中华猕猴桃(Actinidia chinensis)原产于中国,庭院栽培历史距今至少有1200 a(年)[1]。猕猴桃果实在发育过程中,内果皮呈现出不同的颜色,比如浅绿色、深绿、黄色、橙色、红色、紫色或黄绿色[2],内果皮红颜色差异的主要原因是花青苷(anthocyanin,AC)和原花青素(proanthocyanidin,PA)种类和含量的不同。目前已经鉴定出630 多种花青苷和680 多种原花青素[3]。花青苷以3,5,7-三羟基-2-苯基苯并吡喃为基本结构基元,由于R1、R2 取代基的不同,衍生出多种花青苷,常见的花青苷有6 大类:矢车菊素类(cyanidin)、天竺葵素类(pelargonidin)、飞燕草素类(delphindin)、芍药素类(paeoniflorin)、矮牵牛色素类(petunidin)和锦葵色素类(malvidin)[4-5]。花青苷是一种水溶性色素,在植物细胞质中产生并存储在液泡中,故在植物有色部位中含量最为丰富,是许多水果、蔬菜以及花卉色泽的主要组成成分之一[6-7]。
红心猕猴桃品种红阳是从中华猕猴桃中人工实生选育出来的优良品种[8],果心红色中花青苷种类主要是矢车菊素半乳糖苷[9],红心猕猴桃色泽艳丽、营养价值高,备受消费者青睐,市场需求不断加大。但是,红阳猕猴桃引入中国长江中下游平原地区栽植时,高温环境会使果心出现褪色或着色较浅的现象。近年来,安徽农业大学从红阳实生后代培育出来的猕猴桃新品种平原红,在相同的条件下,果心着色比红阳更好,并且不易受高温环境的影响[10]。笔者在本试验中以红阳猕猴桃为对照,对平原红猕猴桃不同发育阶段果实(5 个时期)花青苷含量开展测定,同时将相应时期样品开展转录组测序分析,以期筛选出与花青苷代谢相关的关键基因、解析猕猴桃新品种平原红在高温下着色更好的分子机制。
1 材料和方法
1.1 材料及采集方法
试验材料选用红阳和平原红猕猴桃果实,两个品种栽培条件相同、树龄均为5 a,砧木为美味猕猴桃实生砧木,株行距为1.5 m×3.0 m。均取自安徽省霍邱县皖西猕猴桃研究所。2020 年5—9 月(花后30~150 d),选择生长状况良好的果实(图1),每30 d取样1 次,遵循随机取样原则,每时期、每个样品取10 个果实,3 次重复(A1、A2、A3)。采样完成后,将果实用湿纱布包裹置于冰盒中运回安徽农业大学园艺作物品质生物学安徽省重点实验室。果实去皮后打浆混匀,用液氮速冻处理并保存在-80 ℃超低温冰箱中备用。

图1 平原红和红阳猕猴桃花后不同天数果实横切图
Fig.1 Cross-sectional view of the fruits of Pingyuanhong and Hongyang kiwifruit on different days after full bloom
1.2 花青苷含量的测定
样品处理,进样条件和标准液配制参照前人研究方法略有改进[8]。将矢车菊-3-O-半乳糖苷和矢车菊-3-O-葡萄糖苷标准品,分别配制成50 mg·mL-1的溶液,逐步稀释为25 μg·mL-1、12.5 μg·mL-1、6.25 μg·mL-1、3.13 μg·mL-1、1.56 μg·mL-1的标准混合液。
将矢车菊-3-O-半乳糖苷和矢车菊-3-O-葡萄糖苷标准液用0.22 μm 滤膜过滤到进样瓶中,按照设定程序上机进样,结果显示矢车菊-3-O-半乳糖苷的出峰时间在6.7 min,矢车菊-3-O-葡萄糖苷的出峰时间在8.0 min,不同质量浓度对应的峰面积如表1 所示。以标准混合液浓度(x)对峰面积(y)求线性回归方程,矢车菊-3-O-半乳糖苷对应的标准曲线是y=9.062 8x-4.290 1,相关系数R2=0.998 7;矢车菊-3-O-葡萄糖苷对应的标准曲线是y=12.296x-6.09,相关系数R2=0.999 5。
表1 不同质量浓度花青苷标准样品对应的峰面积
Table 1 Peak area corresponding to different concentrations of anthocyanin standards
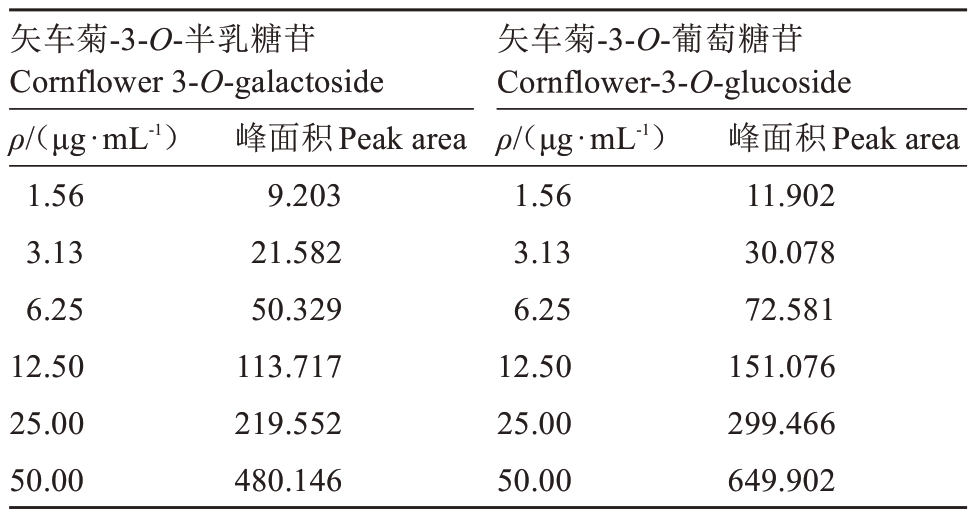
矢车菊-3-O-半乳糖苷Cornflower 3-O-galactoside ρ/(μg·mL-1)1.56 3.13 6.25 12.50 25.00 50.00峰面积Peak area 9.203 21.582 50.329 113.717 219.552 480.146矢车菊-3-O-葡萄糖苷Cornflower-3-O-glucoside ρ/(μg·mL-1)1.56 3.13 6.25 12.50 25.00 50.00峰面积Peak area 11.902 30.078 72.581 151.076 299.466 649.902
1.3 转录组测序与分析
本试验的转录组测序部分委托深圳华大基因股份有限公司完成。以红阳猕猴桃果实作为对照组,平原红果实作为处理组,样品分组如表2所示。
表2 样品分组设置
Table 2 Sample grouping settings
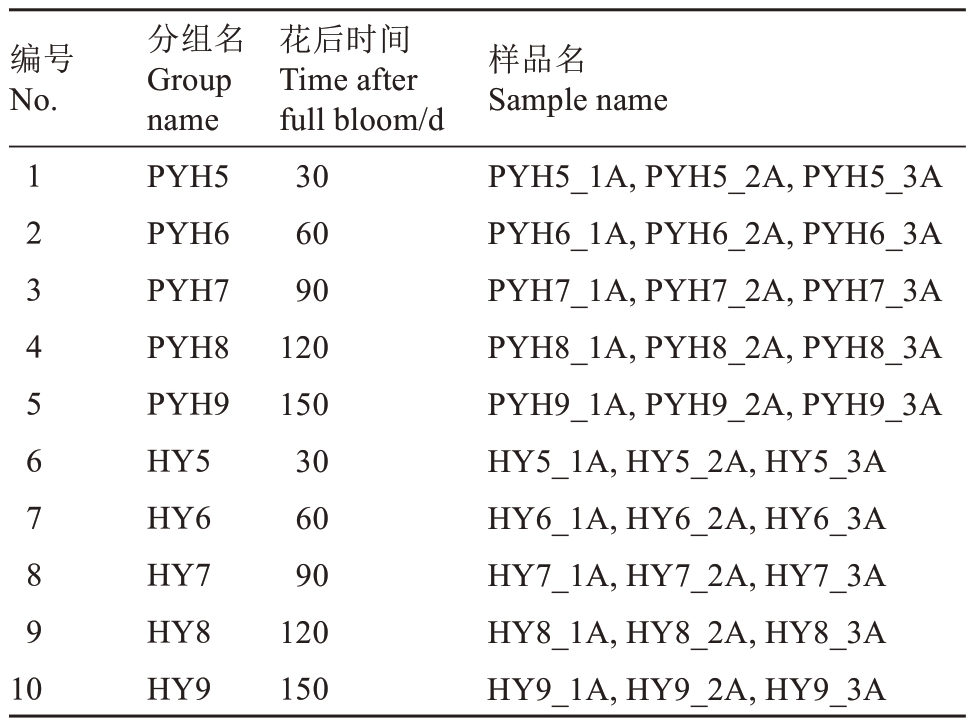
注:HY.红阳;PYH.平原红。
Note:HY.Hongyang;PYH.Pingyuanhong.
