柑橘是我国和世界的第一大果树作物,属芸香科(Rutaceae)真正柑橘果树组(true citrus fruit tree group)[1]。常见的柑橘作物分布于柑橘属(Citrus)、枳属(Poncirus)和金柑属(Fortunella)[2]。柑橘是蜜源植物,缺乏种间甚至属间的生殖隔离,通过自然或人工授粉易产生种间甚至属间杂交种。橘柚(tange‐lo)属于一类杂种宽皮柑橘。一些橘柚是葡萄柚(C. paradisi Macf.)或柚[C. maxima(Burm.)Merr.]和橘(C. reticulata Blanco)的杂种[3],例如奥兰多(Orlando)和明尼奥拉(Minneola)橘柚就是邓肯(Duncan)葡萄柚和丹西(Dancy)红橘的杂种后代。广义上,具有橘柚风味特征的橘柚杂种后代,例如费尔柴尔德[Fairchild,奥兰多橘柚与克里曼丁橘(Cle‐mentine)的杂种],都可被认为是橘柚。此外,日本杂柑春香(Haruka)也被认为是橘柚,这是因为春香是日向夏(Yuganatsu,C. tamurana Hort. ex Tanaka)和夏蜜柑(Natsudaidai,C. natsudaidai Hayata)的杂种后代[4],其父本夏蜜柑在柑橘斯文格(Swingle)分类系统中属于柚类。由于橘柚兼具橘和柚的特征,风味独特,较易剥皮,近年来越来越受到消费者的欢迎和柑橘育种工作者的重视。
中国的橘柚育种工作起步晚,但目前已有所突破。笔者所在的研究团队在爱媛28 号[Ehime Kashi No. 28,为南香(Nanko)与天草(Amakusa)橘橙的杂交后代]与春香橘柚的杂交后代中,筛选出了两个性状优良、具有商业价值的株系,分别命名为阳光1号和阳光2号橘柚,其中阳光1号已获得植物新品种权,目前在中国的栽培面积约为3300 hm2,并正在迅速推广。
分子标记(molecular marker)是植物基因分型(genotyping)、品种鉴定、遗传多样性分析和辅助育种的重要工具[5-8]。在柑橘研究中,除了广泛应用的SSR(simple sequence repeats)[9-24]和SNP(single nu‐cleotide polymorphism)[18,24-30]标记之外,InDel(inser‐tion-deletion)因含量丰富且具共显性的特点,也受到了人们的关注。在较早的研究中,Garcia-Lor等[24]对45 种真正柑橘果树组植物的27 个基因进行桑格(Sanger)测序,获得了50 个InDel 标记,结合SSR 和SNP,他们可有效应用于对真正柑橘果树组植物的系统发育和种间遗传结构分析。Fang 等[31]根据已公布的多种柑橘的全基因组测序数据,共挖掘到1958 个InDel 标记,其中268 个为30~200 bp 的大片段InDel,这些标记可区分柑橘属、枳属和金柑属植物。日本学者Noda等[32]利用119个适用于琼脂糖凝胶电泳检测的InDel,成功地将温州蜜柑(satsuma)的合子胚从珠心胚群体中筛选出来;后又从中筛选了28 个标记,成功地区分了31 种杂种柑橘[33]。汤雨晴等[34]鉴定了金兰柚的全基因组InDel,并筛选出2 个能将金兰柚与其他47种柚类区分的InDel标记。
笔者首先对阳光1 号和阳光2 号橘柚及其亲本爱媛28号和春香进行全基因重测序,从中发掘出48个适用于普通琼脂糖电泳检测的InDel标记,并进一步评价了这些InDel 在51 份种质资源的遗传多样性,用主坐标分析(PCoA)展示了他们的遗传距离。基于这48 个InDel标记的基因分型是区分和鉴定橘柚品种的有效简易方法,也可应用于真正柑橘果树组植物的遗传多样性和群体分类分析。
1 材料和方法
1.1 试验材料
材料枸橼(C.medica L.)为普通枸橼和佛手;金柑(Fortunella spp.)为宁波罗浮和四季橘;橘类为立花橘、汕头酸橘、武隆酸橘、克里曼丁橘、尾张、宫川早生、粗皮狗屎柑、莽山野橘、芦柑、椪柑、大红袍、清见、春见、砂糖橘、中柑所5 号、无核纪州柑、本地早、伊予柑、药香柑和爱媛28 号;橘柚(C. reticulata Blanco)为春香、阳光1号、阳光2号、奥兰多、明尼奥拉、甜春、科肯、诺瓦、费尔柴尔德和清峰;柠檬[C.li‐mon(L.)Burm.f.]为里斯本、尤力克和北京柠檬;葡萄柚为马叙和胡柚;甜橙[C.sinensis(L.)Osbeck]为塔罗科、北碚447、纽荷尔、哈姆林和伏令夏橙;香橙(C.junos Siebold.ex Tanaka)为资阳香橙和蟹橙;柚为强得勒和琯溪蜜柚;枳(P. trifoliata Raf.)为孝感枳、湖北早实枳和飞龙枳,共51 份种质资源,其中阳光1号和阳光2号橘柚来自重庆奔象果业有限公司,其他种质资源来自国家柑橘种质资源圃。
1.2 基因组DNA提取
使用RaPure Plant DNA Kit(美基生物,广州)提取健康成熟叶片的基因组DNA。使用Nanodrop 2000分光光度计(Thermo Scientific,美国)检测所提取DNA浓度和质量。当DNA质量浓度大于10 ng·μL-1且OD260/280和OD260/230在1.8~2.1 时,可用于后续试验的建库测序和PCR扩增。
1.3 全基因组重测序及InDel挖掘
用Bioruptor Pico系统(Diagenode,比利时)对所提基因组DNA进行剪切,形成约350 bp的片段。使用TrueLib DNA Library Rapid Prep Kit(依科赛生物,深圳)建库,用NovaSeq 6000 Sequencer(Illumi‐na,美国)进行平均深度为10×的双端测序。滤去低质量(N 比例大于10%或Phred<10 的比例小于50%)的读长(read)后,用BWA[35]与克里曼丁橘的基因组序列(https://phytozome.jgi.doe.gov/pz/portal.ht‐ml)比对,用GATK[36]获取和过滤InDel。过滤标准:(1)滤去10 bp 内的相邻InDel;(2)变异位点的质量值(QUAL)大于30;(3)变异质量值与覆盖深度得到的比值(QD)大于2.0;(4)比对质量值的均方根(MQ)大于40;(5)费舍尔精确检验(Fisher’s exact test)正负链偏差(FS)小于60;(6)其他GATK 设定的默认值。所得变异用SnpEff[37]进行注释。
1.4 引物设计和PCR扩增
使用BatchPrimer3[38],对变异长度大于30 bp 或小于-30 bp的InDel的两端设计引物,PCR产物长度约为200 bp,引物信息见表1。用MonAmp™2×Taq Mix(莫奈生物,上海)进行PCR,反应程序为94 ℃3 min,然后94 ℃30 s,58 ℃30 s,72 ℃30 s,共35个循环,最后72 ℃反应2 min。PCR产物用含1/10 000 AidRed(艾得莱,北京)的2.5%琼脂糖凝胶电泳分离,在紫外灯下拍照记录。
表1 用于检测48 个InDel 标记的引物
Table 1 Primers for detecting 48 InDel markers

注:a.第一行和第二行分别为上游引物序列和下游引物序列;b.通过克里曼丁橘参考基因组计算PCR 产物的理论长度;c.以InDel 变异基因组为模板的PCR 产物的理论长度。
Note:a.The first and second row indicated the upstream and downstream prime sequence; b.The theoretical PCR size from Clementine reference genome;c.The theoretical PCR size from the genome of InDel variant.
