广义柑橘属(Citrus s.l.)隶属于芸香科(Rutaceae)柑橘族(Citreae),包括柑橘、柚、柠檬等著名水果[1-2]。起初,Swingle 等[2]根据雄蕊数目及汁胞构造将芸香科(Rutaceae)、柑橘族(Citreae)中6 个关系密切的属归为“真柑橘类”(True Citrus Fruit Trees),包括原产中国的枳属(Poncirus Raf.)、金柑属(Fortunella Swingle)、狭义柑橘属(Citrus s.s.),以及产自澳大利亚和新几内亚的Eremocitrus Swingle、Microcitrus Swingle 和Clymenia Swingle。近年来,基于叶绿体基因(cpDNA)或核糖体基因内转录间隔区(ITS)基因片段的研究表明,枳属、金柑属及狭义柑橘属关系密切,且“真柑橘类”是单系的[1,3-7]。然而核基因(nDNA)和全基因组的研究结果有所不同,枳属在“真柑橘类”中是一个独立分支[8-10]。尽管这些狭义属之间存在明显的形态差异,但具有杂交亲和性[2]。狭义柑橘属物种的杂交起源历史已得到基因组学方面的证据[10],但相关狭义属间的关系尚未完全明确。总之,杂交和不完全谱系分选仍然对相关属、种的亲缘关系研究造成困扰[9]。
“常绿枳”是广义柑橘属中特殊的成员,它的指状三出复叶与枳(Citrus trifoliata L.)相似,而常绿(春季花较多的枝条上的叶较少,因此也呈现出“半常绿”的状态)习性又与狭义柑橘属内物种相似。目前,有两种“常绿枳”被确立为独立的物种,包括华中枳(Citrus × pubinervia D. G. Zhang  Z. H.Xiang,Y. Wu)和富民枳(Citrus × polytrifolia Govaerts)[11-12]。富民枳的分类地位存在一定争议。起初,研究者根据形态特征的交叉及假定亲本的杂交亲和性将其判定为杂交种[13],亦有RAPD 分子的研究支持富民枳为杂交种[14]。然而cpDNA 或nDNA片段系统发育分析、SSR 分子标记、核型、花粉微形态和叶片同工酶比较均认为富民枳为独立的物种[15-19]。华中枳是发表于2021 年的新名称,形态比较和cpDNA 的系统发育分析表明,其与枳、富民枳发生了分化,是一个可区分的物种[11],但还未见nDNA 方面的研究报道。华中枳通常分布于海拔900~1400 m 的山区,具有常绿和良好的抗寒特性,与富民枳生物学特性相似[20],是潜在的柑橘砧木资源。二者的野生资源数量十分有限[11,21],富民枳在2022年被列为国家二级重点保护野生植物(农业资源)。考虑到杂交起源和不完全谱系分选的影响,笔者在本研究中参考Ramadugu等[9]的报道,挑选了其中4 个没有明显连锁关系的nDNA 片段(CTV.4,HYB,LGT,P12),进一步分析这2 个物种的分类地位,以期为它们的保护和利用提供理论依据。