编号No.1 2 3 4 5 6 7 8 9 10分组名Group name PYH5 PYH6 PYH7 PYH8 PYH9 HY5 HY6 HY7 HY8 HY9花后时间Time after full bloom/d 30 60 90 120 150 30 60 90 120 150样品名Sample name PYH5_1A,PYH5_2A,PYH5_3A PYH6_1A,PYH6_2A,PYH6_3A PYH7_1A,PYH7_2A,PYH7_3A PYH8_1A,PYH8_2A,PYH8_3A PYH9_1A,PYH9_2A,PYH9_3A HY5_1A,HY5_2A,HY5_3A HY6_1A,HY6_2A,HY6_3A HY7_1A,HY7_2A,HY7_3A HY8_1A,HY8_2A,HY8_3A HY9_1A,HY9_2A,HY9_3A
提取果肉RNA 构建cDNA 文库,使用DNBSEQ平台测序,以中华猕猴桃Red5为参考基因组(https://kiwifruitgenome.org/)。使用BLAST 程序将DEGs与非冗余蛋白数据(non-redundant protein sequence,Nr)、基因本体(gene ontology,GO)、京都基因与基因组百科全书(kyoto encyclopedia of genes and ge‐nomes,KEGG)数据库中的已知序列进行比对,进而获得相关的注释信息。使用PlantTFDB 数据库进行转录因子注释。根据GO 和KEGG 注释结果,对差异基因进行功能分类与富集分析。
1.4 统计分析
使用IBM SPSS Statistics 25和GraphPad Prism 9软件分别对数据进行统计分析及作图,并进行差异性分析和相关性分析。结果用平均值±标准差表示。使用Tukey 的多重比较检验评估平均值之间的差异,统计学显著性设定为p<0.05。
2 结果与分析
2.1 猕猴桃果实发育过程中花青苷种类及含量变化
2.1.1 猕猴桃果实发育过程中花青苷的种类 花后30 d 和60 d,在两个品种果实中未检测出花青苷。花后90 d,红阳猕猴桃(HY7)中只检测到矢车菊-3-O-半乳糖苷,而平原红(PYH7)中同时检测到矢车菊-3-O-半乳糖苷和矢车菊-3-O-葡萄糖苷,但后者检出量甚微(图2)。花后120 d和150 d,两个品种中都只检测到矢车菊-3-O-半乳糖苷这一种花青苷。表明红阳和平原红猕猴桃果实中的花青苷以矢车菊-3-O-半乳糖苷为主,平原红果实发育过程中有少量矢车菊-3-O-葡萄糖苷的产生。
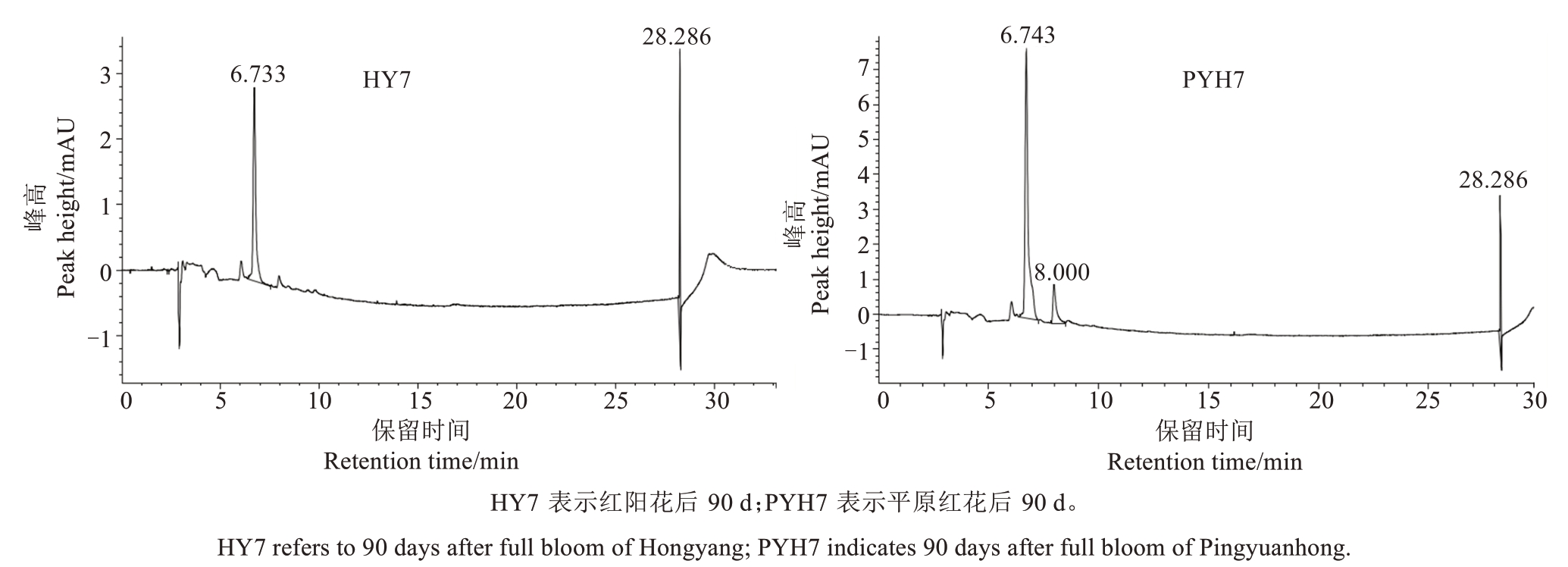
图2 猕猴桃果实花后不同时间花青苷含量的高效液相色谱分析