名称Name片段框架Scaffold位置Position引物序列a Sequence PCR产物长度(参考)b PCR product size(reference)/bp PCR产物长度(变异)c PCR product size(variant)/bp I1_767 74767 187 220 I1_191 19397191 183 153 I1_846 21522846 223 183 I1_400 27287400 174 232 I1_184 28202184 213 178 I1_639 28494639 208 176 I2_691 3793691 192 231 I2_248 17711248 186 140 I2_815 28696815 221 190 I2_137 35384137 234 270 I3_744 173744 180 256 I3_068 879068 257 217 I3_321 17751321 208 157 I3_311 20566311 175 208 I3_326 20885326 273 231 I3_890 31542890 210 169 I3_175 42136175 185 155 I4_604 764604 259 206 I4_690 1095690 192 239 I4_085 7106085 241 289 I4_513 9625513 237 175 I4_457 23930457 237 201 I5_453 1369453 207 175 I5_676 2274676 250 287 I5_322 1 1 1 1 1 1 2 2 2 2 3 3 3 3 3 3 3 4 4 4 4 4 5 5 5 29398322 AACTAAACAGCCCCGCATTT AGCCATTTCAAGAGGAAGTAGAA AAGCCACGAGAGTTACAGCAA TTTCAACGACAAACACCTCCT TTTCCAGGCTCAAGTTACAGC GCACAATAGCGTTTACAATAATCAC GCAACAGCGAATACCTCAGA CTTATGGTGGCGAGTTTCCT TACATTGCGTCCTGTCTTGG GCATCAACATCTCCATAAACGA GCAAGTTCAGGTCATCTCCAG TCAAGCAGCAGACATTCAGTT CGTCACCCTGGCAAATCTA GCAAATCCAAGTATCACGAAAA TCCACGAGTAAGGCTAAGGAA GGGGAATGGGTAATGTTCAC TGGGGGTTAGGTTCATTTTG TCCTGTTTTCCGATTATGTCAA TAAGAGCAAAGGCAACAAGC TTCACCAGAAAGATGATTGGA CTGTTTTGTCACTGCCCGTA TACACCCCGAACCCCACT AATTTCCTCCTGCAGCAACC TTCAACTGAAGCATTGCCGG TCTGAACAGGCAATGATGAGC AGGAGATTTCTGGTGCAAAGC ACTTTGCTTGTTGTGCTCACT CGTTCGAAGCCTAGACCTCT GCGGGAATATTTGAACTCACCA GGTCATTGCTGCCATTTCCA CCGAGCAGCACAGAACAT CACGACGGGGAGAGAATAAA CAGGGTGATTGCTTGCCTC GAGAAATTGGCTCCATGAATTGC TGTCAAATTCGCTATGCTCGT CCGAGATGCACAAAATTGAACC TGGGCAGTTTTGGTTGTGTG TCCCGCGATAACAGATACCT GATTGGCTCAACAGGTCCAC TGACAAGCAAACTCGAATCTGA CGGAGATTGACACAGGAGTG AGATTGGTGGGTTGAGAAGC ACTGGTCCCTGTGATGCTAA ACCCAAAGTCTATCGAGCCC AAACCGCCCTGGATTTGT GCAACCCTGATTTCTTCCTG GGCAGCAAAGGCAATAAAAC TGGTCTACAGGAGGTGTCCA CTCATTTGCGTCCCTTCAAC GCTTCTGTTTCCAGCCCTAA 235 193
表1 (续) Table 1 (Continued)