Z. H.Xiang,Y. Wu)和富民枳(Citrus × polytrifolia Govaerts)[11-12]。富民枳的分类地位存在一定争议。起初,研究者根据形态特征的交叉及假定亲本的杂交亲和性将其判定为杂交种[13],亦有RAPD 分子的研究支持富民枳为杂交种[14]。然而cpDNA 或nDNA片段系统发育分析、SSR 分子标记、核型、花粉微形态和叶片同工酶比较均认为富民枳为独立的物种[15-19]。华中枳是发表于2021 年的新名称,形态比较和cpDNA 的系统发育分析表明,其与枳、富民枳发生了分化,是一个可区分的物种[11],但还未见nDNA 方面的研究报道。华中枳通常分布于海拔900~1400 m 的山区,具有常绿和良好的抗寒特性,与富民枳生物学特性相似[20],是潜在的柑橘砧木资源。二者的野生资源数量十分有限[11,21],富民枳在2022年被列为国家二级重点保护野生植物(农业资源)。考虑到杂交起源和不完全谱系分选的影响,笔者在本研究中参考Ramadugu等[9]的报道,挑选了其中4 个没有明显连锁关系的nDNA 片段(CTV.4,HYB,LGT,P12),进一步分析这2 个物种的分类地位,以期为它们的保护和利用提供理论依据。
1 材料和方法
1.1 采样
枳的样品采于华中枳模式产地附近3 个不同县域,华中枳的样品来源于模式产地的3 个不同居群,富民枳样品来源于昆明植物研究所种质园(引种自富民县)。宜昌橙(C.cavaleriei)样品采于湘西吉首市与鄂西神农架林区。外类群和其他同属近源类群的DNA序列(含克隆测序数据)根据文献[9]从NCBI下载。近源类群种类在不同基因分析中并不能完全保持一致,但都涵盖了广义柑橘属的大部分物种。样品信息见表1,相关凭证标本保存于吉首大学植物标本馆(JIU),外类群及其他近源类群的DNA序列信息见附表。
表1 试验材料的采集信息及GenBank 序列号
Table 1 Collection information and GenBank accession numbers of experimental materials
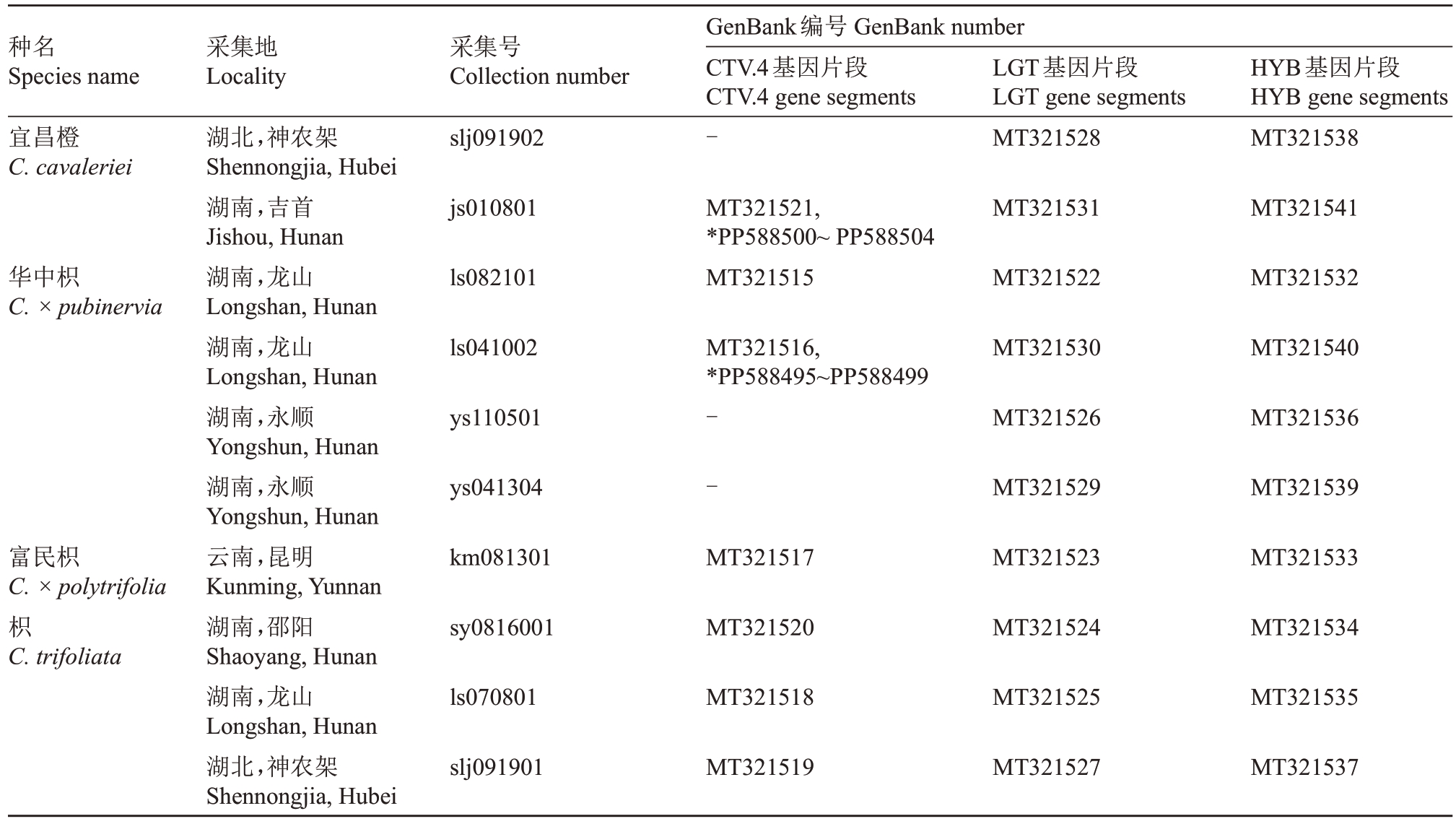
注:*标记克隆测序结果,-表示数据缺失。
Note:*Representative cloning sequencing results,-Indicates data is missing.