Fig.2 Anthocyanin content in kiwifruit fruit at different days after full bloom determined by the high performance liquid chromatography
2.1.2 猕猴桃果实发育过程中花青苷含量的变化在果实发育的不同时期,红阳中的花青苷含量均显著低于平原红。花后90 d,平原红中花青苷含量是红阳的3.34倍。花后120 d,平原红猕猴桃果肉的花青苷含量是红阳猕猴桃的2.15倍。至花后150 d(果实采收时),两个品种猕猴桃果肉中的花青苷含量都有所下降,平原红猕猴桃果肉的花青苷含量达到红阳猕猴桃的3.97倍。果实不同发育阶段,平原红和红阳猕猴桃花青苷含量均呈现“先升后降”的变化趋势(图3-A)。
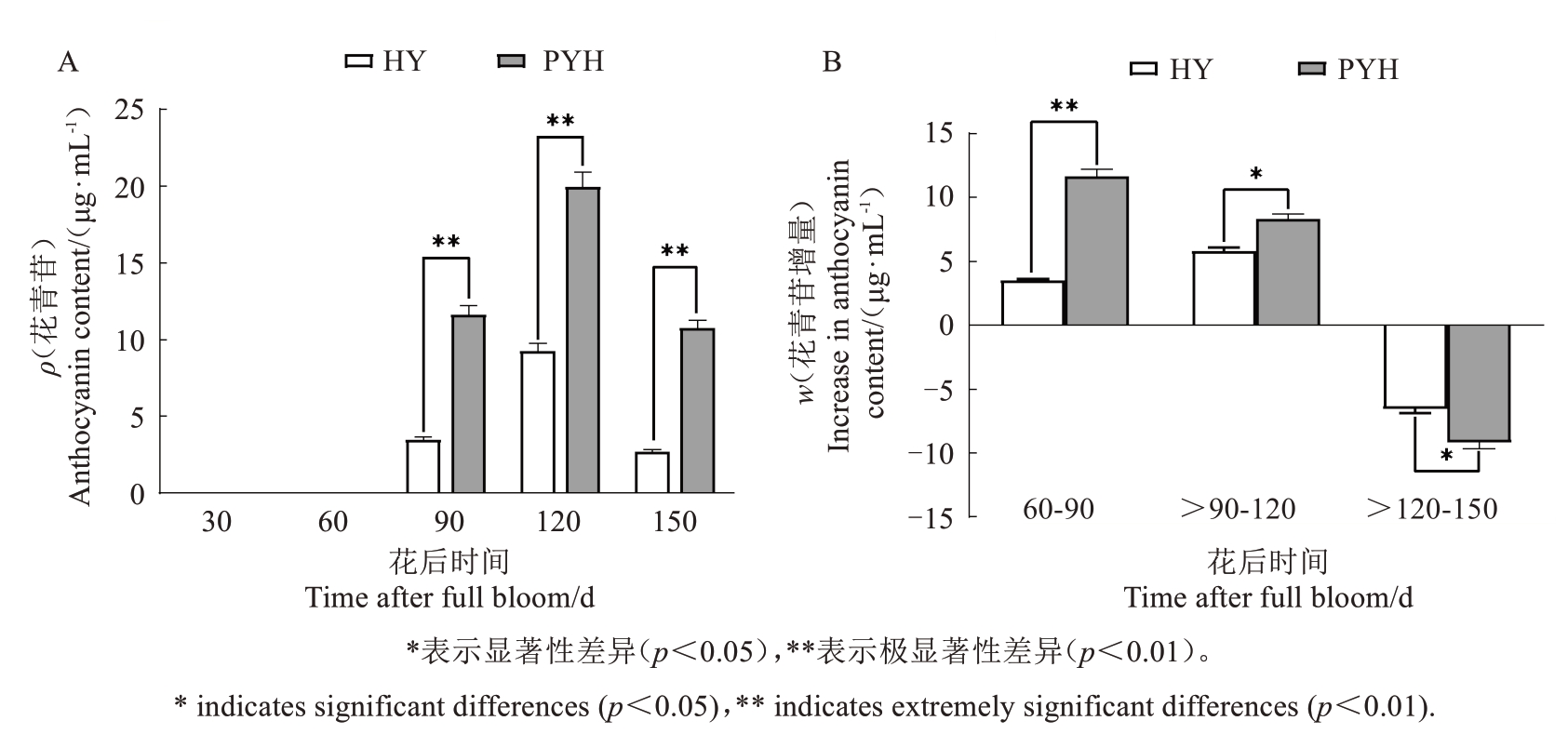
图3 猕猴桃果实花后不同时间的花青苷含量(A)和增量(B)
Fig.3 Anthocyanin content(A)and increment(B)of kiwifruit fruit at different days after full bloom
花后60~90 d,两个猕猴桃品种果肉中花青苷增量差距最大,达到极显著水平;花后90~120 d,平原红果肉中花青苷增量比红阳高43.3%,达到显著水平;花后120~150 d,两个猕猴桃品种果肉中花青苷均有部分降解,平原红花青苷降解速率为46.1%,显著低于红阳70.7%的降解速率(图3-B)。
2.2 猕猴桃果实中基因表达转录组测序结果
2.2.1 不同样本的差异表达基因 花后30 d,两个品种果肉中差异基因数量最少;花后60、90和120 d,DEGs 的数目差别不大,但下调差异基因多于上调差异基因;花后150 d,上调、下调及总DEGs 数是5个时期中最多的(表3)。
表3 猕猴桃果肉样本DEGs 数量统计
Table 3 Statistics of the number of differentially expressed genes in kiwifruit fruit samples
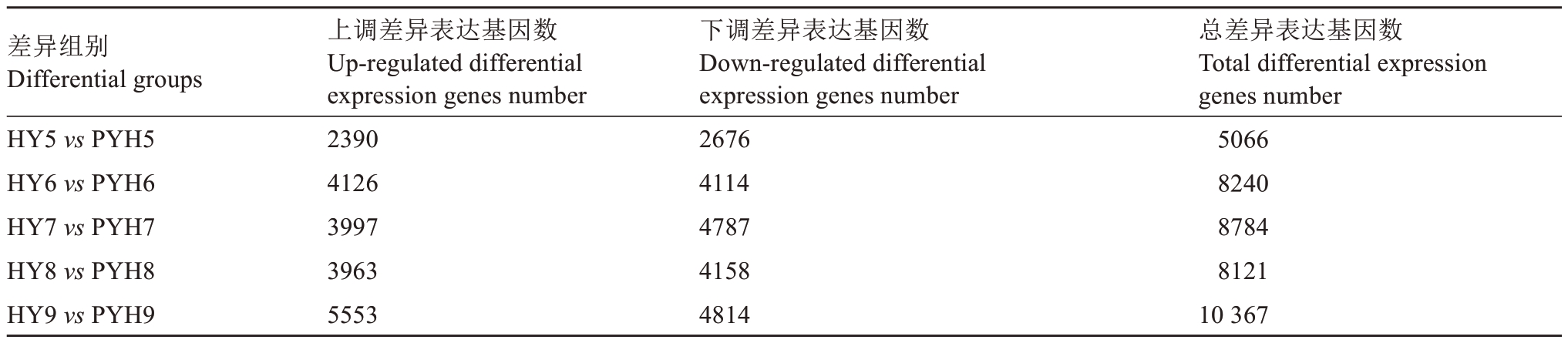
注:“HY5、6、7、8、9”表示红阳猕猴桃花后30、60、90、120、150 d;“PYH5、6、7、8、9”表示平原红猕猴桃花后30、60、90、120、150 d。
Note:“HY5,6,7,8,9”means 30,60,90,120,150 days after full bloom of Hongyang;“PYH5,6,7,8,9”indicates 30,60,90,120,150 days after full bloom of Pingyuanhong.