名称Name片段框架Scaffold位置Position引物序列a Sequence PCR产物长度(参考)b PCR product size(reference)/bp PCR产物长度(变异)c PCR product size(variant)/bp I5_445 43058445 229 189 I6_283 2190283 238 275 I6_552 7580552 276 235 I6_309 9762309 233 288 I6_502 10107502 221 182 I6_890 17090890 226 260 I6_401 20009401 204 170 I6_280 20088280 244 208 I6_681 25481681 262 210 I7_101 468101 235 203 I7_449 963449 269 331 I7_272 2253272 231 201 I7_826 2259826 264 224 I7_845 2627845 244 193 I7_170 19369170 256 287 I8_554 949554 219 189 I8_516 18035516 217 273 I8_533 20194533 177 146 I9_200 955200 161 192 I9_272 3463272 231 201 I9_459 6543459 185 231 I9_669 14389669 255 211 I9_386 5 6 6 6 6 6 6 6 6 7 7 7 7 7 7 8 8 8 9 9 9 9 9 24807386 TCCACTTACCACTTCTTCCAAC AACTTCACCACCCCAGACAG GAGAGAGAGAGAGGGAGAGAATGA ACCTTGGGATTGTTGTGTCA GAACTTTGACCATTTTCCCGTA CTTCCCCATTTTGCCTGAG AAGAGGCGAGGAGCAGTTG TGTTGTTTTGTTGGCTATTTCC GTGGTTGTGAATAAGGGCATT CGGACCCAAACGAATCTAAA GAGCAAGGAGCCATTTTGAG GCCATCTTCTACACCAAGCA CGTGTATGCTCTCAAAGTCTCAA GATGGAAACCCGATTACGAA TCATGCTCTCCAAACTCTCCA GGATTGGCATTCATATTCCACCA CCGCAAAACACGCAAAAA AGAAGTAACAATGACAGGACACG AAACCTCTCCAACTTTCATCG GTCGGCTTTTGCTTTCTCAT TGTGTTACGGCAGGTATGGT TGCGTCTCCAGTTCTATGAGG TCCCCAGTCCCCATAAGTAA AATCCGCAACAAATGAGAGG TTTGCGGTGTAAGGAACAGT CCATCATTGGCTCCCACA GAGAGCATTGTTTGGGGGTA GGGAGTCGCAGAACCTTGTA TCAGTGTTTGGGCTTGAGAA GATAGATTAGTTGATGTGTGGCGTA GCCAACGAGAGAAGAAGGAT CATTGCTGCCTGGTGAGTT AACCAATCTAAAAACCCAGCA AGGAGCAAACTGCCTTGTGT TCTCTGAACCAATAATCACTACCG ATGCTTTCTTCTGCCAAGTCT TCACCGAAACATCATAACCTTCA TTTCACCGCCTTGATGTTCA TTCTACCAACCTTATCCCAACTG TCTGCTACACTTATCCCCTACGA GAAAAAGGGGCACGGATT GCAAGGGGCTGAAAGACTAC GGCTGAGAGATGAGGGAGTTC CGAAATAACAAAAGGCACCAA ACTTGGAGTTGCTGGATGCT TTAGCGGTGCTCATTTCCTC 223 187
1.5 遗传多样性分析和主坐标分析(PCoA)
使用PowerMarker(V3.25)计算群体遗传学参数,包括基因型数量(Ng)、等位基因数量(Na)、有效等位位点数量(Ne)、最大等位基因频率(MAF)、期望杂合度(He)、观测杂合度(Ho)和多态信息含量(PIC)。主坐标分析基于51 份种质资源的欧氏遗传距离,使用R 语言ade4 程序包,用ggplot2 程序包绘制散点图。
2 结果与分析
2.1 全基因组InDel的挖掘
阳光1 号、阳光2 号、爱媛28 号和春香的基因组重测序原始数据(fastq 格式)已上传至中国国家基因库(https://db.cngb.org,项目号CNP0001863)。每一个品种都产生了超过1000万个高质量读长(read)和30亿个碱基,Q20和Q30均分别大于96%和92%,正确定位到克里曼丁橘参考基因组的读长比例均大于85%,1×基因组覆盖度均超过89.9%,5×基因组覆盖度超过67.95%,平均深度超过7×。以上结果表明,测序数据质量良好,可用于后续的变异检测及相关分析。