种名Species name宜昌橙C.cavaleriei华中枳C.×pubinervia富民枳C.×polytrifolia枳C.trifoliata采集地Locality湖北,神农架Shennongjia,Hubei湖南,吉首Jishou,Hunan湖南,龙山Longshan,Hunan湖南,龙山Longshan,Hunan湖南,永顺Yongshun,Hunan湖南,永顺Yongshun,Hunan云南,昆明Kunming,Yunnan湖南,邵阳Shaoyang,Hunan湖南,龙山Longshan,Hunan湖北,神农架Shennongjia,Hubei采集号Collection number slj091902 js010801 ls082101 ls041002 ys110501 ys041304 km081301 sy0816001 ls070801 slj091901 GenBank编号GenBank number CTV.4基因片段CTV.4 gene segments-MT321521,*PP588500~PP588504 MT321515 MT321516,*PP588495~PP588499--MT321517 MT321520 MT321518 MT321519 LGT基因片段LGT gene segments MT321528 MT321531 MT321522 MT321530 MT321526 MT321529 MT321523 MT321524 MT321525 MT321527 HYB基因片段HYB gene segments MT321538 MT321541 MT321532 MT321540 MT321536 MT321539 MT321533 MT321534 MT321535 MT321537
1.2 DNA的提取与扩增
总DNA的提取使用改良的CTAB法[22],材料为硅胶快速干燥的新鲜叶片。对CTV.4、HYB、LGT、P12的DNA片段进行扩增,将条带清晰的扩增产物用于双向测序,扩增与测序使用的引物与文献[9]相同。鉴于CTV.4 基因树的特殊性,选取华中枳和宜昌橙为代表对CTV.4 基因进行克隆测序。克隆使用擎科TSV-007VS-pClone007试剂盒,将目的DNA片段连入载体、转入受体细胞后进行培养,最后挑取5个单克隆进行双向测序。将双向测序结果进行手动组装后,用于后续分析。
1.3 系统发育分析
采用最大似然法(ML)和贝叶斯法(BI)分别构建系统发育树。使用MAFFT 7.0 将序列对齐[23]。最佳核酸替代模型使用PhyloSuite 中集成的ModelFinder计算[24-25]。最大似然法使用IQtree2.2.0[26]进行10 000 次bootstrap 运算。 贝叶斯法使用MrBayes 3.1.2[27],以马尔科夫链-蒙特卡罗算法(MCMC)进行2 次独立重复运算,参数设置为4 条平行热链运行100 000 代,以平均标准误差(<0.01)来判断数据收敛性,舍弃25%老化样本后估算后验概率。
2 结果与分析
HYB基因树拓扑结构如图1所示。枳、富民枳和华中枳在广义柑橘属中形成第一个支持率较强的独立分支(LP=100%,PP=1.00),即枳属分支。枳属进一步分化成“落叶枳”分支(LP=76%,PP=0.81),和包含了富民枳和华中枳的“常绿枳”分支(LP=81%,PP=0.76)。HYB 基因树不能区分2 种“常绿枳”。曾有学者认为华中枳是与宜昌橙有关的杂交种[11],但宜昌橙在HYB基因树上与“常绿枳”分支2个物种相距甚远。