总差异表达基因数Total differential expression genes number 5066 8240 8784 8121 10 367差异组别Differential groups HY5 vs PYH5 HY6 vs PYH6 HY7 vs PYH7 HY8 vs PYH8 HY9 vs PYH9上调差异表达基因数Up-regulated differential expression genes number 2390 4126 3997 3963 5553下调差异表达基因数Down-regulated differential expression genes number 2676 4114 4787 4158 4814
2.2.2 红阳和平原红猕猴桃果实中DEGs 的GO 与KEGG 分析 花后60 d,两个品种DEGs主要参与了细胞质、核糖体等细胞成分,具备与RNA 结合、结构分子活性等分子功能,参与细胞酰胺代谢、细胞内转运等生物工程;花后90 d DEGs主要参与了细胞质膜、液泡等细胞成分,具备磷酸转移酶活性、跨膜转运蛋白活性、水解酶活性、蛋白激酶活性等分子功能,参与小分子代谢、有机酸代谢、羧酸代谢等生物过程;花后120 d DEGs主要参与了细胞质、质体等细胞成分,具备小分子结合、辅酶结合、磷酸酯水解酶活性、氧化还原酶活性等分子功能,参与有机酸代谢、羧酸代谢、对温度刺激的响应等生物过程;花后150 d DEGs主要参与了细胞质、质体、叶绿体等细胞成分,具备磷酸酯水解酶活性、核苷结合等分子功能,参与了小分子代谢、有机酸代谢等生物过程(图4)。
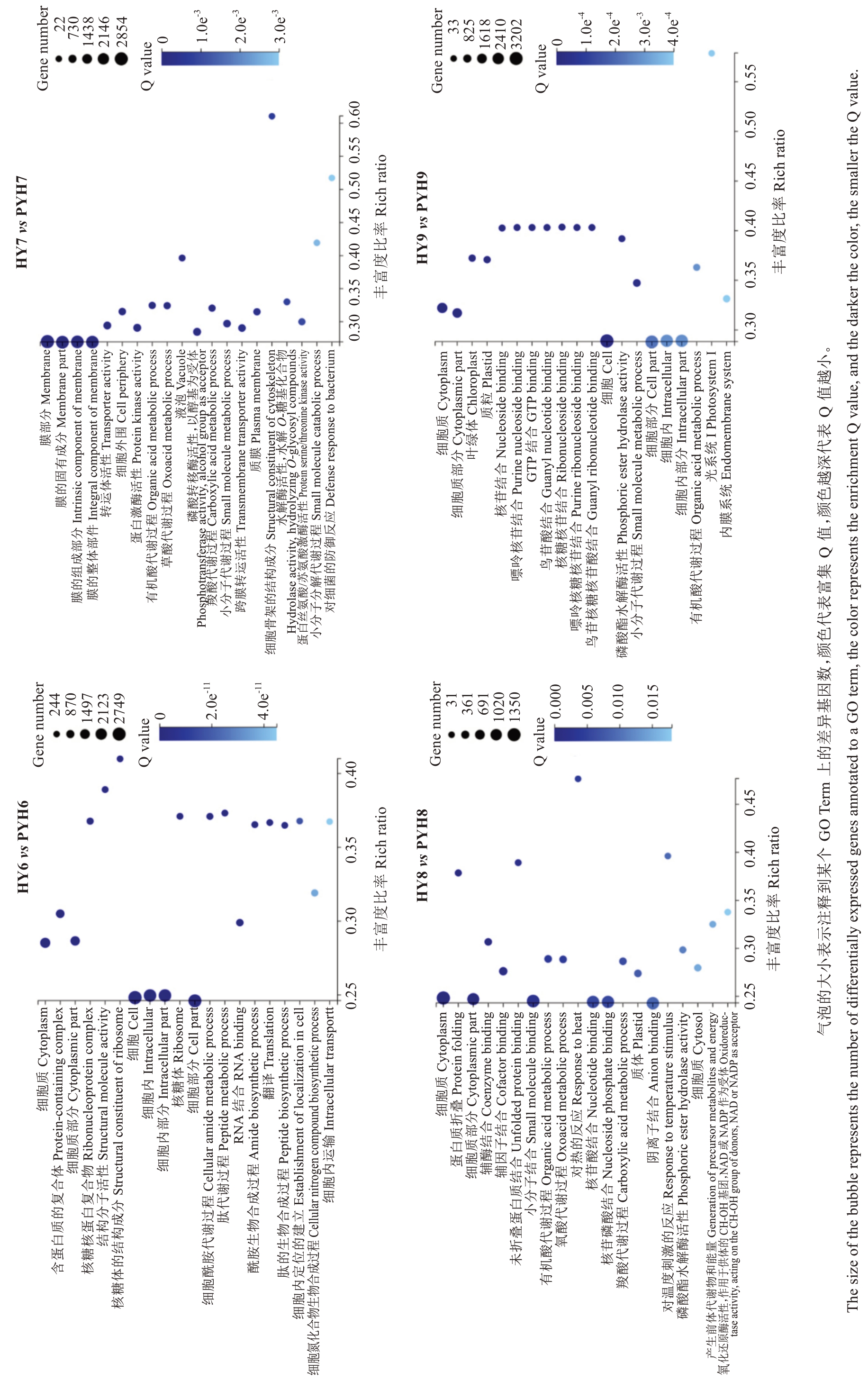
图4 红阳和平原红猕猴桃果实发育不同时期 DEGs 的 GO 功能富集
Fig. 4 GO functional enrichment of differentially expressed genes in different periods of fruit development of Hongyang and Pingyuanhong kiwifruits
DEGs的KEGG富集分析表明,花后60 d,DEGs主要参与了核糖体代谢、半乳糖代谢、谷胱甘肽代谢、苯丙氨酸和色氨酸的生物合成、苯丙氨酸代谢等代谢途径;花后90 d,DEGs 主要参与了氨基酸的生物合成、半乳糖代谢、丙酮酸代谢、类固醇生物合成、异黄酮类生物合成等代谢途径;花后120 d,DEGs主要参与了碳代谢、氨基糖和核苷酸糖代谢、半乳糖代谢、卟啉和叶绿素代谢、酮体的合成与降解等代谢途径;花后150 d,DEGs主要参与了内质网中的蛋白质加工、氨基酸的生物合成、内吞作用、半胱氨酸和蛋氨酸代谢、丙氨酸和谷氨酸代谢等代谢途径(图5)。

图5 红阳和平原红猕猴桃果实发育不同时期 DEGs 的 KEGG 富集
Fig. 5 Enrichment of KEGG metabolic pathway of differentially expressed genes in different periods of fruit development of Hongyang and Pingyuanhong kiwifruit
此外,有6 个MYB 家族转录因子(Acc01631、Acc0876、Acc25854、Acc27724、Acc30277、Acc32342)被富集到与花青苷合成相关的苯丙烷类生物合成途径(ko00940)。
2.2.3 花青苷合成通路中的关键DEGs 根据花青苷合成途径中基因差异表达量,筛选出平原红中表达量显著高于红阳的8 个基因,作为差异表达候选关键基因。其中,1 个香豆酸辅酶A 连接酶基因(Acc23647),1个查尔酮合酶基因(Acc02540.1),1个查尔酮异构酶基因(Acc03638),4 个黄烷酮3-羟化酶基因(Acc01302、Acc02579、Acc10395、Acc20991),1个二氢黄酮醇-4-还原酶基因(Acc23632)。
花后30 d,红阳和平原红猕猴桃果肉中都没有Acc23647(Acc4CL)表达,花后60~150 d,红阳果肉中Acc4CL 表达量显著低于平原红猕猴桃。Acc10395(AccF3H)在花后60 d 的红阳猕猴桃果肉中有少量表达,其他时间段在红阳猕猴桃中均没有表达。Acc02540.1(AccCHS)、Acc03638(AccCHI)在两个猕猴桃品种的果肉中表达模式相同,花后30 d表达量最高。
然而,Acc23647(Acc4CL)、Acc23632(AccDFR)主要在花后60~150 d 猕猴桃果肉中表达,花后30 d的果肉中表达量甚微,这两个基因表达模式与果肉中花青苷含量变化趋势相吻合,可作为关键DEGs(图6)。
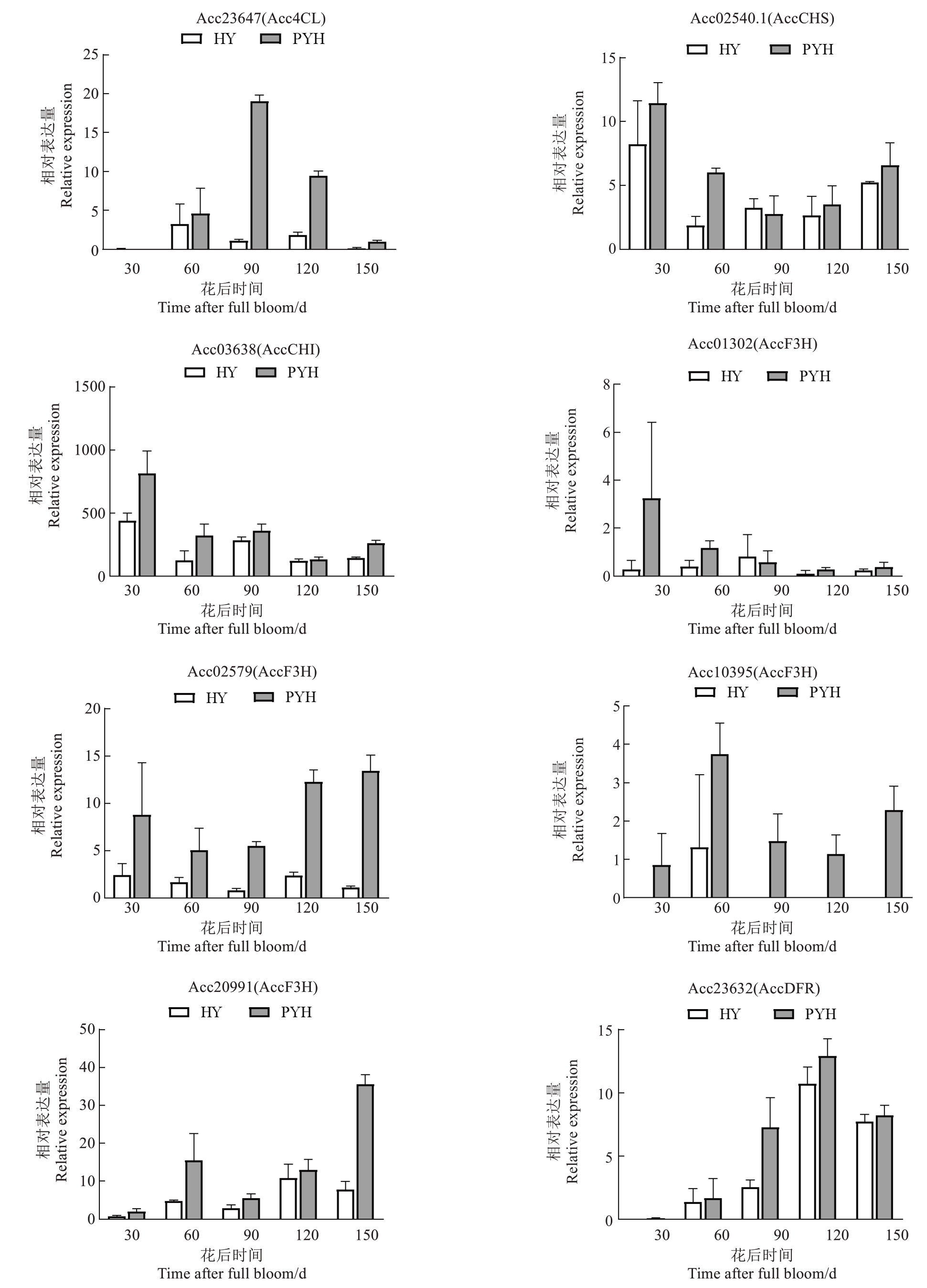
