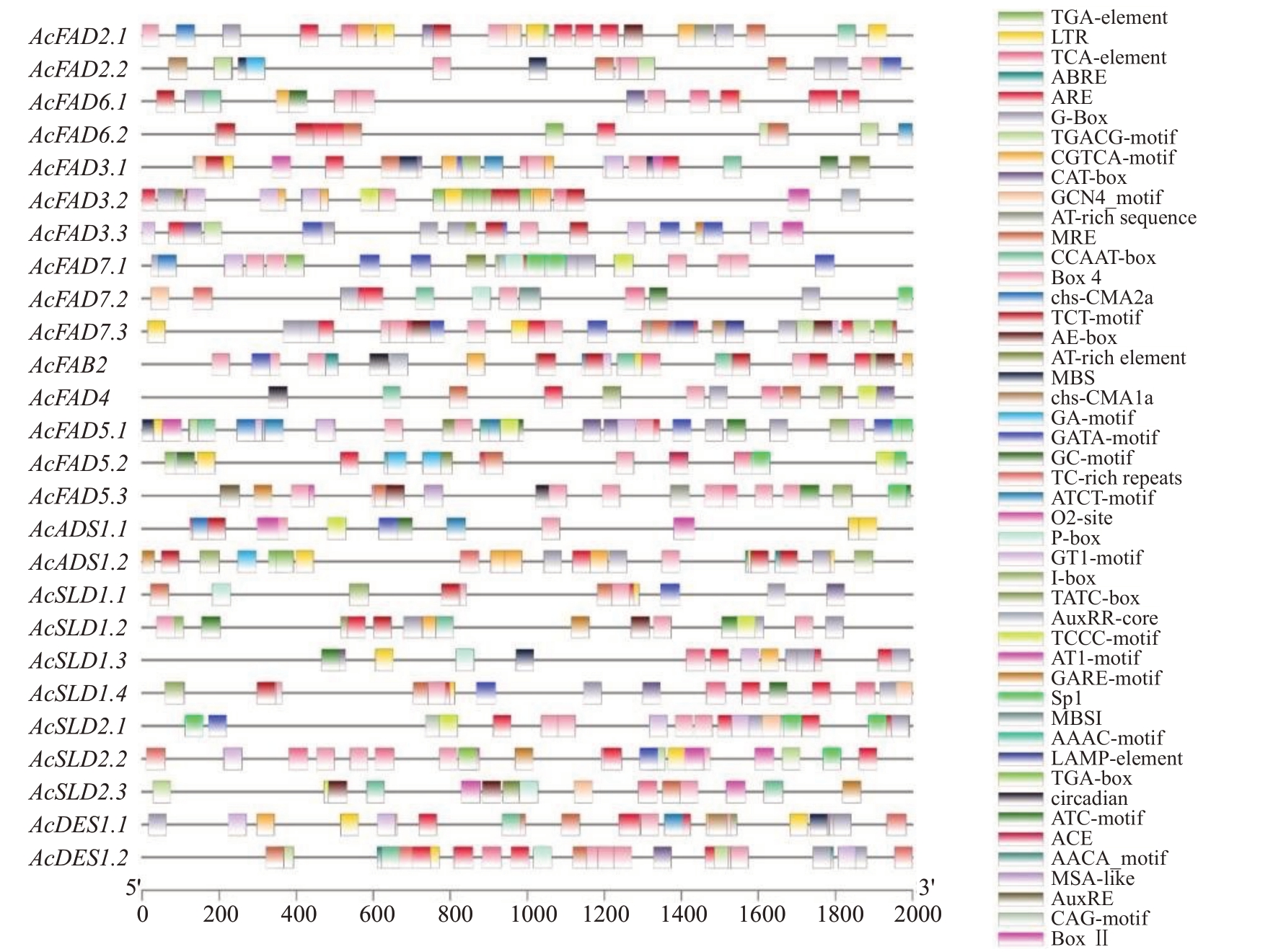
图6 猕猴桃FAD 基因家族启动子顺式作用元件
Fig.6 Cis-elements of kiwifruit FAD gene family promoters
2.6 猕猴桃FAD基因家族表达模式分析
为了了解猕猴桃FAD 基因在采后成熟过程中的表达模式,对红阳猕猴桃采后0、4、10 d 果肉样品转录组数据进行了热图绘制以表征其表达丰度。结果(图7)显示,AcFAD3.1、AcFAD3.3、AcSLD1.4 和AcDES1.1这4个基因成员在猕猴桃整个采后成熟过程中均不表达;AcFAD6.1、AcFAD7.1、AcSLD1.2 和AcSLD2.1 在采后成熟早期(常温贮藏0 d)表现出最高的表达水平,随着后熟时间的增加其表达水平也随之下降,在成熟后期(常温贮10 d)达到最低的表达水平;AcFAD3.2、AcFAD5.3、AcADS1.1、AcADS1.2和AcFAD6.2 同样在采后成熟早期表现出最高的表达水平,但在采后成熟中期(常温贮4 d)表现出最低的表达水平,后期其表达水平又有所提高;Ac-FAD7.3、AcFAD5.1 在成熟早期和后期的表达水平一致,中期略有下降;AcFAD4 仅在成熟早期和中期表达且表达水平较低;AcSLD2.3 在成熟中期和后期表达水平基本一致,早期表达水平最低;Ac-SLD1.1 在成熟后期表达水平最高,中期表达水平最低;AcFAD2.1 只在成熟后期表达且表达水平极低;AcDES1.2、AcSLD1.3、AcFAD5.2、AcFAD7.2、Ac-SLD2.2、AcFAB2 和AcFAD2.2 的表达水平均随着成熟而不断提高,且值得注意的是AcFAD2.2的表达水平极高且各样点间表达水平增加1.4倍左右,后期相比于早期增加了2 倍。因此,猕猴桃FAD 基因家族各成员在猕猴桃采后成熟过程中有不同的表达模式,暗示着其在成熟过程中的各阶段发挥着不同的作用。
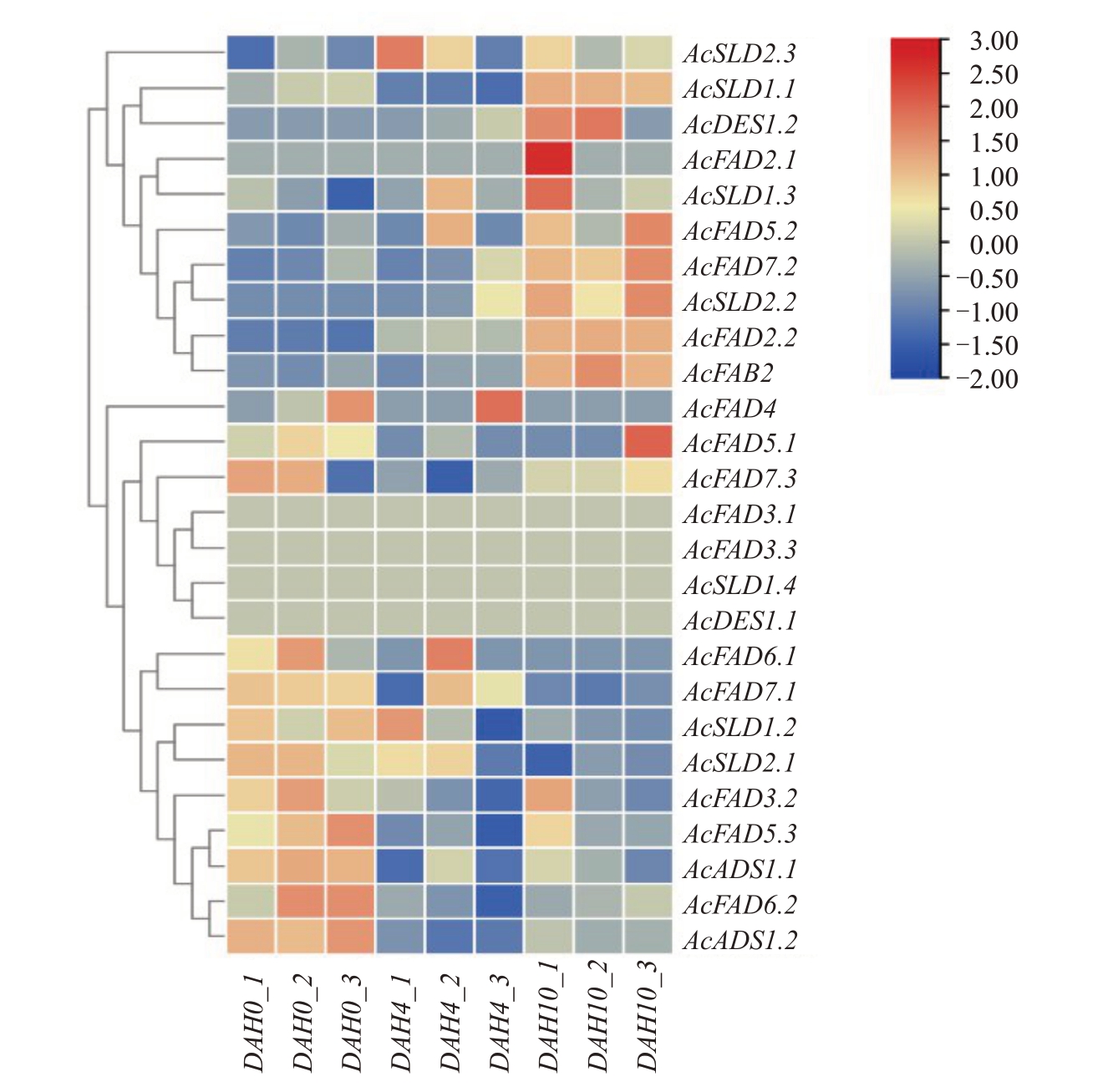
图7 猕猴桃FAD 基因家族表达热图
Fig.7 Heat map of kiwifruit FAD gene family expression
2.7 猕猴桃果实成熟过程中硬度和不饱和脂肪酸含量变化
在猕猴桃采后成熟过程中硬度呈下降趋势,并于采后10 d达到了可食用成熟度(图8-A)。不饱和脂肪酸是猕猴桃香气合成的重要前体物质,对采后猕猴桃果实中不饱和脂肪酸含量进行测定,发现油酸(OA,18∶1)含量随着猕猴桃采后成熟过程不断下降,亚油酸(LA,18∶2)则表现出与之相反的趋势,亚麻酸(LeA,18∶3)含量在成熟早期和中期无显著差别,而在后期含量显著降低(图8-B~D)。
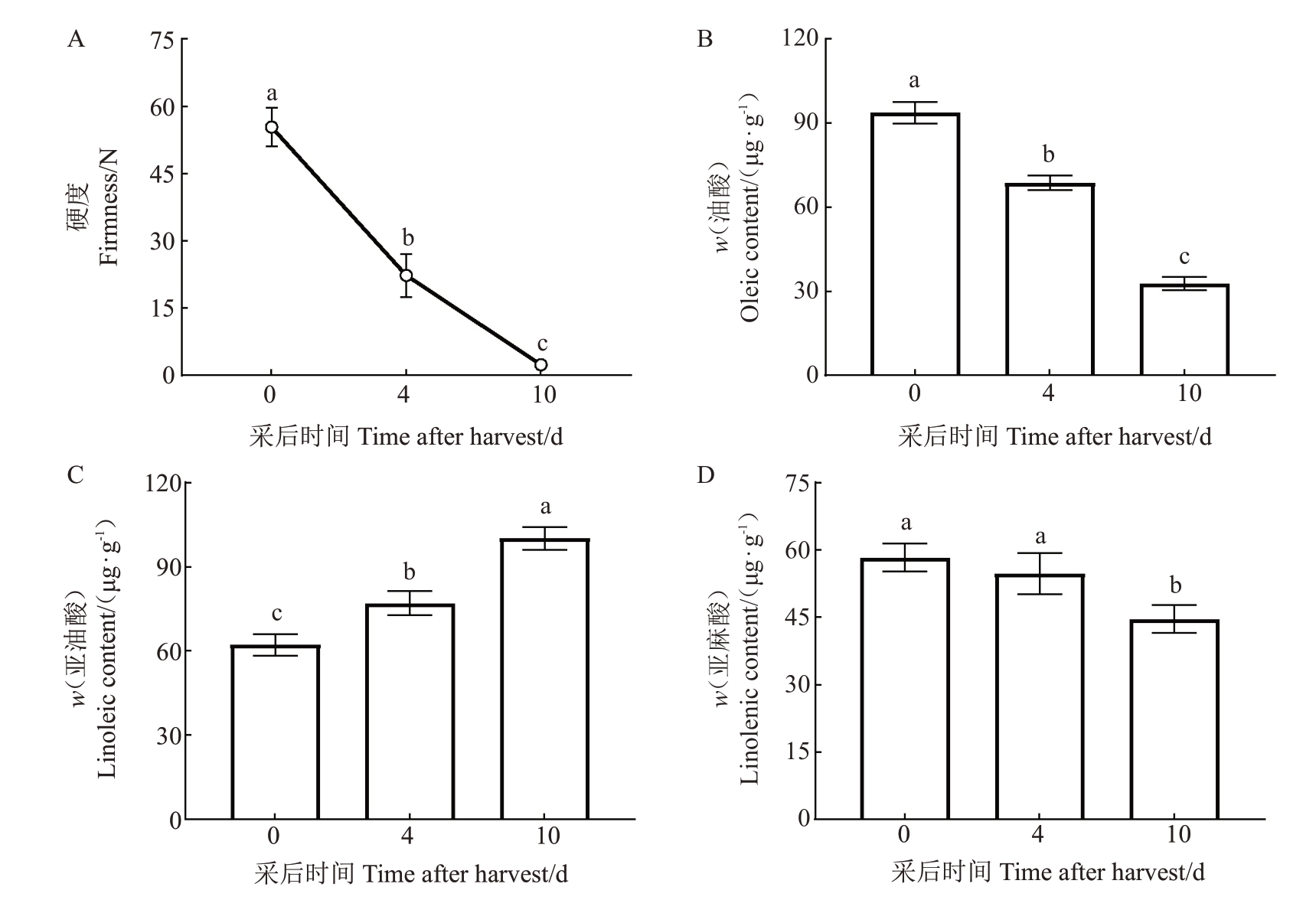
图8 猕猴桃采后成熟过程中的硬度(A)及油酸(B)、亚油酸(C)和亚麻酸(D)含量的变化