通过与克里曼丁橘的基因组对比以及GATK 软件对变异的提取和过滤,笔者在4 个品种的基因组中分别获得了285 757(爱媛28号)、338 510(春香)、320 991(阳光1 号)和296 648 个(阳光2 号)InDel 位点(图1-A)。作为杂交后代,阳光1 号和阳光2 号的InDel位点数目介于两个亲本之间。爱媛28号的In‐Del 数目明显低于春香,这是因为爱媛28 号是南香(Nanko)和天草(Amakusa)的杂交后代,而南香的母本正是克里曼丁橘,即与春香相比,爱媛28 号与克里曼丁橘有更近的亲缘关系。在剔除4 个品种共有的相同InDel 后,鉴定到607 376 个InDel 位点,共有657 414 种变异方式。绝大多数(92.5%,561 914 个)位点只有一种变异方式,其余41 112、4127、220 和3个位点则存在2、3、4、5 种变异。19.6%(119 007 个)的InDel 位点位于基因内,80.4%(488 369 个)的In‐Del 位点位于基因间。前者有83 983个(70.6%)位于内含子,其余35 024 个(29.4%,占总InDel 位点的5.77%)与基因编码相关,包括编码区(1.64%,9986个)、剪切相关区(0.36%,2169 个)、5'-UTR(1.49%,9021 个)、3'-UTR(2.25%,13 640 个);后者有192 203和133 980个分别处于基因上游和下游5 kb以内(图1-B)。变异长度为1 或2 的InDel 最为丰富,而大片段的InDel较少(图1-C),其中有2.1%~2.3%(5312~6402个)的InDel片段大于30 bp或小于-30 bp。这些大片段的InDel 有潜力适用于琼脂糖凝胶电泳检测的分子标记的开发。位于非编码区以及变异长度短的InDel 数量比例低,反映这些InDel 对基因功能的影响较小,承受较小的选择压力。
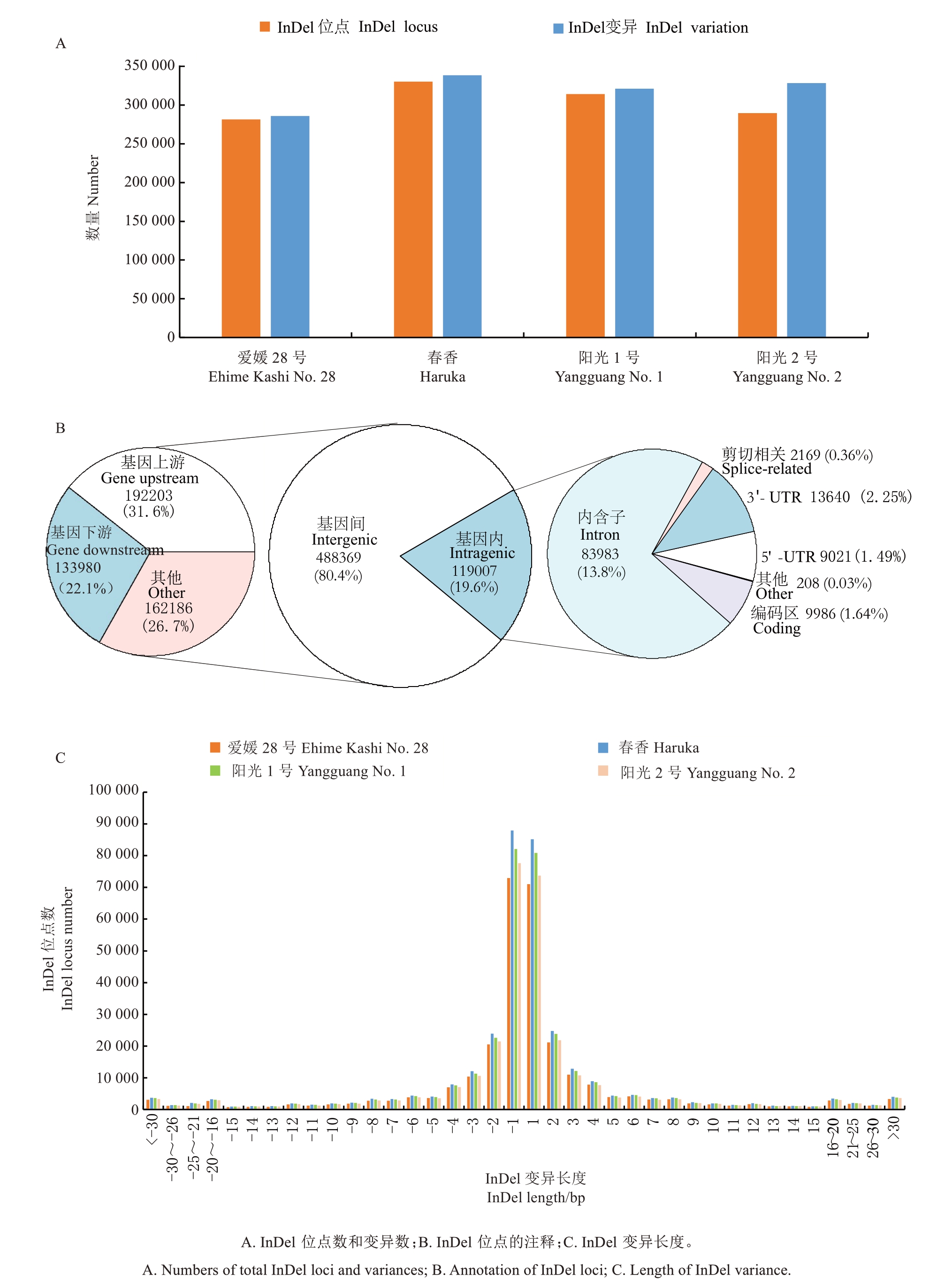
图1 两个橘柚品种(阳光1 号和阳光2 号)及其亲本(爱媛28 号和春香)的全基因组InDel
Fig.1 Whole-genome InDel of the two tangelos(Yanggaung No.1 and Yangguang No.2)and their parents(Ehime Kashi No.28 and Haruka)
2.2 适用于琼脂糖凝胶电泳检测的InDel分子标记的开发
根据上述大于30 bp 或小于-30 bp 的InDel 位点的旁邻序列,笔者共设计60 对InDel 引物,分散在基因组的9 个片段框架(scaffold)中,其中有48对引物可对这10 个橘柚品种和爱媛28 号的基因组扩增出97 条清晰条带。其中多数标记只产生两条带,说明只有一种变异方式,其条带位置与通过重测序数据和引物位置计算的理论值一致;而I9_459在明尼奥拉和清峰[清见(Kiyomi)橘橙×明尼奥拉]中出现了第3 条带,说明很可能产生除理论值之外的第2 种InDel 变异方式。图2 展示了I3_744、I7_170、I8_516、I8_533、I9_669 和I9_459 的PCR 扩增电泳图谱,扩增条带清晰可见,多态性明显。图3概括了由所有48 个标记所产生的基因型,能有效地对这10 个橘柚品种以及爱媛28 号进行区分和鉴定。其中奥兰多和明尼奥拉最为接近,在12 个位点上有15 条带的差异;阳光1 号和阳光2 号差别最大,在42 个位点上有47 条带的差异。奥兰多和明尼奥拉皆为邓肯葡萄柚和丹西红橘的杂种姐妹系,遗传关系近,基因型较为接近。虽然阳光1号和阳光2号也是姐妹系,但是笔者对InDel位点的选择并非完全随机,偏向选择在阳光1 号、阳光2 号、爱媛28 号和春香相互之间具有差异的位点,因而差异较大的基因型并不能反映阳光1 号和阳光2 号之间真实的遗传距离。

图2 对爱媛28 号和10 个橘柚品种的PCR 扩增Fig.2 PCR amplification for 10 tangelo varieties and Ehime Kashi No.28
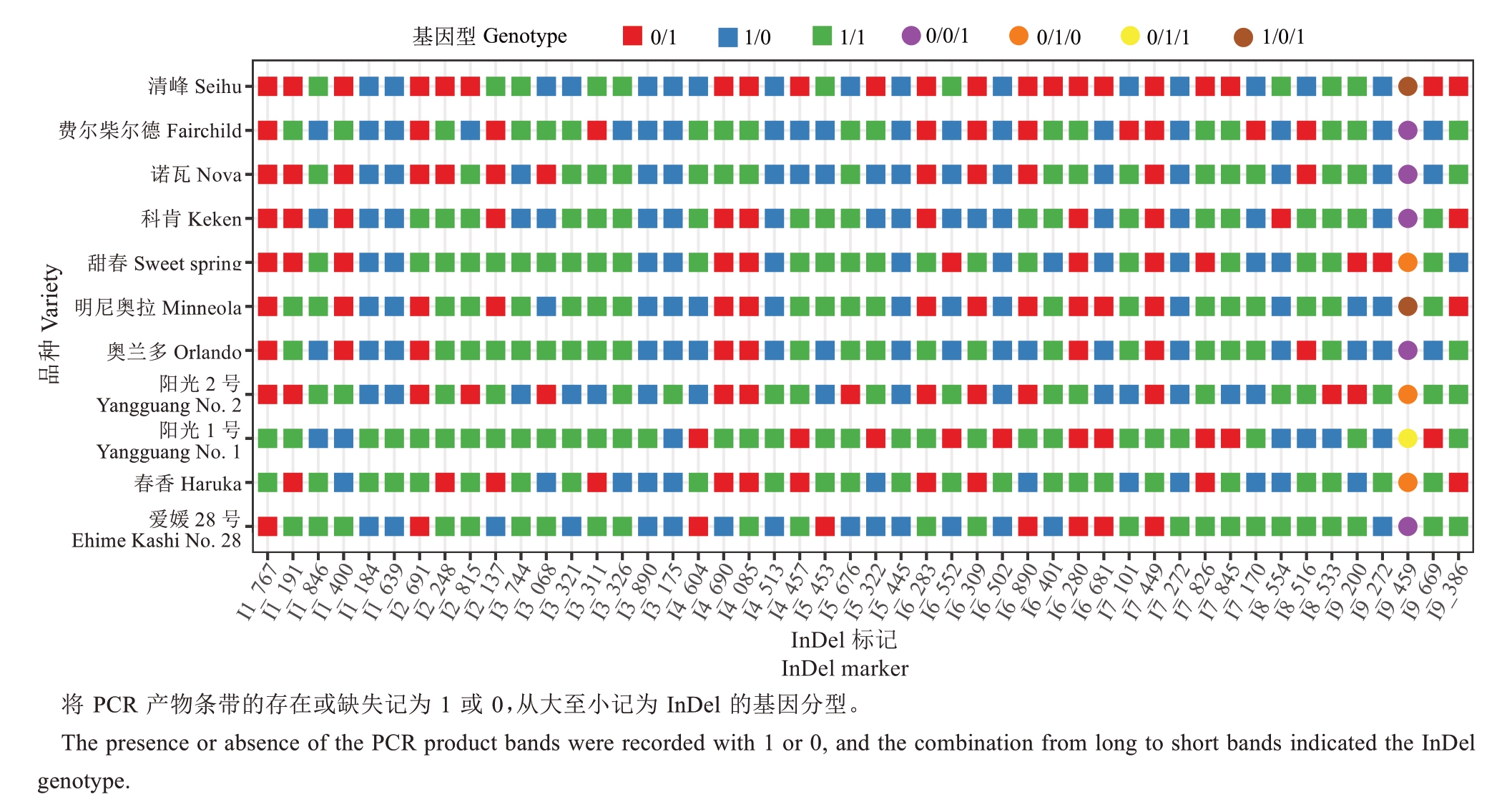
图3 用48 个InDel 分子标记对爱媛28 号和10 个橘柚品种的基因分型
Fig.3 Genotyping 10 tangelo varieties and Ehime Kashi No.28 based on the 48 InDel markers
2.3 对48 个InDel 标记在51 份种质资源中的遗传多样性分析
鉴于这些InDel标记可有效地对上述10 个橘柚品种和爱媛28号进行基因分型和区分鉴定的事实,笔者将研究范围扩大到51 份种质资源。InDel标记在这51 份种质资源中遗传多样性分析结果可参见表2。其PIC 介于0.055(I1_639、I1_767 和I7_449)和0.450(I8_516)之间,平均0.281;MAF 介于0.510(I6_401)和0.971(I1_639)之间,平均0.739;Ho 介于0.039(I1_184、I5_445、I6_502 和I6_681)和0.647(I1_846)之间,平均0.306;He 介于0.059(I1_767、I1_639 和I7_449)和0.546(I8_516)之间,平均0.350。一些标记(例如I3_744、I7_170、I8_516、I8_533 和I9_669)具有较高的He/PIC 和低MAF,表明他们在群体中有较高的遗传多样性,对柑橘作物的遗传多样性分析较为重要。
表2 48 个InDel 标记在51 份种质资源中的遗传多样性
Table 2 Genetic diversity of the 48 InDel markers in the 51 Citrus germplasm resources