图6 红阳和平原红猕猴桃果肉中花青苷合成途径DEGs 的表达水平
Fig.6 The differentially expressed genes of Hongyang and Pingyuanhong kiwifruit fruits in the anthocyanin synthesis pathway
2.3 平原红猕猴桃实花青苷合成关键基因实时定量表达验证
对筛选出的花青苷合成两个关键基因Acc23647(Acc4CL)和Acc23632(AccDFR),通过实时荧光定量PCR 验证其在平原红猕猴桃果肉中实际表达水平。检验结果显示(图7),转录组分析基因表达的FPKM 值与RT-qPCR 相对表达量变化趋势一致,表明这两个关键基因转录组数据真实可靠。
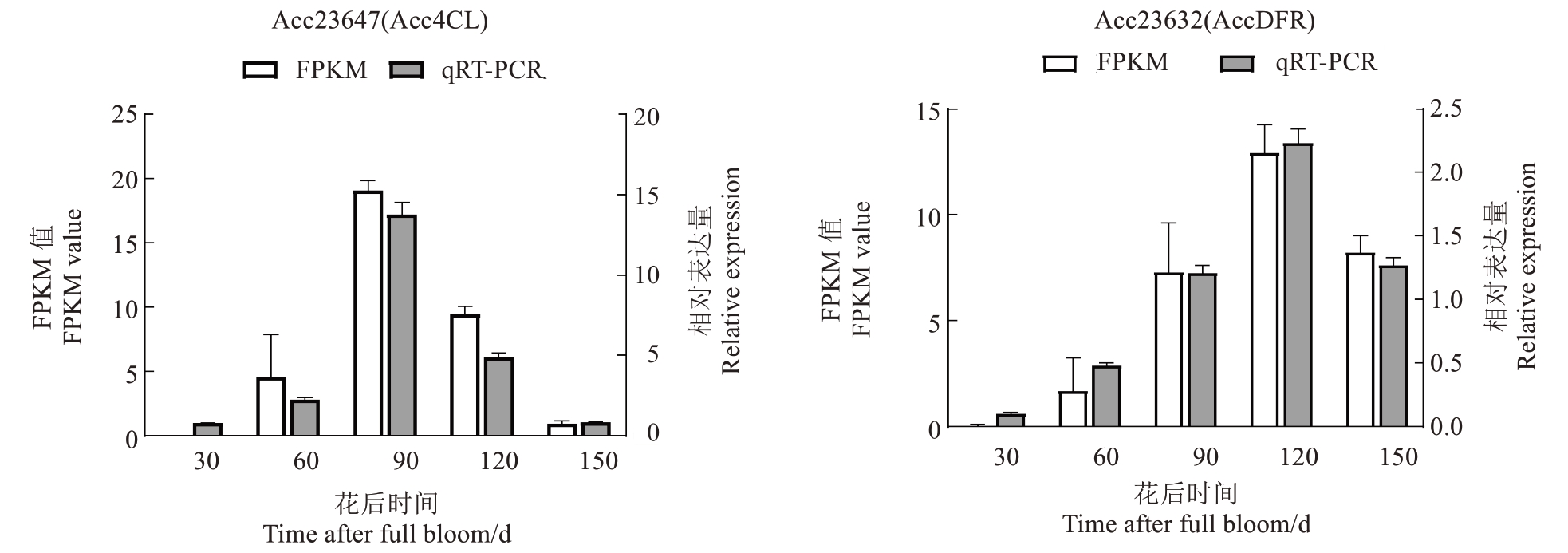
图7 平原红猕猴桃果肉花青苷合成途径DEGs 的RT-qPCR 验证
Fig.7 RT-qPCR verification of differentially expressed genes in anthocyanin synthesis pathway in Pingyuanhong samples
3 讨 论
3.1 平原红和红阳猕猴桃果实花青苷积累规律差异
Montefiori 等[11]初步鉴定出红心猕猴桃内的3种主要的花青素是花青素3-O-半乳糖苷、花青素3-O-葡萄糖苷和花青素3-O-木糖(1-2)-半乳糖苷。Comeskey等[9]从猕猴桃中鉴定出花青素-3-[2-(木糖基)半乳糖苷]、花青素-3-半乳糖苷、花青素-3-葡萄糖苷、飞燕草素-3-[2-(木糖基)半乳糖苷]和飞燕草素-3-半乳糖苷。猕猴桃的遗传特性决定了不同品种间花青苷的糖基取代位置、种类及花青苷含量的明显不同[12]。Liu 等[13]在2019 年的研究中,通过对6个红心和6 个绿肉猕猴桃品种进行花青苷含量的测定,结果在红心猕猴桃中发现5 种花青苷,其中还包括新鉴定出的锦葵色素和天竺葵素,而在绿肉猕猴桃中并未检测到花青苷。研究发现,在中华猕猴桃中,红心品种的黄酮类含量高于黄肉品种,即在红心猕猴桃中花青苷含量和类黄酮含量均高于黄肉猕猴桃;而在软枣猕猴桃中,绿肉品种的黄酮类含量高于紫肉品种,即在紫色品种中倾向于合成花青苷,而在绿色品种中倾向于合成黄酮类物质[14]。通过对几种猕猴桃果实的花青苷种类进行鉴定与分析,发现果实呈现的是红色还是紫色主要取决于矢车菊素和飞燕草素的相对含量[15]。根据笔者在本试验中得到的结果推测,平原红猕猴桃采收时果肉中花青苷含量高于红阳,不仅是其内果皮中花青苷合成量显著增多,而且其花青苷降解的速率也显著降低。关于平原红猕猴桃采收期花青苷降解的速率降低的分子机制,尚待进一步研究。
3.2 关于猕猴桃花青苷代谢途径关键基因
通过对红阳和平原红猕猴桃花后不同天数的果肉转录组进行分析,对8个候选关键DEGs进一步筛选,得到Acc23647(Acc4CL)和Acc23632(AccDFR)这两个上调基因,而且在两个品种猕猴桃果肉中的表达量变化趋势与花青苷含量变化趋势一致。DFR表达上调促进二氢槲皮素转化为无色花青苷[16],4CL 表达促进肉桂酰辅酶A 和对香豆酰辅酶A 的合成[17],从而促进花青苷生物合成。在蛇根草中,OjD‐FR1 编码活性DFR 蛋白,催化二氢黄酮醇还原为白花青素[18]。在石榴果实中,根据AMP 结构域分类,其中Pg2CL4与类黄酮合成有关[19]。
花青苷的主要合成场所是内质网,为了防止被氧化分解,之后需要转移到液泡中贮藏。转运的方式主要是通过内质网中产生的囊泡转移到液泡[20],或者通过谷胱甘肽S 转移酶(GST)从内质网中转移到液泡中,部分花青苷也可通过膜上的转运蛋白完成花青苷的转运[21-22]。
笔者在本试验中通过对红阳和平原红猕猴桃果肉DEGs 分析发现,两个猕猴桃品种果肉在发育过程中,DEGs 主要富集在与花青苷合成、分解相关的功能和代谢途径中;在与花青苷转运、贮藏相关的功能和代谢途径中,也存在部分DEGs。此外,糖类、有机酸等代谢物,在猕猴桃发育过程中也发生了变化。
3.3 转录因子参与调控花青苷合成