Fig.8 Changes in firmness(A),oleic(B),linoleic(C)and linolenic(D)contents of kiwifruit during postharvest ripening
不同小写字母表示在不同成熟阶段存在显著差异(p<0.05)。下同。
Different small letters indicate significant differences at different stages of maturation(p<0.05).The same below.
2.8 猕猴桃FAD基因家族荧光定量分析
从猕猴桃转录组热图中挑选出在成熟后期差异表达上调的8 个基因(AcSLD1.1、AcSLD1.3、Ac-SLD2.2、AcSLD2.3、AcFAD5.2、AcFAD7.2、AcFAB2 和AcFAD2.2)进行了qPCR 验证。结果(图9)显示,在猕猴桃整个采后成熟过程中AcSLD1.3 表达量无显著变化;AcSLD2.2表达量先上调后下调但变化幅度不大;AcSLD1.1、AcSLD2.3和AcFAB2表达量在成熟早期和中期无显著变化,在后期显著上调,但表达量较低;与早期相比,AcFAD5.2在后期表达量显著上调,中期表达量相对于早期和后期无显著变化;AcFAD7.2和AcFAD2.2表达量都是随着成熟而不断显著上调,但AcFAD7.2表达量并不是很高,中期表达量相比于早期上调2.42 倍,后期上调量是早期的13.09 倍,而AcFAD2.2中期表达量比早期上调8.44倍,后期表达量比中期上调9.05 倍,后期上调量是早期的76.43倍,这与转录组数据的变化模式一致,因此推测Ac-FAD2.2 极有可能是参与编码猕猴桃采后成熟过程中ω-6 脂肪酸去饱和酶的关键候选基因,使得单不饱和脂肪酸转化为双不饱和脂肪酸,从而导致了油酸的减少和亚油酸的积累。
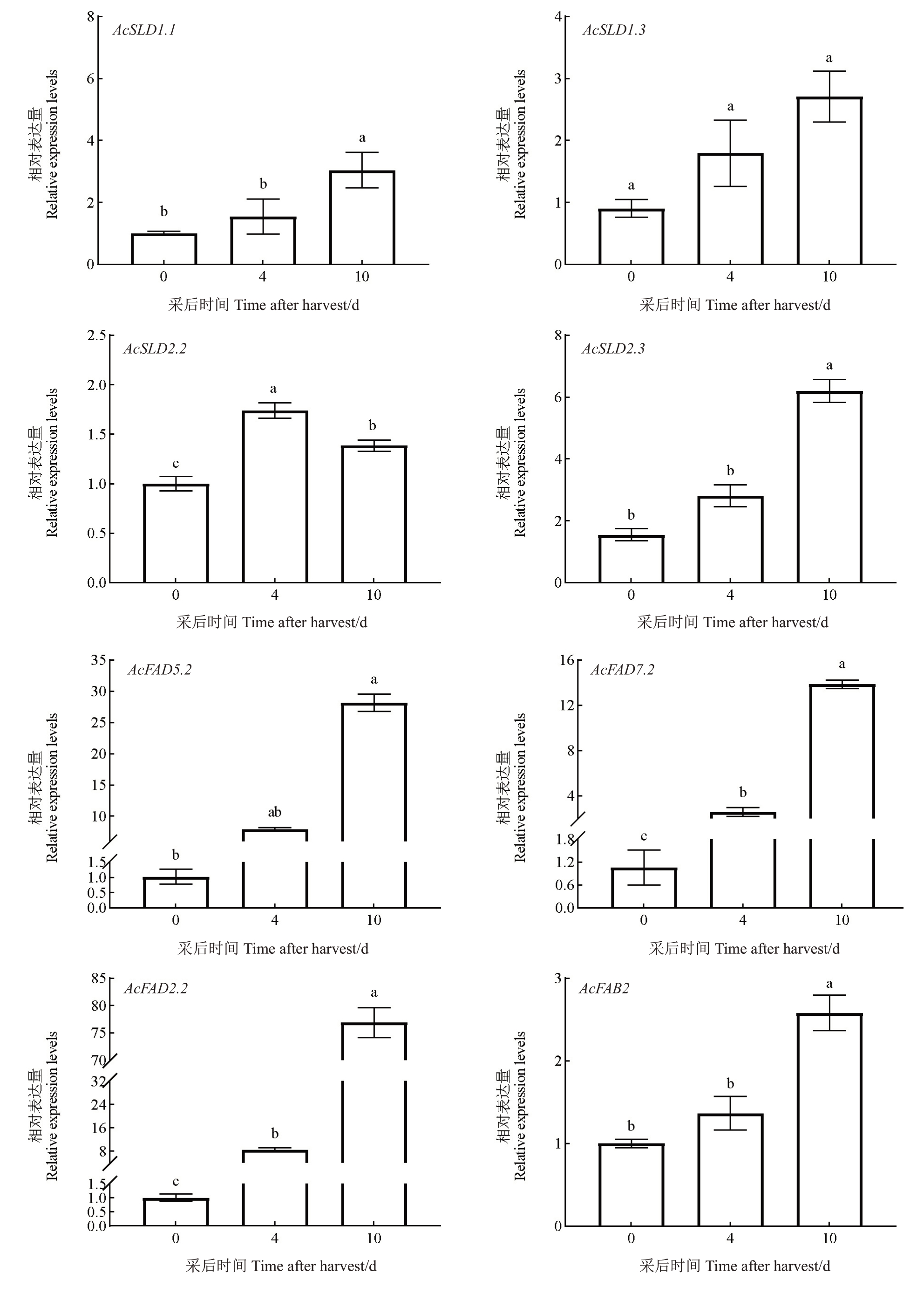
图9 猕猴桃FAD 基因家族荧光定量PCR
Fig.9 Fluorescence quantitative PCR of kiwifruit FAD gene family
3 讨 论
脂肪酸去饱和酶对植物生长发育、抵抗生物与非生物胁迫和品质形成等方面具有正向的积极作用[1-4]。近年来,已经在多种植物中对FAD基因家族进行了鉴定,如亚麻中有43个[37]、茄子有38个[19]、核桃中有24个[38]、桃中有6个FAD基因家族成员[39],而本研究中笔者首次在猕猴桃中鉴定出了26个FAD基因。由此可见,不同物种间FAD成员数量存在一定差异,这可能是其基因组大小不同和进化过程中某些基因的复制、分化或缺失造成的。对猕猴桃FAD家族蛋白序列进行理化性质分析发现,其分子质量与氨基酸数量成正比,而在茄子[19]、番茄[20]和亚麻[37]等中也有同样的规律。