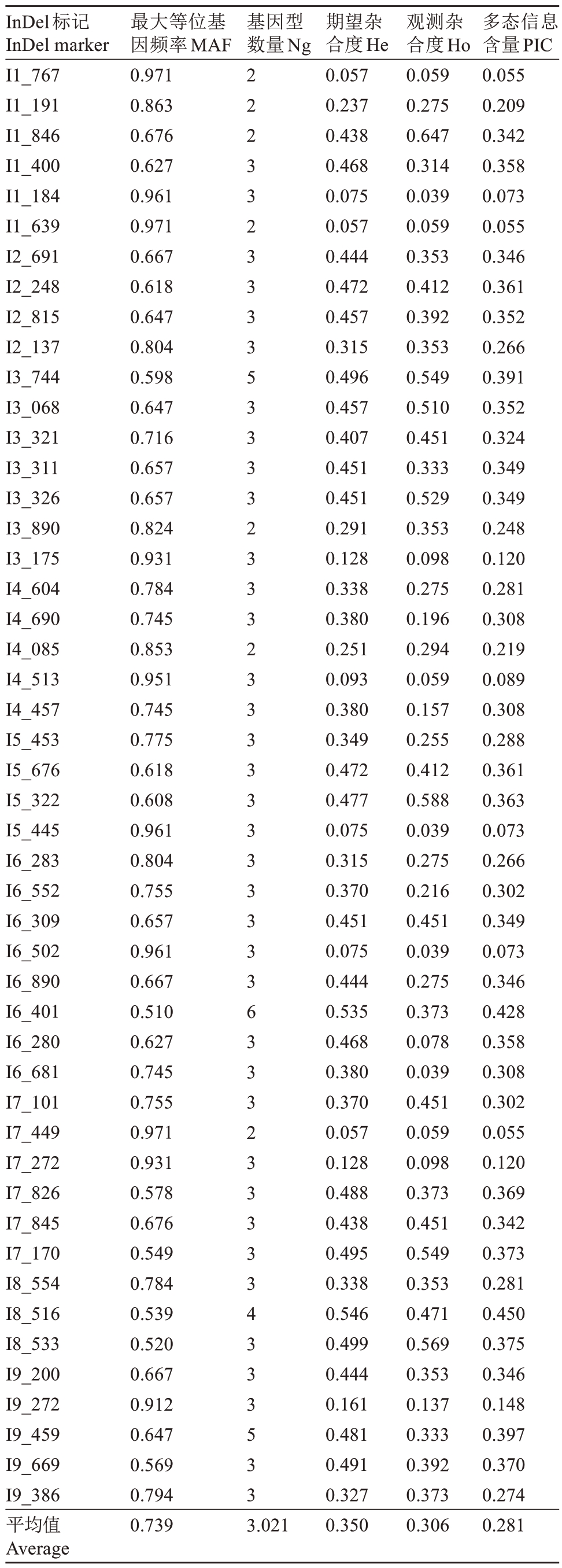
InDel标记InDel marker I1_767 I1_191 I1_846 I1_400 I1_184 I1_639 I2_691 I2_248 I2_815 I2_137 I3_744 I3_068 I3_321 I3_311 I3_326 I3_890 I3_175 I4_604 I4_690 I4_085 I4_513 I4_457 I5_453 I5_676 I5_322 I5_445 I6_283 I6_552 I6_309 I6_502 I6_890 I6_401 I6_280 I6_681 I7_101 I7_449 I7_272 I7_826 I7_845 I7_170 I8_554 I8_516 I8_533 I9_200 I9_272 I9_459 I9_669 I9_386平均值Average最大等位基因频率MAF 0.971 0.863 0.676 0.627 0.961 0.971 0.667 0.618 0.647 0.804 0.598 0.647 0.716 0.657 0.657 0.824 0.931 0.784 0.745 0.853 0.951 0.745 0.775 0.618 0.608 0.961 0.804 0.755 0.657 0.961 0.667 0.510 0.627 0.745 0.755 0.971 0.931 0.578 0.676 0.549 0.784 0.539 0.520 0.667 0.912 0.647 0.569 0.794 0.739基因型数量Ng 2 2 2 3 3 2 3 3 3 3 5 3 3 3 3 2 3 3 3 2 3 3 3 3 3 3 3 3 3 3 3 6 3 3 3 2 3 3 3 3 3 4 3 3 3 5 3 3 3.021期望杂合度He 0.057 0.237 0.438 0.468 0.075 0.057 0.444 0.472 0.457 0.315 0.496 0.457 0.407 0.451 0.451 0.291 0.128 0.338 0.380 0.251 0.093 0.380 0.349 0.472 0.477 0.075 0.315 0.370 0.451 0.075 0.444 0.535 0.468 0.380 0.370 0.057 0.128 0.488 0.438 0.495 0.338 0.546 0.499 0.444 0.161 0.481 0.491 0.327 0.350观测杂合度Ho 0.059 0.275 0.647 0.314 0.039 0.059 0.353 0.412 0.392 0.353 0.549 0.510 0.451 0.333 0.529 0.353 0.098 0.275 0.196 0.294 0.059 0.157 0.255 0.412 0.588 0.039 0.275 0.216 0.451 0.039 0.275 0.373 0.078 0.039 0.451 0.059 0.098 0.373 0.451 0.549 0.353 0.471 0.569 0.353 0.137 0.333 0.392 0.373 0.306多态信息含量PIC 0.055 0.209 0.342 0.358 0.073 0.055 0.346 0.361 0.352 0.266 0.391 0.352 0.324 0.349 0.349 0.248 0.120 0.281 0.308 0.219 0.089 0.308 0.288 0.361 0.363 0.073 0.266 0.302 0.349 0.073 0.346 0.428 0.358 0.308 0.302 0.055 0.120 0.369 0.342 0.373 0.281 0.450 0.375 0.346 0.148 0.397 0.370 0.274 0.281
此外,基于这些InDel 标记的基因分型,笔者进一步用PCoA 分析这51 份种质资源的遗传距离。PCoA 第一坐标和第二坐标可分别解释18.29%和15.72%的总变异。在PCoA二维散点图中(图4),相同类型的柑橘聚集在一起,其中5 个甜橙品种或3个枳品种只呈现出单点,说明这些甜橙或枳品种的基因型完全一致,不同类型的柑橘则距离较远。两种柚类(强得勒和琯溪蜜柚)第一坐标方向远离其他柑橘,说明他们有较少的来自其他柑橘类型的基因渗入。枸橼、枳、金柑、柠檬分布于散点图的偏右下侧。金柑(四季橘和宁波罗浮)相距较远,这是因为四季橘是有宽皮柑橘血统的金柑。相比于宁波罗浮,四季橘在散点图中更靠近橘类。橘类总体位于左下侧,但分布较为离散,这是由于一些橘类渗入其他类型柑橘的血统,例如伊予柑(酸橙×丹西红橘[4])、中柑所5 号(爱媛28 号×砂糖橘)、清见(温州蜜柑×甜橙[4]),导致在第二坐标方向上与橘柚和甜橙类靠近。橘柚主要分布于散点图的左侧中上部,在第一坐标方向上与橘类一致,第二坐标方向上则与柚和葡萄柚接近。总体上,基于这48 个InDel 标记的PCoA清楚地展示了这51份种质资源的遗传距离,他们在二维散点图的分布结果与已知的柑橘类型划分总体一致。