花青素生物合成在很大程度上受特定转录因子控制[23-24],尤其是R2R3-MYB 转录因子[25][26]。苗立祥等[27]的研究表明MYB10 在调节草莓果肉花青苷合成中起关键作用,且花青苷的积累与MYB10 基因的表达变化趋势一致,而林源秀[28]在有关草莓的研究中发现FaMYB10 并非通过自身表达调控花青苷的积累,而是通过调控合成相关结构基因FaF3’H的表达参与到果实中花青苷的合成积累。基因PpMYB10.1在红肉桃种质花青苷合成途径中起关键作用[29],桃中MYB10.1、MYB10.3 基因的表达与花青苷合成通路中的关键结构基因CHS、F3H 及UFGT的表达呈正相关;梨中PbMYB10b 和PbMYB9 正向调控果肉中花青苷和原花青素的合成。此外,Pb‐MYB10b可通过上调结构基因PbDFR的表达影响原花青素的合成[30];MdCEP1 在拟南芥中异位表达,依旧能够促进拟南芥中花青苷积累,同时提高其花青苷合成相关基因的表达水平[31]。在猕猴桃中,Ac‐MYB75 可直接作用于前期合成基因ANS 启动子序列而促进花青苷的积累[32]。Petunia PhMYB27 是一种R2R3-MYB 阻遏因子,可作为牵牛花花青苷生物合成过程中的负调节因子,其过表达导致花瓣和叶片中的花青素减少[33]。蒺藜MtMYB2 负调控PA 和花青素含量,MtMYB2 突变株系在叶片中增加花青素积累,种子中PA 含量增加[34]。MYB 家族的转录因子部分可以与花青苷代谢途径中的酶结合促进花青苷合成,也有一部分可以作为一种蛋白阻遏因子,负面调控花青苷的生成。
笔者在本试验研究中发现,猕猴桃花后90~150 d,在共有差异表达的MYB 家族基因中,有2 个是花后90、120、150 d 三个时期都上调的,有7 个在这三个时期都是下调的,有6 个差异表达的MYB 家族转录因子被富集到苯丙烷类生物合成途径中,说明这些转录因子也可能参与调控猕猴桃果肉花青苷代谢相关的基因表达。
4 结 论
平原红和红阳果肉中的花青苷,主要以矢车菊素3-O-半乳糖苷的形式存在。花后90、120、150 d,平原红猕猴桃果肉花青苷含量分别为红阳的3.34、2.15、3.97 倍。平原红内果皮着色好于红阳的主要原因,不仅是花青苷合成量增多,而且其花青苷降解的速率也较低。香豆酸辅酶A 连接酶基因(Acc23647)、二氢黄酮醇-4-还原酶基因(Acc23632)在平原红花青苷的合成过程中起正调控作用,MYB家族的转录因子参与了花青苷合成代谢。
[1] 孙雷明,方金豹.我国猕猴桃种质资源的保存与研究利用[J].植物遗传资源学报,2020,21(6):1483-1493.SUN Leiming,FANG Jinbao.Conservation,research and utiliza‐tion of kiwifruit germplasm resources in China[J]. Journal of Plant Genetic Resources,2020,21(6):1483-1493.
[2] 张计育,莫正海,宣继萍,贾晓东,刘永芝,郭忠仁.猕猴桃果肉颜色相关色素代谢研究进展[J]. 中国农学通报,2013,29(13):77-85.ZHANG Jiyu,MO Zhenghai,XUAN Jiping,JIA Xiaodong,LIU Yongzhi,GUO Zhongren.Advance of research on flesh color re‐lated pigment metabolism in kiwifruit[J]. Chinese Agricultural Science Bulletin,2013,29(13):77-85.
[3] CHO K,CHO K S,SOHN H B,HA I J,HONG S Y,LEE H,KIM Y M,NAM M H.Network analysis of the metabolome and transcriptome reveals novel regulation of potato pigmentation[J].Journal of Experimental Botany,2016,67(5):1519-1533.
[4] 葛翠莲,黄春辉,徐小彪. 果实花青素生物合成研究进展[J].园艺学报,2012,39(9):1655-1664.GE Cuilian,HUANG Chunhui,XU Xiaobiao. Research on an‐thocyanins biosynthesis in fruit[J]. Acta Horticulturae Sinica,2012,39(9):1655-1664.
[5] ZHAO D Q,TAO J. Recent advances on the development and regulation of flower color in ornamental plants[J]. Frontiers in Plant Science,2015,6:261.
[6] 张震.基于转录组学和代谢组学研究梨花青苷和原花青苷合成调控机制[D].泰安:山东农业大学,2020.ZHANG Zhen. Transcriptomic and metabolomic analysis pro‐vides insights into anthocyanin and procyanidin accumulation in pear[D].Tai’an:Shandong Agricultural University,2020.