猕猴桃FAD的亚细胞定位结果发现FAD2s 和多数SLDs/DESs 都定位在内质网中,FAD6s定位在叶绿体中,FAD3s/FAD7s定位在叶绿体和内质网中,说明不同成员在这些不同的特定部位中发挥作用,与前人的研究结果一致[9-10,12]。猕猴桃与19个拟南芥FAD同样都可以分为6大亚族,在FAB2和FAD4亚族中这两个物种都只有一个FAD成员,而在其他亚族中成员数量分布存在较大的差异(尤其是SLD和FAD3/FAD7/FAD8亚族),该现象与番茄FAD家族相似[20],暗示着这些亚族基因在进化过程中可能发生了特异性扩增。本研究在猕猴桃FAD家族中鉴定出9对串联重复基因和22对片段重复基因,同样地在甘蓝型油菜FAD 家族中存在3 对串联重复和25 对片段重复基因[8]、在小麦中形成26对串联重复和126 对片段重复基因[40],因此推测FAD基因家族的扩增主要是基因片段引起的。具有相似的外显子-内含子基因结构的AcFAD成员通常聚为同一分支,且其编码的蛋白具有相似的基序组成,但也有部分成员(AcFAD4 和AcADS1.2)丢失了保守基序,在茄子FAD相应亚族中也发现此现象[19],说明FAD 家族在物种间具有相对保守的进化趋势。对猕猴桃FAD启动子序列进行分析,发现大量的光响应元件、植物激素响应元件(茉莉酸甲酯,脱落酸,生长素,水杨酸,赤霉素)、逆境胁迫响应元件(厌氧,低温,干旱)和生长发育相关元件,尤其是逆境胁迫响应元件遍布在每个成员中,说明猕猴桃FAD 的表达受到光的调控,并广泛参与了植物生长发育、成熟及抗逆过程,在其他呼吸跃变型果实中(如番茄和香蕉等)也发现了相同的顺式作用元件[18,20]。
果实风味作为水果重要的品质性状,是在成熟过程中形成的品质之一,也是影响其销量以及消费者偏好的重要因素,果实的风味取决于糖酸以及挥发性芳香化合物的种类和含量。猕猴桃作为典型的呼吸跃变型果实,在成熟过程中,存在明显的呼吸跃变并促发内源乙烯大量合成,从而加速各种挥发性化合物的合成,逐渐产生独特的果香。在LOX途径中,亚油酸和亚麻酸首先被氧化为脂肪酸氢过氧化物,随后被氢过氧化物裂解酶裂解形成己醛和己烯醛。然后,C6醛通过醇脱氢酶还原为相应的C6醇,随后通过醇酰基转移酶转化为酯[30]。哈密瓜亚麻酸含量在贮藏过程中均有所增加,贮藏前期油酸含量急剧增多而中后期则呈下降趋势,亚油酸含量与之相反,呈上升趋势[41],这与本研究中发现猕猴桃在采后成熟过程中油酸含量随成熟进程而显著下降,亚油酸表现出与之负相关的趋势,亚麻酸含量在早中期无显著变化,而在后期有所下降的现象一致。同时,也有研究表明油酸在油棕果实采后成熟阶段含量均减少,而亚油酸只在成熟早期阶段减少,却在后期阶段显著增加[42]。番茄果实成熟阶段亚油酸含量最高,油酸含量下降,亚油酸含量逐渐升高,亚麻酸含量无显著变化[43]。以上研究表明大多数果实成熟过程往往伴有亚油酸含量的增加和油酸含量的下降。但也有研究发现了与本研究不一致的现象,如杧果采后成熟过程中亚油酸含量逐渐下降,亚麻酸含量逐渐升高[44],在香蕉果实的成熟进程中亚麻酸增加,呼吸跃变前期亚麻酸含量增加缓慢,至呼吸跃变上升期迅速增加,而亚油酸含量则有所下降[45]。这可能是热带水果和亚热带水果差异及其芳香物质种类不同所致。