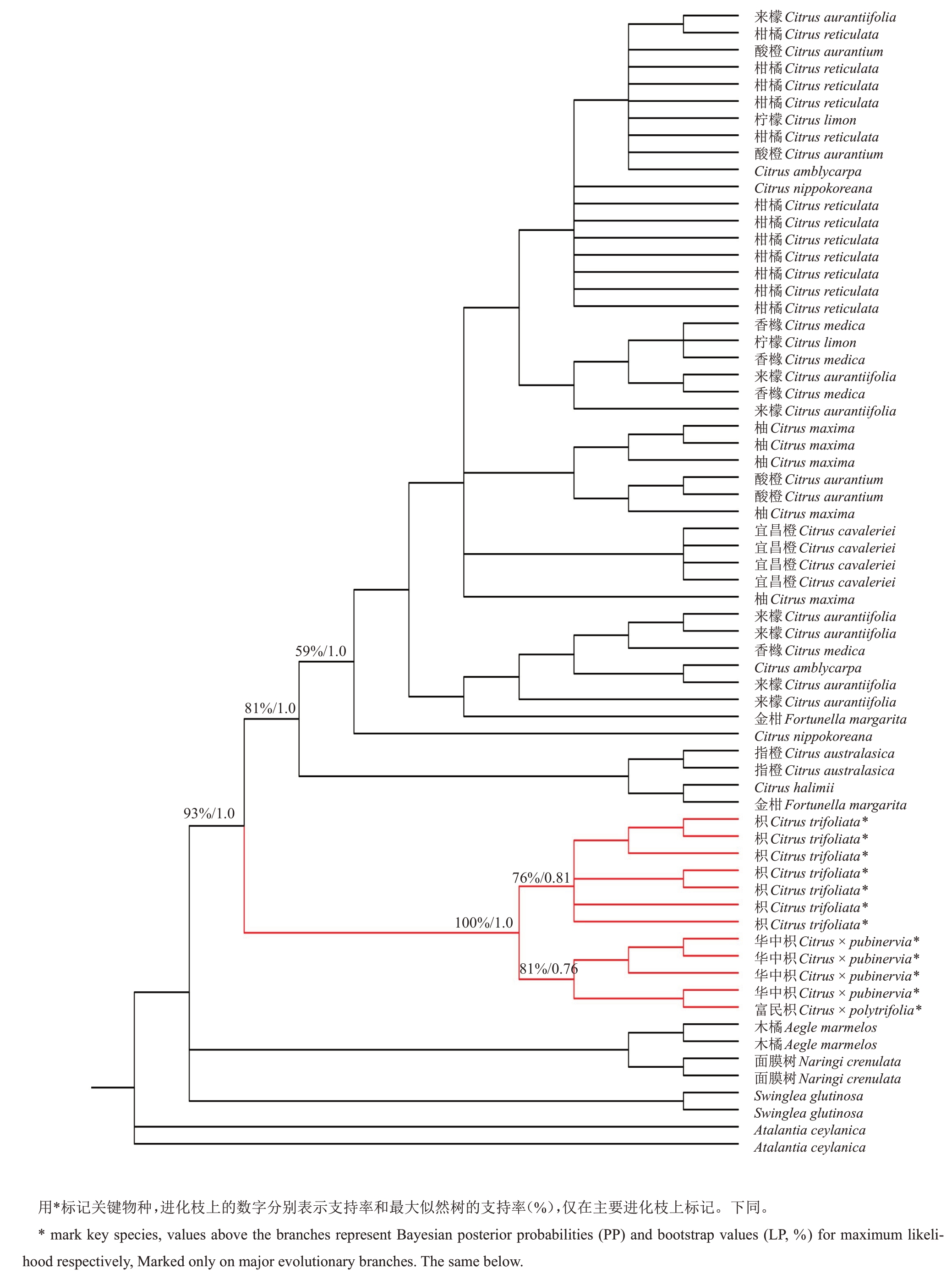
图1 HYB 贝叶斯系统发育树的拓扑结构
Fig.1 The phylogram of BI tree of the HYB
如图2 所示,在LGT 基因树上,枳与2 种“常绿枳”同样在广义柑橘属中形成一个独立的分支(LP=100%,PP=1.00)。但与HYB基因树不同,LGT基因树不能区分“落叶枳”和“常绿枳”。宜昌橙在LGT基因树上仍然处于狭义柑橘属中。LGT 与HYB 基因树的拓扑没有明显的不一致,但LGT 比HYB 更保守,对物种的区分度更低。
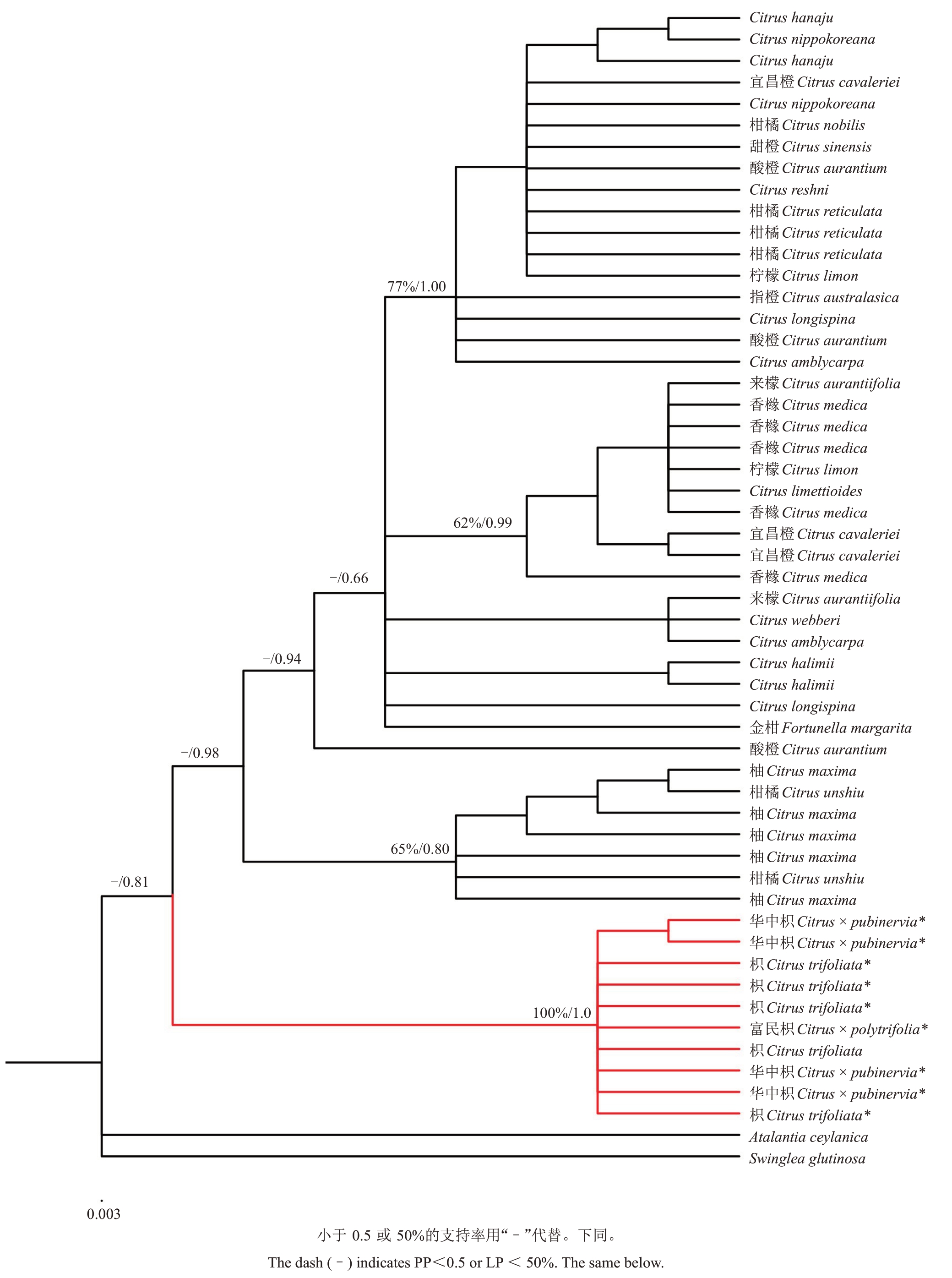
图2 LGT 贝叶斯系统发育树的拓扑结构
Fig.2 The phylogram of BI tree of the LGT
如图3 所示,P12 基因树拓扑结构与前两者不一致。将枳属分支嵌入广义柑橘属中,与金柑属同处一个支持率较低的大分支(LP=0,PP=0.66)。P12基因树不能区分枳属分支的3 个物种,且对广义柑橘属的其他物种的区分能力有限,整体呈现“梳子状”。

图3 P12 贝叶斯系统发育树的拓扑结构
Fig.3 The phylogram of BI tree of the P12
如图4 所示,CTV.4 基因树的拓扑结构与前3者的明显不同,2种“常绿枳”与枳发生了分离,而与柚[C. maxima(Burman)Merrill]以及酸橙[C. × aurantium Siebold 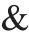 Zucc. ex Engl.]聚集在一个支持率较弱的大分支上(LP=52%,PP=0.63)。这一大分支进一步分化为2 个具有较强支持率的小分支,即“柚”分支(LP=98%,PP=1.00)和“常绿枳”分支(LP=97%,PP=1.00),这2 个小分支之间产生了明显的分化。CTV.4 基因树不能将柚与酸橙区分为2 个物种,也不能将华中枳与富民枳区分为两个物种。笔者补充了华中枳的克隆测序来验证这一现象,结果显示它的3条克隆序列位于枳属分支,2条克隆序列与富民枳一同位于“柚”分支中,表现出杂交起源的特性。宜昌橙直接测序及克隆测序序列形成一个具有较强支持率的分支(LP=98%,PP=1.00),与枳属分支相邻而与“常绿枳”距离较远。
Zucc. ex Engl.]聚集在一个支持率较弱的大分支上(LP=52%,PP=0.63)。这一大分支进一步分化为2 个具有较强支持率的小分支,即“柚”分支(LP=98%,PP=1.00)和“常绿枳”分支(LP=97%,PP=1.00),这2 个小分支之间产生了明显的分化。CTV.4 基因树不能将柚与酸橙区分为2 个物种,也不能将华中枳与富民枳区分为两个物种。笔者补充了华中枳的克隆测序来验证这一现象,结果显示它的3条克隆序列位于枳属分支,2条克隆序列与富民枳一同位于“柚”分支中,表现出杂交起源的特性。宜昌橙直接测序及克隆测序序列形成一个具有较强支持率的分支(LP=98%,PP=1.00),与枳属分支相邻而与“常绿枳”距离较远。
图4 CTV.4 贝叶斯系统发育树的拓扑结构
Fig.4 The phylogram of BI tree of the CTV.4
3 讨 论
3.1 富民枳和华中枳的分类地位