[7] 曹琳娇,李晓杰,焦棒棒,梁毅,马长生.蔬菜花青苷生物合成及转录调控的研究进展[J].中国瓜菜,2019,32(12):1-7.CAO Linjiao,LI Xiaojie,JIAO Bangbang,LIANG Yi,MA Changsheng.Advances in biosynthesis and transcriptional regu‐lation of anthocyanin in vegetables[J].China Cucurbits and Veg‐etables,2019,32(12):1-7.
[8] 余敏.猕猴桃花青苷着色:MYB 调节基因的鉴定及其功能解析[D].北京:中国科学院大学,2020.YU Min. Mechanisms underlying the regulation of anthocvan‐in coloration synthesis in kiwifruit[D]. Beijing:University of Chinese Academy of Sciences,2020.
[9] COMESKEY D J,MONTEFIORI M,EDWARDS P J B,MC‐GHIE T K.Isolation and structural identification of the anthocy‐anin components of red kiwifruit[J]. Journal of Agricultural and Food Chemistry,2009,57(5):2035-2039.
[10] 贾兵,王克灿,叶振风,王谋才,刘莉,朱立武.猕猴桃新品种平原红的选育[J].果树学报,2021,38(8):1403-1406.JIA Bing,WANG Kecan,YE Zhenfeng,WANG Moucai,LIU Li,ZHU Liwu.Breeding of kiwifruit new cultivar Pingyuanhong[J].Journal of Fruit Science,2021,38(8):1403-1406.
[11] MONTEFIORI M,MCGHIE T K,COSTA G,FERGUSON A R.Pigments in the fruit of red-fleshed kiwifruit(Actinidia chinensis and Actinidia deliciosa)[J]. Journal of Agricultural and Food Chemistry,2005,53(24):9526-9530.
[12] 涂美艳.猕猴桃果肉颜色差异机理及相关EST-SSR 分子标记开发[D].雅安:四川农业大学,2019.TU Meiyan.Studies on the mechanism of flesh color various and development of EST-SSR molecular markers of kiwifruit[D].Yaan:Sichuan Agricultural University,2019.
[13] LIU Y F,QI Y W,CHEN X,HE H H,LIU Z D,ZHANG Z,REN Y M,REN X L.Phenolic compounds and antioxidant activ‐ity in red- and in green-fleshed kiwifruits[J]. Food Research In‐ternational,2019,116:291-301.
[14] LI Y K,FANG J B,QI X J,LIN M M,ZHONG Y P,SUN L M,CUI W. Combined analysis of the fruit metabolome and tran‐scriptome reveals candidate genes involved in flavonoid biosyn‐thesis in Actinidia arguta[J]. International Journal of Molecular Sciences,2018,19(5):1471.
[15] XIONG Y,HE J Y,LI M Z,DU K,LANG H Y,GAO P,XIE Y.Integrative analysis of metabolome and transcriptome reveals the mechanism of color formation in yellow-fleshed kiwifruit[J].International Journal of Molecular Sciences,2023,24(2):1573.