基于FAD 家族在猕猴桃成熟各阶段的表达模式,发现了AcFAD7.2 和AcFAD2.2 这两个表达量均随着成熟而不断显著上调的家族成员。前人研究显示,在油茶和拟南芥中分别同源和异源过表达ω-3脂肪酸去饱和酶CoFAD7 后,亚麻酸的含量显著增加并且提高了亚麻酸/亚油酸的比值[46]。此外,在烟草中分别过量表达桃PpFAD3-1 和PpFAD3-2 后发现亚油酸含量大幅下降,亚麻酸含量显著上升[47]。但在本研究中并未发现亚麻酸含量增多,反而出现下降,说明猕猴桃成熟过程中亚麻酸的合成速率可能低于其作为前体物质参与芳香物质合成的速率,造成亚麻酸积累量降低,因此编码ω-3 脂肪酸去饱和酶基因AcFAD7.2 在猕猴桃采后成熟不饱和脂肪酸合成和积累中发挥的作用并不是很显著。对大肠杆菌中异源表达桃的PpFAD2-1 和PpFAD2-2 及其酶活性分析,发现这两个ω-6 脂肪酸去饱和酶基因都能促进油酸向亚油酸的去饱和[48];对橄榄进行Oe-FAD2-2 和OeFAD2-5 超量表达,发现其具有将油酸转化为亚油酸并提高亚油酸含量的能力[49];在番茄中进行异源酵母表达SlFAD2-1 和SlFAD2-2 能将油酸转化生成亚油酸并显著增加了亚油酸含量[50]。在本研究中也同样发现了油酸含量减少和亚油酸含量增多的现象,作为与番茄SlFAD2-1和SlFAD2-2高度同源的ω-6 脂肪酸去饱和酶基因,AcFAD2.2 的表达水平也随着成熟而不断攀高,其表达模式与其发挥作用的两种不饱和脂肪酸(油酸和亚油酸)含量变化存在明显的正相关,说明AcFAD2.2是参与猕猴桃采后成熟过程中脂肪酸代谢的关键基因,在不饱和脂肪酸合成与积累过程中发挥着极为重要的作用。
4 结 论
笔者在猕猴桃中鉴定出了26 个FAD 基因家族成员,根据其发挥作用的部位被定位到细胞的不同部位,因此绝大多数成员定位于内质网或叶绿体中,这些成员随机分布在19条不同染色体上,片段重复事件是猕猴桃FAD扩增和进化的主要动力,在猕猴桃FAD启动子序列上发现大量的光响应元件、植物激素响应元件(茉莉酸甲酯、脱落酸、生长素、水杨酸、赤霉素)、逆境胁迫响应元件(厌氧、低温、干旱)和生长发育相关元件。猕猴桃采后成熟过程中油酸含量下降、亚油酸含量增多,并且AcFAD2.2 表达水平随着成熟而不断显著上调,由此可以推断出Ac-FAD2.2是油酸去饱和生成亚油酸的关键酶基因,并且可能会通过光、激素等响应方式在猕猴桃采后成熟过程中表达来调控不饱和脂肪酸的合成与积累。因此,今后可借助分子生物学技术对此关键候选基因进行克隆、功能验证和上游调控试验,深入解析猕猴桃FAD 基因在采后成熟过程中不饱和脂肪酸积累和香气物质合成中的功能机制和调控网络。