CTV.4 与其他3 个nDNA 单基因树之间的拓扑结构明显不一致,显示富民枳和华中枳可能经历过杂交事件。在CTV.4 基因树中,作为与柚有关的杂交种,酸橙与柚关系十分亲密,在“柚”分支中不分彼此;而“常绿枳”与“柚”分支发生了明显的遗传分化,表明其起源应该要远远早于酸橙,而且经历了独立的演化进程。更早的cpDNA 的系统发育分析显示[11],枳、富民枳和华中枳在叶绿体基因水平上发生了分化,是3 个可以区分的物种。因此富民枳和华中枳可能起源于枳(♀)与其他广义柑橘属物种(♂)的自然杂交,而且柚是最有可能成为它们母本的物种。
由于广义柑橘属经历了复杂的网状进化,柚被认为参与了其他杂交物种的形成[9-10](如酸橙),所以“常绿枳”的父本并不能通过少数基因片段的研究而下定论,其父本可能是柚或含有柚血缘的自然杂交种。但这一推论并不说明2种“常绿枳”不是分类学意义上的独立物种,因为自然杂交事件是物种形成,尤其是许多广义柑橘属物种形成的重要方式之一。这2 个物种是狭义柑橘属和枳属之间的桥梁,对研究广义柑橘属的系统发育关系和柑橘育种具有重要价值,应受到更多的关注和保护。建议将华中枳列为地方保护物种或与富民枳同等级的保护物种。
3.2 对枳属分类地位的建议
在不同类型和不同深度的基因研究中,枳属分支的位置不一致。枳属分支在9个叶绿体基因系统发育树中嵌入广义柑橘属中,位于“南半球为主(澳大利亚、新几内亚、新喀里多尼亚、新爱尔兰)的分支”与“北半球分支”之间[1]。在本研究的4个nDNA单基因树中,枳属分支的位置不一致。在HYB 与LGT基因树中,枳属分支是广义柑橘属中第一个较独立的分支,而在P12 和CTV.4 单基因树中枳属分支嵌入广义柑橘属中。在全基因组水平上的分析表明,枳属分支是广义柑橘属最外侧较独立的分支[8,10],它的起源应早于广义柑橘属南北半球分支的形成[10]。
总体上来说,枳属分支是广义柑橘属中较独立的一个分支,但它更像一个没能完成独立演化的古老“演化残枝”,通过共同起源、基因交流或趋同进化等复杂因素与广义柑橘属其他物种在遗传上产生复杂联系。因为枳属分支与“北半球分支”有共同的祖先、相似的气候环境以及分布区重叠,不排除在演化过程中与“北半球分支”产生基因交流,它们之间实际上也存在杂交亲和性[2]。富民枳和华中枳的存在也为它们之间存在基因交流提供了证据。因此枳属分支可以归为广义柑橘属的一个亚属,“常绿枳”位于枳和广义柑橘属的其他物种之间,属于中间产物。
4 结 论
笔者在对枳和常绿枳广泛采集样本的基础上,选取了4 个无明显连锁关系的核DNA 片段进行直接或克隆测序,重建了广义柑橘属的系统发育树。结果显示,富民枳和华中枳可能起源于枳(♀)与柚的杂交。二者在遗传上与推定亲本产生了分化,应是经历了独立演化后的自然杂交物种。这一研究结果为富民枳和华中枳的分类地位提供了新的见解,为枳属与狭义柑橘属之间存在自然状态下的基因交流提供了证据。“常绿枳”是狭义柑橘属和枳属之间的中间产物,对研究广义柑橘属的系统发育关系和柑橘育种具有重要价值。因此,建议将华中枳列为地方保护物种或与富民枳同等级的保护物种。
[1] BAYER R J,MABBERLEY D J,MORTON C,MILLER C H,SHARMA I K,PFEIL B E,RICH S,HITCHCOCK R,SYKES S. A molecular phylogeny of the orange Subfamily(Rutaceae:Aurantioideae) using nine cpDNA sequences[J].American Journal of Botany,2009,96(3):668-685.