[16] 于婷婷,倪秀珍,高立宏,韩国军,朱长甫,盛彦敏.高等植物二氢黄酮醇4-还原酶基因研究进展[J]. 植物研究,2018,38(4):632-640.YU Tingting,NI Xiuzhen,GAO Lihong,HAN Guojun,ZHU Changfu,SHENG Yanmin. Advances in study of dihydroflavo‐nol 4-reductase(DFR) genes of higher plants[J]. Bulletin of Bo‐tanical Research,2018,38(4):632-640.
[17] LAVHALE S G,KALUNKE R M,GIRI A P. Structural,func‐tional and evolutionary diversity of 4-coumarate-CoA ligase in plants[J].Planta,2018,248(5):1063-1078.
[18] SUN W,ZHOU N N,FENG C,SUN S Y,TANG M,TANG X X,JU Z G,YI Y. Functional analysis of a dihydroflavonol 4-re‐ductase gene in Ophiorrhiza japonica (OjDFR1) reveals its role in the regulation of anthocyanin[J].PeerJ,2021,9:e12323.
[19] WANG Y Y,GUO L H,ZHAO Y J,ZHAO X Q,YUAN Z H.Systematic analysis and expression profiles of the 4-coumarate:CoA ligase (4CL) gene family in pomegranate (Punica grana‐tum L.)[J].International Journal of Molecular Sciences,2022,23(7):3509.
[20] 赵文军,张迪,马丽娟,柴友荣.原花青素的生物合成途径、功能基因和代谢工程[J].植物生理学通讯,2009,45(5):509-519.ZHAO Wenjun,ZHANG Di,MA Lijuan,CHAI Yourong. Bio‐synthetic pathway,functional genes and metabolic engineering of proanthocyanidins[J]. Plant Physiology Communications,2009,45(5):509-519.
[21] 刘艳飞. 红阳猕猴桃果实花青苷生物合成及其转录调控研究[D].杨凌:西北农林科技大学,2019.LIU Yanfei. Anthocyanin biosynthesis and its transcriptional regulation in kiwifruit (Actinidia chinensis cv. Hongyang)[D].Yangling:Northwest A&F University,2019.
[22] LIU Y F,QI Y W,ZHANG A L,WU H X,LIU Z D,REN X L.Molecular cloning and functional characterization of AcGST1,an anthocyanin-related glutathione S-transferase gene in kiwi‐fruit (Actinidia chinensis)[J]. Plant Molecular Biology,2019,100(4/5):451-465.
[23] LASHBROOKE J,ADATO A,LOTAN O,ALKAN N,TSIM‐BALIST T,RECHAV K,FERNANDEZ-MORENO J P,WIDE‐MANN E,GRAUSEM B,PINOT F,GRANELL A,COSTA F,AHARONI A.The tomato MIXTA-like transcription factor coor‐dinates fruit epidermis conical cell development and cuticular lipid biosynthesis and assembly[J]. Plant Physiology,2015,169(4):2553-2571.
[24] 彭亚丽,高倩,董文,熊安平,秦玉芝,林原,熊兴耀,胡新喜.MYB 转录因子调控蔬菜花青素生物合成的研究进展[J].中国瓜菜,2020,33(12):1-7.PENG Yali,GAO Qian,DONG Wen,XIONG Anping,QIN Yu‐zhi,LIN Yuan,XIONG Xingyao,HU Xinxi.Advances of MYB transcription factors regulating vegetable anthocyanins biosyn‐thesis[J].China Cucurbits and Vegetables,2020,33(12):1-7.
[25] OSORIO S,ALBA R,DAMASCENO C M B,LOPEZ-CASA‐DO G,LOHSE M,ZANOR M I,TOHGE T,USADEL B,ROSE J K C,FEI Z J,GIOVANNONI J J,FERNIE A R. Sys‐tems biology of tomato fruit development:Combined transcript,protein,and metabolite analysis of tomato transcription factor(nor,rin) and ethylene receptor (Nr) mutants reveals novel regu‐latory interactions[J].Plant Physiology,2011,157(1):405-425.
[26] CAO Y P,HAN Y H,LI D H,LIN Y,CAI Y P. MYB transcrip‐tion factors in Chinese pear (Pyrus bretschneideri Rehd.):Ge‐nome-wide identification,classification,and expression profil‐ing during fruit development[J]. Frontiers in Plant Science,2016,7:577.
[27] 苗立祥,荣宁宁,张豫超,杨肖芳,张琴,蒋桂华.草莓花青苷分子调控机理的初步研究[J]. 浙江农业学报,2015,27(8):1373-1380.MIAO Lixiang,RONG Ningning,ZHANG Yuchao,YANG Xiaofang,ZHANG Qin,JIANG Guihua. Preliminary study on molecular regulation mechanism of anthocyanin biosynthesis in strawberry[J]. Acta Agriculturae Zhejiangensis,2015,27(8):1373-1380.
[28] 林源秀.草莓果实花青素苷转录调控机制研究[D].雅安:四川农业大学,2018.LIN Yuanxiu. Research on transcriptional regulation of anthocy‐anin in red-and white-fleshed strawberry[D].Ya’an:Sichuan Ag‐ricultural University,2018.
[29] 丁体玉,曹珂,方伟超,朱更瑞,陈昌文,王新卫,王力荣.红肉桃两类花色素苷积累模式与相关基因表达差异[J].中国农业科学,2017,50(13):2553-2563.DING Tiyu,CAO Ke,FANG Weichao,ZHU Gengrui,CHEN Changwen,WANG Xinwei,WANG Lirong. The difference of anthocyanin accumulation pattern and related gene expression in two kinds of red flesh peach[J]. Scientia Agricultura Sinica,2017,50(13):2553-2563.
[30] 翟锐.芽变‘红早酥’梨的花青苷与类黄酮合成机理解析[D].杨凌:西北农林科技大学,2019.ZHAI Rui. The biosynthesis pattern of anthocyanin and flavo‐noid in bud sport‘Red Zaosu’pear[D]. Yangling:Northwest A&F University,2019.
[31] 李睿,安建平,由春香,王小非,郝玉金.过表达苹果多肽激素基因MdCEP1 促进花青苷积累[J]. 园艺学报,2018,45(6):1031-1040.LI Rui,AN Jianping,YOU Chunxiang,WANG Xiaofei,HAO Yujin. Overexpression of apple peptide hormone gene MdCEP1 promotes the accumulation of anthocyanins[J]. Acta Horticul‐turae Sinica,2018,45(6):1031-1040.
[32] LI W B,DING Z H,RUAN M B,YU X L,PENG M,LIU Y F. Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthe‐sis[J].Scientific Reports,2017,7(1):16861.
[33] ALBERT N W,LEWIS D H,ZHANG H B,SCHWINN K E,JAMESON P E,DAVIES K M. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegeta‐tive pigmentation patterning[J]. The Plant Journal,2011,65(5):771-784.
[34] JUN J H,LIU C G,XIAO X R,DIXON R A.The transcriptional repressor MYB2 regulates both spatial and temporal patterns of proanthocyandin and anthocyanin pigmentation in Medicago truncatula[J].The Plant Cell,2015,27(10):2860-2879.