[1] HE M,QIN C X,WANG X,DING N Z.Plant unsaturated fatty acids:Biosynthesis and regulation[J].Frontiers in Plant Science,2020,11:390.
[2] FAN R S,LI L,CAI G,YE J,LIU M H,WANG S H,LI Z Q.Molecular cloning and function analysis of FAD2 gene in Idesia polycarpa[J].Phytochemistry,2019,168:112114.
[3] 王利民,符真珠,高杰,董晓宇,张晶,袁欣,蒋卉,王慧娟,李艳敏,师曼,张和臣.植物不饱和脂肪酸的生物合成及调控[J].基因组学与应用生物学,2020,39(1):254-258.WANG Limin,FU Zhenzhu,GAO Jie,DONG Xiaoyu,ZHANG Jing,YUAN Xin,JIANG Hui,WANG Huijuan,LI Yanmin,SHI Man,ZHANG Hechen.Molecular mechanism of unsaturated fatty acids synthesis and regulation in plant[J].Genomics and Applied Biology,2020,39(1):254-258.
[4] 薛晓梦,李建国,白冬梅,晏立英,万丽云,康彦平,淮东欣,雷永,廖伯寿.花生FAD2 基因家族表达分析及其对低温胁迫的响应[J].作物学报,2019,45(10):1586-1594.XUE Xiaomeng,LI Jianguo,BAI Dongmei,YAN Liying,WAN Liyun,KANG Yanping,HUAI Dongxin,LEI Yong,LIAO Boshou.Expression profiles of FAD2 genes and their responses to cold stress in peanut[J].Acta Agronomica Sinica,2019,45(10):1586-1594.
[5] KUGLER A,ZORIN B,DIDI- COHEN S,SIBIRYAK M,GORELOVA O,ISMAGULOVA T,KOKABI K,KUMARI P,LUKYANOV A,BOUSSIBA S,SOLOVCHENKO A,KHOZIN-GOLDBERG I.Long-chain polyunsaturated fatty acids in the green microalga Lobosphaera incisa contribute to tolerance to abiotic stresses[J].Plant and Cell Physiology,2019,60(6):1205-1223.