[2] SWINGLE W T,REECE P C. The botany of citrus and its wild relatives of the orange subfamily[M]. Berkeley,California,USA:University of California,1967.
[3] 谢让金,周志钦,邓烈.真正柑橘果树类植物基于AFLP 分子标记的分类与进化研究[J]. 植物分类学报,2008,46(5):682-691.XIE Rangjin,ZHOU Zhiqin,DENG Lie.Taxonomic and phylogenetic relationships among the Genera of the True Citrus Fruit Trees Group (Aurantioideae,Rutaceae) based on AFLP markers[J]. Journal of Systematics and Evolution,2008,46(5):682-691.
[4] MORTON C M. Phylogenetic relationships of the Aurantioideae(Rutaceae) based on the nuclear ribosomal DNA ITS region and three noncoding chloroplast DNA regions,atpB-rbcL spacer,rps16,and trnL- trnF[J]. Organisms Diversity  Evolution,2009,9(1):52-68.
Evolution,2009,9(1):52-68.
[5] PENJOR T,YAMAMOTO M,UEHARA M,IDE M,MATSUMOTO N,MATSUMOTO R,NAGANO Y. Phylogenetic relationships of Citrus and its relatives based on matK gene sequences[J].PLoS One,2013,8(4):e62574.
[6] PENJOR T,ANAI T,NAGANO Y,MATSUMOTO R,YAMAMOTO M. Phylogenetic relationships of Citrus and its relatives based on rbcL gene sequences[J]. Tree Genetics  Genomes,2010,6(6):931-939.
Genomes,2010,6(6):931-939.
[7] MORTON C M,GRANT M,BLACKMORE S.Phylogenetic relationships of the Aurantioideae inferred from chloroplast DNA sequence data[J]. American Journal of Botany,2003,90(10):1463-1469.
[8] GARCIA-LOR A,CURK F,SNOUSSI-TRIFA H,MORILLON R,ANCILLO G,LURO F,NAVARRO L,OLLITRAULT P. A nuclear phylogenetic analysis:SNPs,indels and SSRs deliver new insights into the relationships in the‘true Citrus fruit trees’group (Citrinae,Rutaceae) and the origin of cultivated species[J].Annals of Botany,2013,111(1):1-19.
[9] RAMADUGU C,PFEIL B E,KEREMANE M L,LEE R F,MAUREIRA-BUTLER I J,ROOSE M L. A six nuclear gene phylogeny of Citrus (Rutaceae) taking into account hybridization and lineage sorting[J].PLoS One,2013,8(7):e68410.
[10] WU G A,TEROL J,IBANEZ V,LÓPEZ-GARCÍA A,PÉREZROMÁN E,BORREDÁ C,DOMINGO C,TADEO F R,CARBONELL-CABALLERO J,ALONSO R,CURK F,DU D L,OLLITRAULT P,ROOSE M L,DOPAZO J,GMITTER F G,ROKHSAR D S,TALON M.Genomics of the origin and evolution of Citrus[J].Nature,2018,554(7692):311-316.
[11] WU Y,LIU Q,XIANG Z H,ZHANG D G.Citrus×pubinervia,a new natural hybrid species from Central China[J]. Phytotaxa,2021,523(3):239-246.
[12] 丁素琴,张显努,暴卓然,梁明清.中国枳属一新种[J].云南植物研究,1984,6(3):292-293.DING Suqin,ZHANG Xiannu,BAO Zhuoran,LIANG Mingqing. A new species of Poncirus from China[J]. Acta Botanica Yunnanica,1984,6(3):292-293.
[13] ZHANG D X,MABBERLEY D J. Citrus[M]//WU Z Y,Raven P H. Flora of China. Beijing:Science Press and St. Louis,Missour:Missouri Botanical Garden Press,2008,11:90-96.
[14] 吴兴恩,范眸天,高峻,李文祥,龙雯虹,许明辉.枳、富民枳与枳橙RAPD 分析与分类(亲缘)关系探讨[J].云南农业大学学报,2003,18(3):277-280.WU Xing’en,FAN Moutian,GAO Jun,LI Wenxiang,LONG Wenhong,XU Minghui.Studies on the RAPD analysis and taxonomic relationship of trifoliate orange,Fuming trifoliate orange and citrange[J]. Journal of Yunnan Agricultural University,2003,18(3):277-280.