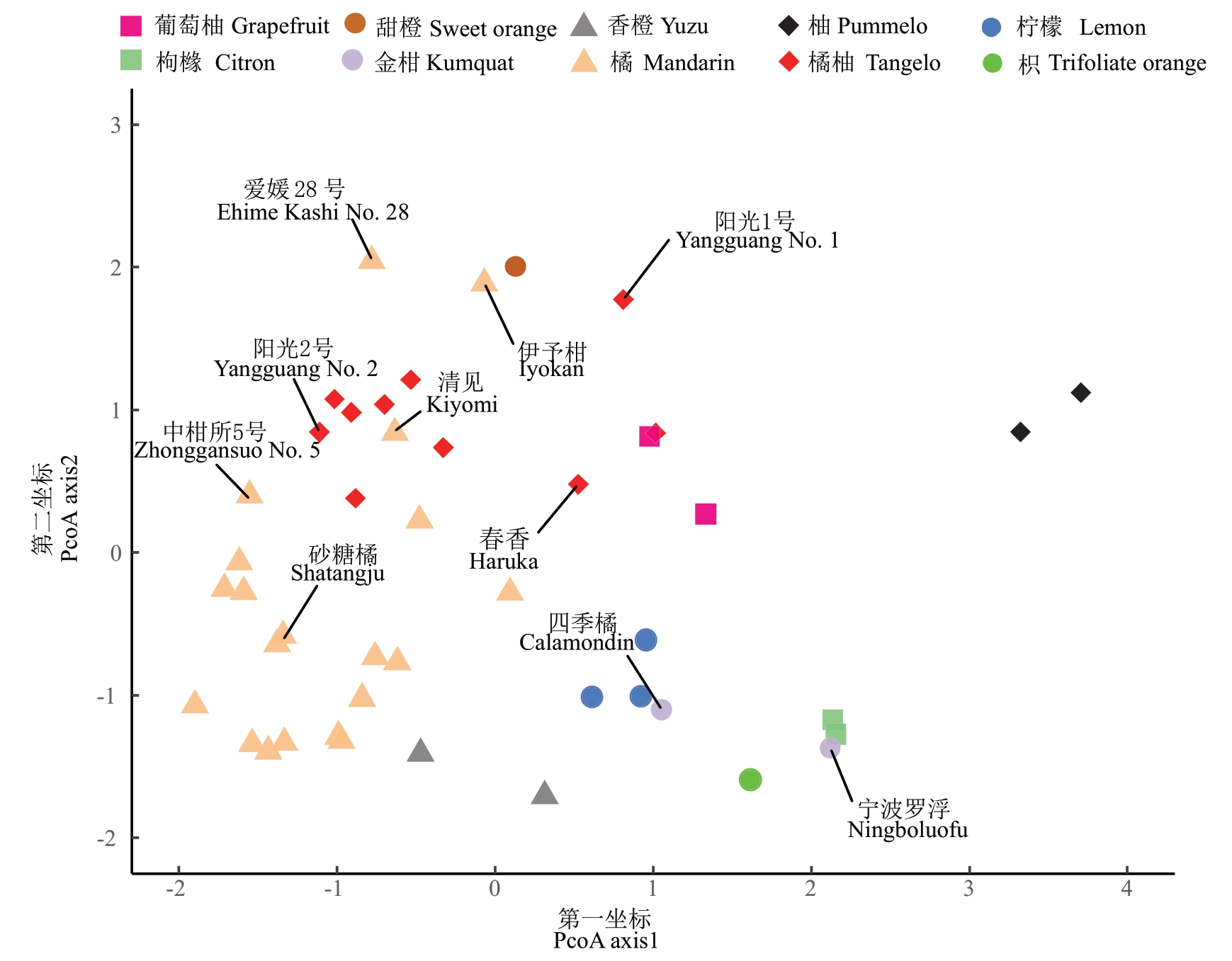
图4 基于48 个InDel 标记基因分型的51 份种质资源的PCoA
Fig.4 PCoA of 51 citrus germplasm resources based on the genotypes with the 48 InDel markers
3 讨 论
橘柚风味独特,日益受到消费者欢迎,已经得到柑橘育种工作者的重视。准确的品种鉴定不仅是新品种保护的前提,也有利于苗木真实性鉴别。基于形态特征的DUS[特异性(distinctness)、一致性(uni‐formity)、稳定性(stability)]测试仍然是新品种鉴定的公认方法[39],但在果树作物中存在周期长、近似品种难筛选的挑战。分子标记技术能准确确定品种的遗传特征,是品种鉴定的重要手段[5-8]。InDel标记有密度大、多态性丰富、准确性高、变异稳定性高和检测简便等优点,已经在柑橘中有所发掘。现有的柑橘InDel 标记主要应用于真正柑橘果树组植物的整体系统发育和遗传多样性分析[25,31],或仅针对柚类[34]、温州蜜柑[32]和某些特定杂柑[33]的品种鉴定,很难直接应用于对橘柚品种的区别和鉴定。笔者通过对两个新橘柚品种及其亲本的全基因组重测序,针对性地发掘和筛选适合琼脂糖电泳凝胶检测的大片段InDel 标记。基于这些InDel 标记的基因型不仅能有效区分和鉴定橘柚,而且操作简单,辨识度高,成本低。此外,如果将这些InDel标记联合形态特征鉴定,不仅能提高鉴定的精确度,也可适用于其他柑橘的鉴定。笔者利用其中33 个InDel 标记,结合部分DUS 测试,成功地把5 个未知品种鉴定为无核金诺[40]。
早期的InDel 挖掘来自于对部分基因的低通量测序,通过对45种真正柑橘组植物的27个基因的桑格测序,Garcia-Lor 等[24]获得了50 个InDel 标记。第二代测序技术的发展使得高密度、全基因组范围的InDel发掘成为可能。植物基因组中的InDel含量丰富,仅次于SNP。笔者在4 种柑橘基因组中共发掘到20 万~30 万的InDel 位点,其总量与金兰柚[33]、茶叶[41]和鹰嘴豆[42]相似,其密度为1.7~2.4 个/kb,远远高于水稻的15.8~21.2 个/Mb[43]和茶叶的84.5 个/Mb[41]。InDel 的产生频率、测试品种与参考基因组品种的亲缘关系、测序深度和数据过滤标准都直接影响所挖掘的InDel密度。本研究较大的InDel密度可能反映了这4 种柑橘与参考物种克林曼丁橘有相对较远的血缘关系。阳光1 号和阳光2 号的母本爱媛28 号(天草×清见)具有葡萄柚和甜橙的血缘,而其父本春香为日向夏和夏蜜柑的杂种后代[4],他们与克林曼丁橘都属不同物种。
在主坐标分析中,不同类型的柑橘总体分布于不同区间。然而,他们的遗传距离并不完全符合主流的柑橘分类观点[1],例如金柑和枳分别属于金柑属和枳属,在主坐标分析中,两者却与柑橘属的枸橼靠近,但又与柑橘属的柚类和葡萄柚相距较远,即并未呈现出与其他柑橘异属的特征。其主要原因可能有:(1)笔者选择的InDel 标记需要优先区别和鉴定阳光1号、阳光2号、春香和爱媛28号,具有偏向性;(2)考虑到真正柑橘果树组植物复杂的遗传背景(跨属、多物种、存在种间和属间杂种),48 个InDel标记在数量上不足以分析精确的遗传关系。在多种类型的柑橘基因组中随机挖掘数量充足的大片段InDel标记将有助于更加精确地分析柑橘的分类和遗传进化。
4 结 论
通过全基因组重测序数据,笔者发掘了48 个适合普通琼脂糖电泳检测的InDel 标记,能清晰地对10 个橘柚品种进行基因分型,可准确快速区分和鉴定不同橘柚品种,且操作简单,辨识度高,成本低。在51 份种质资源中,一些高PIC 和He、低MAF的InDel 标记(例如I3_744、I7_170、I8_516、I8_533、I9-669)具有较丰富的遗传多样性。基于这48 个InDel的基因分型,主坐标分析展示了不同柑橘品种的遗传距离,其结果总体符合已知的不同类型的柑橘分类,但不足以分析其精确的遗传关系。
致谢:感谢袁高鹏博士对本文的修改提出的宝贵意见。
[1] 周开隆,叶荫民.中国果树志:柑橘卷[M].北京:中国林业出版社,2010.ZHOU Kailong,YE Yinmin. Fruit trees in China:Volume cit‐rus[M]. Beijing:China Forestry Publishing House,2010.
[2] 周常勇.柑橘:中国果树科学与实践[M].西安:陕西新华出版传媒集团,2020.ZHOU Changyong.Citrus:Fruit science and practice in China[M].Xi’an:Shaanxi Xinhua Publishing and Media Group,2020.