[6] 蔡洪芳.1-MCP/NO/MeJA 对采后桃果实脂肪酸途径香气物质的调控研究[D].南京:南京农业大学,2020.CAI Hongfang.Study on the regulation of 1-MCP/NO/MeJA on aroma volatiles in fatty acid pathway of postharvest peach fruit[D].Nanjing:Nanjing Agricultural University,2020.
[7] 赵训超,魏玉磊,丁冬,刘梦,盖胜男,张今杰,邵文静,李嘉欣,徐晶宇.甜荞麦脂肪酸脱氢酶基因(FeFAD)家族的鉴定与分析[J].东北农业科学,2021,46(1):36-41.ZHAO Xunchao,WEI Yulei,DING Dong,LIU Meng,GAI Shengnan,ZHANG Jinjie,SHAO Wenjing,LI Jiaxin,XU Jingyu.Genome-wide identification and bioinformatics analysis of fatty acid desaturase gene (FeFAD) family in common buckwheat[J].Journal of Northeast Agricultural Sciences,2021,46(1):36-41.
[8] XUE Y F,CHEN B J,WANG R,WIN A N,LI J N,CHAI Y R.Genome-wide survey and characterization of fatty acid desaturase gene family in Brassica napus and its parental species[J].Applied Biochemistry and Biotechnology,2018,184(2):582-598.
[9] DU C,CHEN Y Y,WANG K,YANG Z,ZHAO C Z,JIA Q L,TAYLOR D C,ZHANG M.Strong co-suppression impedes an increase in polyunsaturated fatty acids in seeds overexpressing FAD2[J].Journal of Experimental Botany,2019,70(3):985-994.
[10] PENG Z Y,RUAN J,TIAN H Y,SHAN L,MENG J J,GUO F,ZHANG Z M,DING H,WAN S B,LI X G.The family of peanut fatty acid desaturase genes and a functional analysis of four ω-3 AhFAD3 members[J].Plant Molecular Biology Reporter,2020,38(2):209-221.
[11] SHANKLIN J,CAHOON E B.Desaturation and related modifications of fatty acids[J].Annual Review of Plant Physiology and Plant Molecular Biology,1998,49:611-641.
[12] 李昊远,郝翠翠,陈明娜,陈娜,王冕,潘丽娟,王通,禹山林,侯艳华,迟晓元.花生鞘脂Δ8 去饱和酶基因(AhSLD2)的克隆与表达分析[J].花生学报,2018,47(2):24-29.LI Haoyuan,HAO Cuicui,CHEN Mingna,CHEN Na,WANG Mian,PAN Lijuan,WANG Tong,YU Shanlin,HOU Yanhua,CHI Xiaoyuan.Cloning and expression analysis of sphingolipid Δ8 desaturase (AhSLD2) gene in peanut[J].Journal of Peanut Science,2018,47(2):24-29.
[13] MICHAELSON L V,ZÄUNER S,MARKHAM J E,HASLAM R P,DESIKAN R,MUGFORD S,ALBRECHT S,WARNECKE D,SPERLING P,HEINZ E,NAPIER J A.Functional characterization of a higher plant sphingolipid Delta4-desaturase:Defining the role of sphingosine and sphingosine-1-phosphate in Arabidopsis[J].Plant Physiology,2009,149(1):487-498.
[14] ARONDEL V,LEMIEUX B,HWANG I,GIBSON S,GOODMAN H M,SOMERVILLE C R.Map-based cloning of a gene controlling omega-3 fatty acid desaturation in Arabidopsis[J].Science,1992,258(5086):1353-1355.
[15] OKULEY J,LIGHTNER J,FELDMANN K,YADAV N,LARK E,BROWSE J. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis[J].The Plant Cell,1994,6(1):147-158.
[16] IBA K,GIBSON S,NISHIUCHI T,FUSE T,NISHIMURA M,ARONDEL V,HUGLY S,SOMERVILLE C.A gene encoding a chloroplast ω-3 fatty acid desaturase complements alterations in fatty acid desaturation and chloroplast copy number of the fad7 mutant of Arabidopsis thaliana[J].Journal of Biological Chemistry,1993,268(32):24099-24105.
[17] REDDY A S,NUCCIO M L,GROSS L M,THOMAS T L.Isolation of a Δ6-desaturase gene from the Cyanobacterium synechocystis sp.strain PCC 6803 by gain-of-function expression in Anabaena sp.strain PCC 7120[J].Plant Molecular Biology,1993,22(2):293-300.
[18] CHENG C Z,LIU F,SUN X L,WANG B,LIU J P,NI X T,HU C H,DENG G M,TONG Z,ZHANG Y Y,LÜ P T.Genomewide identification of FAD gene family and their contributions to the temperature stresses and mutualistic and parasitic fungi colonization responses in banana[J].International Journal of Biological Macromolecules,2022,204:661-676.
[19] 朱宗文,张爱冬,吴雪霞,查丁石.生物信息学鉴定分析茄子脂肪酸去饱和酶(FAD)基因家族[J].分子植物育种,2023,21(8):2453-2463.ZHU Zongwen,ZHANG Aidong,WU Xuexia,ZHA Dingshi.Identification and bioinformatics analysis of fatty acid desaturase (FAD) gene family in eggplant (Solanum melongena L.)[J].Molecular Plant Breeding,2023,21(8):2453-2463.
[20] 张明亚,庞胜群,刘玉东,苏永峰,牛博文,韩琼琼.番茄FAD基因家族的鉴定与表达分析[J].生物技术通报,2024,40(7):150-162.ZHANG Mingya,PANG Shengqun,LIU Yudong,SU Yongfeng,NIU Bowen,HAN Qiongqiong.Identification and expression analysis of FAD gene family in Solanum lycopersicum[J].Biotechnology Bulletin,2024,40(7):150-162.
[21] 高谢旺,谭安琪,胡信畅,祝孟洋,阮颖,刘春林.利用CRISPR/Cas9 技术创制高油酸甘蓝型油菜新种质[J].植物遗传资源学报,2020,21(4):1002-1008.GAO Xiewang,TAN Anqi,HU Xinchang,ZHU Mengyang,RUAN Ying,LIU Chunlin.Creation of new germplasm of higholeic rapeseed using CRISPR/Cas9[J].Journal of Plant Genetic Resources,2020,21(4):1002-1008.