[15] 张余,龚洵,冯秀彦.利用DNA 片段测序方法探究枳属和富民枳的分类地位[J].广西植物,2021,41(1):114-122.ZHANG Yu,GONG Xun,FENG Xiuyan. Phylogenetic position of Poncirus and Poncirus polyandra by DNA sequencing[J].Guihaia,2021,41(1):114-122.
[16] 龚桂芝,洪棋斌,彭祝春,江东,向素琼.枳属种质遗传多样性及其与近缘属植物亲缘关系的SSR 和cpSSR 分析[J].园艺学报,2008,35(12):1742-1750.GONG Guizhi,HONG Qibin,PENG Zhuchun,JIANG Dong,XIANG Suqiong. Genetic diversity of Poncirus and its phylogenetic relationships with relatives as revealed by nuclear and chloroplast SSR[J]. Acta Horticulturae Sinica,2008,35(12):1742-1750.
[17] 刘利勤,杨静,顾志建.富民枳的核型与分类位置探讨[J].云南植物研究,2007,29(2):198-200.LIU Liqin,YANG Jing,GU Zhijian. Karyomorphology and taxonomic position of Poncirus polyandra (Rutaceae)[J].Acta Botanica Yunnanica,2007,29(2):198-200.
[18] 范眸天,梁明清,浦卫琼.富民枳与枳的花粉形态与分类位置探讨[J].云南农业大学学报,1998,13(3):298-300.FAN Moutian,LIANG Mingqing,PU Weiqiong. Studies on the pollen morphology and taxonomic position of P.trifoliata and P.Polyandra[J]. Journal of Yunnan Agricultural University (Natural Science),1998,13(3):298-300.
[19] FANG D Q. Intra-and intergeneric relationships of Poncirus polyandar:Investigation by leaf isozymes[J]. Journal of Wuhan Botanical Research,1993,11(1):34-40.
[20] 钟瑞芳,王仕玉,梁明清,杜元明,段志宇.富民枳的生物学特性及利用研究初报[J]. 云南农业大学学报,2001,16(3):244-246.ZHONG Ruifang,WANG Shiyu,LIANG Mingqing,DU Yuanming,DUAN Zhiyu. Studies on biological characteristics and application of Poncirus polyandra S.Q.Ding et al[J].Journal of Yunnan Agricultural University,2001,16(3):244-246.
[21] 高雪松,张玉萍,唐金武,顾文辉.浅谈富民枳生存现状与保护发展[J].防护林科技,2017(12):83-85.GAO Xuesong,ZHANG Yuping,TANG Jinwu,GU Wenhui.Survival status and protection development of Fumin Trifoliate Orange[J].Protection Forest Science and Technology,2017(12):83-85.
[22] DOYLE J,DOYLE J L,DOYLE J,DOYLE F J.A rapid DNA isolation procedure for small amounts of fresh leaf tissue[J].Phytochemistry,1987,19(1):11-15.
[23] KATOH K,STANDLEY D M.MAFFT multiple sequence alignment software version 7:Improvements in performance and usability[J]. Molecular Biology and Evolution,2013,30(4):772-780.
[24] KALYAANAMOORTHY S,MINH B Q,WONG T K F,VON HAESELER A,JERMIIN L S. ModelFinder:Fast model selection for accurate phylogenetic estimates[J]. Nature Methods,2017,14(6):587-589.
[25] ZHANG D,GAO F L,JAKOVLIĆ I,ZOU H,ZHANG J,LI W X,WANG G T. PhyloSuite:An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies[J]. Molecular Ecology Resources,2020,20(1):348-355.
[26] NGUYEN L T,SCHMIDT H A,VON HAESELER A,MINH B Q. IQ-TREE:A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies[J]. Molecular Biology and Evolution,2015,32(1):268-274.
[27] HUELSENBECK J P,RONQUIST F. MRBAYES:Bayesian inference of phylogenetic trees[J]. Bioinformatics,2001,17(8):754-755.