[3] OUESLATI A,SALHI-HANNACHI A,LURO F,VIGNES H,MOURNET P,OLLITRAULT P. Genotyping by sequencing re‐veals the interspecific C. maxima/C. reticulata admixture along the genomes of modern citrus varieties of mandarins,tangors,tangelos,orangelos and grapefruits[J]. PLoS One,2017,12(10):e0185618.
[4] SHIMIZU T,KITAJIMA A,NONAKA K,YOSHIOKA T,OH‐TA S,GOTO S,TOYODA A,FUJIYAMA A,MOCHIZUKI T,NAGASAKI H,KAMINUMA E,NAKAMURA Y. Hybrid ori‐gins of citrus varieties inferred from DNA marker analysis of nu‐clear and organelle genomes[J]. PLoS One,2016,11(11):e0166969.
[5] SORIANO J M. Molecular marker technology for crop improve‐ment[J].Agronomy,2020,10(10):1462.
[6] AHMAD R,ANJUM M A,BALAL R M. From markers to ge‐nome based breeding in horticultural crops:An overview[J].Phyton,2020,89(2):183-204.
[7] 陈星,高子厚.DNA 分子标记技术的研究与应用[J].分子植物育种,2019,17(6):1970-1977.CHEN Xing,GAO Zihou. The study and application of DNA molecular marker technique[J].Molecular Plant Breeding,2019,17(6):1970-1977.
[8] AHMAD R,ANJUM M A,NAZ S,BALAL R M.Applications of molecular markers in fruit crops for breeding programs:A re‐view[J].Phyton,2021,90(1):17-34.
[9] 张连峰,何建,冯焱,刘利,郭启高,梁国鲁.金柑属及其近缘属植物亲缘关系的SSR 分析[J]. 果树学报,2006,23(3):335-338.ZHANG Lianfeng,HE Jian,FENG Yan,LIU Li,GUO Qigao,LIANG Guolu. Phylogenetic relationships among Fortunella and its relatives as revealed by SSR markers[J]. Journal of Fruit Science,2006,23(3):335-338.
[10] 杨海健,周心智,谭平,张云贵,弓亚林,杨世勇,钟昭林.利用SSR 和SRAP 标记分析香橙资源的遗传多样性[J].西南农业学报,2014,27(4):1678-1681.YANG Haijian,ZHOU Xinzhi,TAN Ping,ZHANG Yungui,GONG Yalin,YANG Shiyong,ZHONG Zhaolin. Genetic diver‐sity analysis of junos by SRAP and SSR markers[J]. Southwest China Journal of Agricultural Sciences,2014,27(4):1678-1681.
[11] 徐静,谭李梅,符红艳,朱志媚,龙栎冰,胡哲,马先锋,邓子牛.利用分子标记对14 份枸橼种质进行多样性分析[J].分子植物育种,2021,19(14):4726-4737.XU Jing,TAN Limei,FU Hongyan,ZHU Zhimei,LONG Libing,HU Zhe,MA Xianfeng,DENG Ziniu. Genetic diversity analysis of 14 citron genotypes based on molecular markers[J]. Molecu‐lar Plant Breeding,2021,19(14):4726-4737.
[12] 向素琼,何建,何波,汪卫星,李晓林,郭启高,何桥,梁国鲁.沙田柚多倍体遗传差异的SSR 分析[J].果树学报,2009,26(3):382-385.XIANG Suqiong,HE Jian,HE Bo,WANG Weixing,LI Xiaolin,GUO Qigao,HE Qiao,LIANG Guolu.Genetic diversity of poly‐ploids of Shatianyou pomelo by SSR markers[J]. Journal of Fruit Science,2009,26(3):382-385.
[13] 王旭,彭洁,朱延松,杨胜男,张晓楠,余洪,江东,梁大成.基于SSR 分子标记的68 份柚类种质资源亲缘关系分析[J].安徽农业科学,2021,49(4):100-103.WANG Xu,PENG Jie,ZHU Yansong,YANG Shengnan,ZHANG Xiaonan,YU Hong,JIANG Dong,LIANG Dacheng.Analysis of genetic relationship of 68 pummelo germplasm re‐sources based on SSR molecular marker[J]. Journal of Anhui Agricultural Sciences,2021,49(4):100-103.
[14] 庞晓明,胡春根,邓秀新.用SSR 标记研究柑橘属及其近缘属植物的亲缘关系[J].遗传学报,2003,30(1):81-87.PANG Xiaoming,HU Chungen,DENG Xiuxin.Phylogenetic re‐lationships among Citrus and its relatives as revealed by SSR markers[J]. Journal of Genetics and Genomics,2003,30(1):81-87.
[15] 刘勇,吴波,刘德春,孙中海.江西柑橘地方品种资源及野生近缘种SSR 分子标记[J]. 江西农业大学学报,2005,27(4):486-490.LIU Yong,WU Bo,LIU Dechun,SUN Zhonghai.On genetic di‐versity of Jiangxi native citrus and its wild varieties based on SSR markers[J]. Acta Agriculturae Universitatis Jiangxiensis,2005,27(4):486-490.
[16] 陈焱,余歆,曹立,雷天刚,周雪,张晓勇,彭良志,陆智明.杂柑新品种“中柑所5 号”的SSR 分子标记鉴定[J].中国南方果树,2018,47(3):1-4.CHEN Yan,YU Xin,CAO Li,LEI Tiangang,ZHOU Xue,ZHANG Xiaoyong,PENG Liangzhi,LU Zhiming. Identifica‐tion of CRIC 5 by using SSR molecular markers[J]. South Chi‐na Fruits,2018,47(3):1-4.
[17] BISWAS M K,CHAI L J,AMAR M H,ZHANG X L,DENG X X. Comparative analysis of genetic diversity in Citrus germplasm collection using AFLP,SSAP,SAMPL and SSR markers[J].Scientia Horticulturae,2011,129(4):798-803.
[18] AMAR M H,BISWAS M K,ZHANG Z W,GUO W W.Exploi‐tation of SSR,SRAP and CAPS-SNP markers for genetic diver‐sity of Citrus germplasm collection[J]. Scientia Horticulturae,2011,128(3):220-227.
[19] BISWAS M K,CHAI L J,MAYER C,XU Q,GUO W W,DENG X X. Exploiting BAC-end sequences for the mining,characterization and utility of new short sequences repeat (SSR)markers in citrus[J]. Molecular Biology Reports,2012,39(5):5373-5386.
[20] NALIATH R,KUMAR K,ARORA P K,KAUR S,KAUR D,SINGH K. Genetic identification and inference on genetic rela‐tionships of important Citrus rootstocks with microsatellite markers[J].Fruits,2017,72(6):350-362.
[21] SÜLÜ G,AKA KAÇAR Y,POLAT İ,KİTAPCI A,TUR‐GUTOĞLU E,ŞİMŞEK Ö,SATAR G. Identification of genetic diversity among mutant lemon and mandarin varieties using dif‐ferent molecular markers[J]. Turkish Journal of Agriculture and Forestry,2020,44(5):465-478.
[22] LURO F L,COSTANTINO G,TEROL J,ARGOUT X,AL‐LARIO T,WINCKER P,TALON M,OLLITRAULT P,MO‐RILLON R. Transferability of the EST- SSRs developed on Nules clementine (Citrus clementina Hort ex Tan) to other Cit‐rus species and their effectiveness for genetic mapping[J]. BMC Genomics,2008,9:287.