[22] 王庆华,王磊,吴文江,郭家选,沈元月,吴国良.果实香气物质的合成及其激素调控研究进展[J/OL].分子植物育种,2021:1-11(2021-11-02)[2024-09-07].https://kns.cnki.net/kcms/detail/46.1068.S.20211029.1846.006.html.WANG Qinghua,WANG Lei,WU Wenjiang,GUO Jiaxuan,SHEN Yuanyue,WU Guoliang.Advances in aroma compounds biosynthesis and hormone regulation of fruit[J/OL].Molecular Plant Breeding,2021:1-11(2021-11-02)[2024-09-07].https://kns.cnki.net/kcms/detail/46.1068.S.20211029.1846.006.html.
[23] 蔡璨,白玉,韩艺,郭佳欣,沙伟,张梅娟,彭疑芳,马天意.植物多不饱和脂肪酸的研究进展[J].高师理科学刊,2023,43(9):64-69.CAI Can,BAI Yu,HAN Yi,GUOJIA Xin,SHA Wei,ZHANG Meijuan,PENG Yifang,MA Tianyi.Research progress of plant polyunsaturated fatty acids[J].Journal of Science of Teachers’College and University,2023,43(9):64-69.
[24] 陈成,王依,杨勇,阎永齐.采收成熟度对‘金艳’猕猴桃果实品质及香气成分的影响[J].中国农学通报,2020,36(31):28-36.CHEN Cheng,WANG Yi,YANG Yong,YAN Yongqi.Effects of maturity stage on fruit quality and aroma components of‘Jinyan’kiwifruit[J].Chinese Agricultural Science Bulletin,2020,36(31):28-36.
[25] 陈义挺,赖瑞联,冯新,程春振,钟春水,高敏霞,吴如健.不同贮藏条件下猕猴桃香气成分的变化规律研究[J].热带作物学报,2020,41(6):1251-1256.CHEN Yiting,LAI Ruilian,FENG Xin,CHENG Chunzhen,ZHONG Chunshui,GAO Minxia,WU Rujian.Change of aroma components in different storage conditions of kiwifruit[J].Chinese Journal of Tropical Crops,2020,41(6):1251-1256.
[26] COZZOLINO R,DE GIULIO B,PETRICCIONE M,MARTIGNETTI A,MALORNI L,ZAMPELLA L,LAURINO C,PELLICANO M P.Comparative analysis of volatile metabolites,quality and sensory attributes of Actinidia chinensis fruit[J].Food Chemistry,2020,316:126340.
[27] 张波,李鲜,陈昆松.基于EST 库的猕猴桃脂氧合酶基因家族成员的克隆[J].园艺学报,2008,35(3):337-342.ZHANG Bo,LI Xian,CHEN Kunsong.Molecular cloning of lipoxygenase gene family members in kiwifruit based on EST database[J].Acta Horticulturae Sinica,2008,35(3):337-342.
[28] CROWHURST R N,GLEAVE A P,MACRAE E A,AMPOMAH-DWAMENA C,ATKINSON R G,BEUNING L L,BULLEY S M,CHAGNE D,MARSH K B,MATICH A J,MONTEFIORI M,NEWCOMB R D,SCHAFFER R J,USADEL B,ALLAN A C,BOLDINGH H L,BOWEN J H,DAVY M W,ECKLOFF R,FERGUSON A R,FRASER L G,GERA E,HELLENS R P,JANSSEN B J,KLAGES K,LO K R,MACDIARMID R M,NAIN B,MCNEILAGE M A,RASSAM M,RICHARDSON A C,RIKKERINK E H,ROSS G S,SCHRÖDER R,SNOWDEN K C,SOULEYRE E J F,TEMPLETON M D,WALTON E F,WANG D,WANG M Y,WANG Y Y,WOOD M,WU R M,YAUK Y K,LAING W A.Analysis of expressed sequence tags from Actinidia:Applications of a cross species EST database for gene discovery in the areas of flavor,health,color and ripening[J].BMC Genomics,2008,9:351.
[29] ZHANG J Y,HUANG S N,CHEN Y H,WANG G,GUO Z R.Identification and characterization of two waterlogging responsive alcohol dehydrogenase genes (AdADH1 and AdADH2) in Actinidia deliciosa[J].Molecular Breeding,2017,37(4):52.
[30] 陶淑华,陈丽,蒋镇烨,宋倩倩,宋亦超,姜天甲,郑小林.低温贮藏对美味猕猴桃布鲁诺果实主要挥发性物质和脂肪酸代谢的影响[J].核农学报,2020,34(2):288-297.TAO Shuhua,CHEN Li,JIANG Zhenye,SONG Qianqian,SONG Yichao,JIANG Tianjia,ZHENG Xiaolin.Effects of lower temperature on flavor components and fatty acid pathway in harvested kiwifruit(Actinidia deliciosa cv.Bruno)[J].Journal of Nuclear Agricultural Sciences,2020,34(2):288-297.
[31] 李可,袁怀瑜,朱永清,周艳,夏陈,赵楠,李华佳.不同品种猕猴桃籽油脂肪酸组成的PCA 分析[J].中国调味品,2021,46(2):70-74.LI Ke,YUAN Huaiyu,ZHU Yongqing,ZHOU Yan,XIA Chen,ZHAO Nan,LI Huajia.PCA analysis of fatty acid composition of kiwi seed oils with different varieties[J].China Condiment,2021,46(2):70-74.
[32] ZHANG B,YIN X R,LI X,YANG S L,FERGUSON I B,CHEN K S.Lipoxygenase gene expression in ripening kiwifruit in relation to ethylene and aroma production[J].Journal of Agricultural and Food Chemistry,2009,57(7):2875-2881.
[33] 甘武.猕猴桃果实品质和香气成分分析研究[D].南昌:江西农业大学,2018.GAN Wu.Analysis of fruit quality and aroma components of kiwifruit[D].Nanchang:Jiangxi Agricultural University,2018.