附表 用于构建系统发育树的DNA序列信息
Annexed table DNA sequence information for constructing phylogenetic trees

基因Gene CTV.4 HYB LGT P12学名,基因编号Scientific name,gene number Atalantia ceylanica(Am.)Oliv.,EU254134;Citrus amblycarpa Ochse.,EU254151,EU254152;Citrus aurantiifolia(Christm.)Swing.EU254146,EU254147;Citrus aurantium L.,EU254158,EU254159,EU254160;Citrus halimii B.C.Stone,EU254140,EU254141;Citrus hanaju Siebold,EU254168,EU254169;Citrus cavaleriei H.Lév.ex Cavalier(Citrus ichangensis Swing.),EU254167;Citrus limettioides Tanaka,EU254148; Citrus limon (L.) Burm.f.,EU254150; Citrus longispina Wester,EU254144,EU254145; Citrus maxima(Burm.) Merrill,EU254163,EU254164,EU254165,EU254166; Citrus medica L.,EU254135,EU254136,EU254137,EU254138 EU254139;Citrus nippokoreana Tanaka,EU254156,EU254157;Citrus nobilis Lour.,EU254162;Citrus reticulata Blanco,EU254153 EU254154,EU254155; Citrus sinensis (L.) Osbeck,EU254161; Citrus unshiu Marc.,EU254149; Citrus webberi Wester,EU254142 EU254143; Fortunella margarita Swingle,EU254171; Citrus australasica F. Muell. (Microcitrus australasica Swingle),EU254172;Citrus trifoliata L.(Poncirus trifoliata(L.)Rafinesque),EU254170.Aegle marmelos (L.) Corrêa,JN612932, JN612933; Atalantia ceylanica (Am.) Oliv.,GQ892193,GQ892192; Citrus amblycarpa Ochse.,GQ892213,GQ892212; Citrus aurantiifolia (Christm.) Swing.,GQ892242,GQ892241,GQ892229,GQ892228,GQ892227 GQ892209, GQ892208; Citrus aurantium L.,GQ892244,GQ892243,GQ892207,GQ892206; Citrus halimii B.C. Stone,GQ892214;Citrus cavaleriei H. Lév. ex Cavalier (Citrus ichangensis Swing.),GQ892216,GQ892215; Citrus limon (L.) Burm.f.,GQ892224 GQ892223; Citrus maxima (Burm.) Merrill,GQ892234,GQ892233,GQ892232,GQ892231,GQ892230; Citrus medica L.GQ892226,GQ892225,GQ892205,GQ892204; Citrus nippokoreana Tanaka,GQ892218,GQ892217; Citrus reticulata Blanco GQ892246,GQ892245,GQ892240,GQ892239,GQ892238,GQ892237,GQ892236,GQ892235,GQ892222,GQ892221,GQ892220 GQ892219, Fortunella margarita Swingle,GQ892211,GQ892210; Citrus australasica F. Muell. (Microcitrus australasica Swingle)GQ892203,GQ892202;Naringi crenulata(Roxb.)Nicolson,GQ892197,GQ892196;Citrus trifoliata L.(Poncirus trifoliata(L.)Rafinesque),GQ892201,GQ892200,GQ892199,GQ892198;Swinglea glutinosa Merr.,GQ892195,GQ892194.Atalantia ceylanica(Am.)Oliv.,EU254173;Citrus amblycarpa Ochse.,EU254193,EU254192;Citrus aurantiifolia(Christm.)Swing.EU254186,EU254185;Citrus aurantium L.,EU254202,EU254201,EU254200;Citrus halimii B.C.Stone,EU254181,EU254180;Citrus hanaju Siebold,EU254211, EU254212;Citrus cavaleriei H.Lév.ex Cavalier(Citrus ichangensis Swing.),EU254210;Citrus limettioides Tanaka,EU254187;Citrus limon(L.)Burm.f.,EU254191,EU254190;Citrus longispina Wester,EU254184,EU254183;Citrus maxima (Burm.) Merrill,EU254207,EU254209,EU254206,EU254208,EU254205; Citrus medica L.,EU254178,EU254176 EU254175,EU254177,EU254179;Citrus nippokoreana Tanaka,EU254198,EU254197;Citrus nobilis Lour.,EU254204;Citrus reshni Hort.ex Tanaka,EU254199;Citrus reticulata Blanco,EU254196,EU254195,EU254194;Citrus sinensis(L.)Osbeck,EU254203;Citrus unshiu Marc.,EU254189,EU254188;Citrus webberi Wester,EU254182;Fortunella margarita Swingle,EU254215;Citrus australasica F. Muell. (Microcitrus australasica Swingle),EU254216; Citrus trifoliata L. (Poncirus trifoliata (L.) Rafinesque),EU254214 EU254213;Swinglea glutinosa Merr.,EU2541.Atalantia ceylanica (Am.) Oliv., EU254033; Citrus medica L., EU254034, EU254035, EU254036, EU254037; Citrus halimii B.C.Stone, EU254038, EU254039; Citrus webberi Wester, EU254040, EU254041; Citrus longispina Wester, EU254042; Citrus aurantiifolia(Christm.)Swing.,EU254043;Citrus limettioides Tanaka,EU254044,EU254045;Citrus unshiu Marc.,EU254046,EU254047;Citrus limon (L.) Burm.f., EU254048, EU254049; Citrus amblycarpa Ochse., EU254050; Citrus reticulata Blanco, EU254051,EU254052, EU254053, EU254054, EU254055; Citrus nippokoreana Tanaka, EU254056, EU254057; Citrus reshni Hort. ex Tanaka,EU254058; Citrus aurantium L., EU254059, EU254060, EU254061, EU254062; Citrus sinensis (L.) Osbeck, EU254063, EU254064;Citrus nobilis Lour., EU254065, EU254066; Citrus maxima (Burm.) Merrill, EU254067, EU254068, EU254069, EU254070,EU254071; Citrus cavaleriei H. Lév. ex Cavalier (Citrus ichangensis Swing.), EU254072; Citrus hanaju Siebold, EU254073,EU254074;Citrus trifoliata L.(Poncirus trifoliata(L.)Rafinesque),EU254075,EU254076,EU254077,EU254078;Fortunella margarita Swingle,EU254079,EU254080;Citrus australasica F.Muell.(Microcitrus australasica Swingle),EU254081,EU254082.