[23] GARCÍA-LOR A,LURO F,NAVARRO L,OLLITRAULT P.Comparative use of InDel and SSR markers in deciphering the interspecific structure of cultivated citrus genetic diversity:A perspective for genetic association studies[J]. Molecular Genet‐ics and Genomics,2012,287(1):77-94.
[24] GARCIA-LOR A,CURK F,SNOUSSI-TRIFA H,MORILLON R,ANCILLO G,LURO F,NAVARRO L,OLLITRAULT P. A nuclear phylogenetic analysis:SNPs,indels and SSRs deliver new insights into the relationships in the‘true citrus fruit trees’group(Citrinae,Rutaceae)and the origin of cultivated species[J].Annals of Botany,2013,111(1):1-19.
[25] GARCIA-LOR A,ANCILLO G,NAVARRO L,OLLITRAULT P.Citrus (Rutaceae) SNP markers based on competitive allele-spe‐cific PCR;transferability across the Aurantioideae subfamily[J].Applications in Plant Sciences,2013,1(4):apps.1200406.
[26] YU Y,CHEN C X,HUANG M,YU Q B,DU D L,MATTIA M R,JR GMITTER F G. Genetic diversity and population struc‐ture analysis of citrus germplasm with single nucleotide poly‐morphism markers[J].Journal of the American Society for Horti‐cultural Science,2018,143(6):399-408.
[27] JIANG D,YE Q L,WANG F S,CAO L. The mining of citrus EST-SNP and its application in cultivar discrimination[J]. Agri‐cultural Sciences in China,2010,9(2):179-190.
[28] OLLITRAULT P,TEROL J,GARCIA-LOR A,BÉRARD A,CHAUVEAU A,FROELICHER Y,BELZILE C,MORILLON R,NAVARRO L,BRUNEL D,TALON M. SNP mining in C.clementina BAC end sequences;transferability in the Citrus ge‐nus (Rutaceae),phylogenetic inferences and perspectives for ge‐netic mapping[J].BMC Genomics,2012,13:13.
[29] 雷天刚,何永睿,彭爱红,许兰珍,刘小丰,姚利晓,邹修平,江东,陈善春.柑橘CAPS 标记和AS-PCR 引物的开发[J].园艺学报,2012,39(6):1027-1034.LEI Tiangang,HE Yongrui,PENG Aihong,XU Lanzhen,LIU Xiaofeng,YAO Lixiao,ZOU Xiuping,JIANG Dong,CHEN Shanchun. Development of CAPS markers and allele-specific PCR primers in Citrus[J]. Acta Horticulturae Sinica,2012,39(6):1027-1034.
[30] 吴波,杨润婷,朱世平,钟云,姜波,曾继吾,钟广炎.宽皮柑橘单核苷酸多态性的高分辨率熔解曲线分型[J]. 园艺学报,2012,39(4):777-782.WU Bo,YANG Runting,ZHU Shiping,ZHONG Yun,JIANG Bo,ZENG Jiwu,ZHONG Guangyan.Genotyping single nucleo‐tide polymorphisms in mandarin cultivars using high resolution melting analysis[J].Acta Horticulturae Sinica,2012,39(4):777-782.
[31] FANG Q Y,WANG L,YU H W,HUANG Y,JIANG X L,DENG X X,XU Q. Development of species-specific InDel markers in Citrus[J]. Plant Molecular Biology Reporter,2018,36(4):653-662.
[32] NODA T,DAIOU K,MIHARA T,NAGANO Y. Development of Indel markers for the selection of Satsuma mandarin (Citrus unshiu Marc.) hybrids that can be used for low-cost genotyping with agarose gels[J].Euphytica,2020,216(7):115.
[33] NODA T,DAIOU K R,MIHARA T,NAGANO Y.Potential ap‐plication of simple easy-to-use insertion-deletion (InDel) mark‐ers in citrus cultivar identification[J]. Breeding Science,2021,71(5):601-608.
[34] 汤雨晴,杨惠栋,闫承璞,王斯妤,王雨亭,胡钟东,朱方红.基于重测序的‘金兰柚’基因组InDel 标记的开发及应用[J].园艺学报,2023,50(1):15-26.TANG Yuqing,YANG Huidong,YAN Chengpu,WANG Siyu,WANG Yuting,HU Zhongdong,ZHU Fanghong. Development and application of Jinlan Pummelo(Citrus maxima)InDel mark‐ers based on genome re-sequencing[J].Acta Horticulturae Sini‐ca,2023,50(1):15-26.
[35] LI H,DURBIN R. Fast and accurate short read alignment with Burrows-Wheeler transform[J]. Bioinformatics,2009,25(14):1754-1760.
[36] MCKENNA A,HANNA M,BANKS E,SIVACHENKO A,CIBULSKIS K,KERNYTSKY A,GARIMELLA K,ALT‐SHULER D,GABRIEL S,DALY M,DEPRISTO M A.The ge‐nome analysis Toolkit:A MapReduce framework for analyzing next-generation DNA sequencing data[J]. Genome Research,2010,20(9):1297-1303.
[37] CINGOLANI P,PLATTS A,WANG L L,COON M,NGUYEN T,WANG L,LAND S J,LU X Y,RUDEN D M.A program for annotating and predicting the effects of single nucleotide poly‐morphisms,SnpEff:SNPs in the genome of Drosophila melano‐gaster strain w1118;iso-2;iso-3[J].Fly,2012,6(2):80-92.
[38] YOU F M,HUO N X,GU Y Q,LUO M C,MA Y Q,HANE D,LAZO G R,DVORAK J,ANDERSON O D. BatchPrimer3:A high throughput web application for PCR and sequencing primer design[J].BMC Bioinformatics,2008,9:253.
[39] 王燕,田泰,马艳,杨佳铭,陈隆隆,廖翊雯,罗雪文,周小杰,王小蓉.中国果树新品种保护与DUS 测试研究进展[J].江苏农业学报,2022,38(3):849-864.WANG Yan,TIAN Tai,MA Yan,YANG Jiaming,CHEN Longlong,LIAO Yiwen,LUO Xuewen,ZHOU Xiaojie,WANG Xiaorong. Research progress of plant variety protection and test for distinctness,uniformity and stability of fruit trees in China[J].Jiangsu Journal of Agricultural Sciences,2022,38(3):849-864.
[40] 刘小丰,王洪,刘梦雨,朱世平,江东,曹立,余歆.性状鉴别联合InDel 标记检测快速鉴定柑桔品种[J]. 中国南方果树,2024,53(1):1-8.LIU Xiaofeng,WANG Hong,LIU Mengyu,ZHU Shiping,JIANG Dong,CAO Li,YU Xin. Rapid identification of Citrus varieties using DUS testing combined with InDel marker[J].South China Fruits,2024,53(1):1-8.
[41] LIU S R,AN Y L,TONG W,QIN X J,SAMARINA L,GUO R,XIA X B,WEI C L. Characterization of genome-wide genetic variations between two varieties of tea plant (Camellia sinensis)and development of InDel markers for genetic research[J]. BMC Genomics,2019,20(1):935.
[42] JAIN A,ROORKIWAL M,KALE S,GARG V,YADALA R,VARSHNEY R K. InDel markers:An extended marker resource for molecular breeding in chickpea[J]. PLoS One,2019,14(3):e0213999.
[43] WU D H,WU H P,WANG C S,TSENG H Y,HWU K K. Ge‐nome-wide InDel marker system for application in rice breeding and mapping studies[J].Euphytica,2013,192(1):131-143.