[34] DONG C J,CAO N,ZHANG Z G,SHANG Q M.Characterization of the fatty acid desaturase genes in cucumber:Structure,phylogeny,and expression patterns[J].PLoS One,2016,11(3):e0149917.
[35] GAN Z Y,SHAN N,FEI L Y,WAN C P,CHEN J Y.Isolation of the 9-Cis-epoxycarotenoid dioxygenase(NCED)gene from kiwifruit and its effects on postharvest softening and ripening[J].Scientia Horticulturae,2020,261:109020.
[36] 袁馨,徐云鹤,张雨培,单楠,陈楚英,万春鹏,开文斌,翟夏琬,陈金印,甘增宇.猕猴桃后熟过程中ABA 响应结合因子AcAREB1 调控AcGH3.1 的表达[J].园艺学报,2023,50(1):53-64.YUAN Xin,XU Yunhe,ZHANG Yupei,SHAN Nan,CHEN Chuying,WAN Chunpeng,KAI Wenbin,ZHAI Xiawan,CHEN Jinyin,GAN Zengyu.Studies on AcAREB1 regulating the expression of AcGH3.1 during postharvest ripening of kiwifruit[J].Acta Horticulturae Sinica,2023,50(1):53-64.
[37] 侯静静,赵利,王斌.亚麻FAD 基因家族的生物信息学鉴定分析[J].寒旱农业科学,2023(3):246-253.HOU Jingjing,ZHAO Li,WANG Bin.Identification and bioinformatics analysis of FAD gene family in Linum usitatissimum L.[J].Journal of Cold-Arid Agricultural Sciences,2023(3):246-253.
[38] LIU K,ZHAO S G,WANG S,WANG H X,ZHANG Z H.Identification and analysis of the FAD gene family in walnuts (Juglans regia L.) based on transcriptome data[J].BMC Genomics,2020,21(1):299.
[39] 金正楠.转录因子PpNAC1 和表观遗传修饰通过调控PpFAD3-1 表达参与桃果实芳香物质合成[D].杭州:浙江大学,2022.JIN Zhengnan.Transcription factor PpNAC1 and epigenetic modification are involved in the synthesis of volatiles in peach fruits by regulating the expression of PpFAD3-1[D].Hangzhou:Zhejiang University,2022.
[40] HAJIAHMADI Z,ABEDI A,WEI H,SUN W B,RUAN H H,ZHUGE Q,MOVAHEDI A.Identification,evolution,expression,and docking studies of fatty acid desaturase genes in wheat(Triticum aestivum L.)[J].BMC Genomics,2020,21(1):778.
[41] 王静,茅林春,杨璐,李学文,张辉,吕卓,刘彩虹,李乾,侯琛元.草酸处理对采后哈密瓜果实膜脂代谢的影响[J].中国食品学报,2019,19(8):189-198.WANG Jing,MAO Linchun,YANG Lu,LI Xuewen,ZHANG Hui,LÜ Zhuo,LIU Caihong,LI Qian,HOU Chenyuan.Effect of oxalic acid on reduction of membrane lipids metabolism of Hami melon’s fruit in postharvest[J].Journal of Chinese Institute of Food Science and Technology,2019,19(8):189-198.
[42] 吴秋妃,杨程,张淑岩,韦露,冯美利,李睿,周丽霞,曹红星.油棕果实发育和采后脂肪酸合成转录代谢差异分析[J].热带作物学报,2024,45(2):234-246.WU Qiufei,YANG Cheng,ZHANG Shuyan,WEI Lu,FENG Meili,LI Rui,ZHOU Lixia,CAO Hongxing.Differential analysis of fatty acid synthesis,transcriptional metabolism during fruit development and postharvest in oil palm[J].Chinese Journal of Tropical Crops,2024,45(2):234-246.
[43] STAVECKIENĖ J,KULAITIENĖ J,LEVICKIENĖ D,VAITKEVIČIENĖ N.Changes in fatty acid content in Solanum spp.fruits during ripening[J].Plants,2023,12(2):268.
[44] DESHPANDE A B,CHIDLEY H G,OAK P S,PUJARI K H,GIRI A P,GUPTA V S.Data on changes in the fatty acid composition during fruit development and ripening of three mango cultivars (Alphonso,Pairi and Kent) varying in lactone content[J].Data in Brief,2016,9:480-491.
[45] WADE N L.Membrane lipid composition and tissue leakage of pre- and early-climacteric banana fruit[J].Postharvest Biology and Technology,1995,5(1/2):139-147.
[46] 张嘉锡.油茶亚麻酸合成关键基因CoFAD7 的功能分析和优异等位变异挖掘[D].长沙:中南林业科技大学,2024.ZHANG Jiaxi.Functional analysis of CoFAD7,a key gene for the linolenic acid synthesis in Camellia oleifera,and mining of excellent allelic variation[D].Changsha:Central South University of Forestry and Technology,2024.
[47] WANG J J,LIU H R,GAO J,HUANG Y J,ZHANG B,CHEN K S.Two ω-3 FADs are associated with peach fruit volatile formation[J].International Journal of Molecular Sciences,2016,17(4):464.
[48] PENG B,GU Z X,ZHOU Y F,NING Y Z,XU H Y,LI G,NI Y,SUN P P,XIE Z Q,SHI S P,DARK A,SONG Z Z.Potential role of fatty acid desaturase 2 in regulating peach aroma formation[J].Postharvest Biology and Technology,2023,204:112473.
[49] HERNÁNDEZ M L,SICARDO M D,BELAJ A,MARTÍNEZRIVAS J M.The oleic/linoleic acid ratio in olive(Olea europaea L.) fruit mesocarp is mainly controlled by OeFAD2-2 and Oe-FAD2-5 genes together with the different specificity of extraplastidial acyltransferase enzymes[J].Frontiers in Plant Science,2021,12:653997.
[50] LEE M W,PADILLA C S,GUPTA C,GALLA A,PEREIRA A,LI J M,GOGGIN F L.The FATTY ACID DESATURASE2 family in tomato contributes to primary metabolism and stress responses[J].Plant Physiology,2020,182(2):1083-1099.