在植物发育过程中,茎尖分生组织首先产生叶片等营养器官,植株发育到特定阶段后,在外界环境和内源激素等因素的共同影响下,开始由营养生长转向生殖生长。此时,茎尖分生组织分化出花序或花原基,最终发育为花[1]。一朵完全花由四轮花器官组成,由外到内依次为萼片、花瓣、雄蕊和心皮[2]。在“ABC(D)E”花发育调控模型中,每一轮花器官的发育都由相应的基因特异性调控,其中A(AP1 和AP2)+E(SEP)类基因决定萼片身份,A+B(AP3 和PI)+E 类基因共同控制花瓣身份,B+C(AG)+E 类基因共同控制雄蕊的生长发育,C+E 类基因确定心皮的身份[3-4]。
植物花在进化过程中的变异,产生了不同于经典的四轮花器官结构。如板栗、杨树、核桃、小桐子、白桦树等植物为柔荑花序,花单性,无花瓣和萼片。在这些植物中,花瓣和萼片由苞片替代[5]。苞片的发育可能同时受A、B、E基因的共同调控[6]。板栗作为壳斗科植物的典型代表,雌花和雄花形态差异较大。板栗雄花苞片小而薄,花期易于雄蕊伸出。板栗雌花通常3~5 朵聚合为雌花簇,着生于混合花序基部,雌花簇由大量苞片包围。板栗雌花受精后,苞片发育为壳斗。板栗苞片的性二态发育可能同样受MADS-box 基因调控。目前已有研究在板栗和欧洲栗基因组中对MADS-box 基因进行了鉴定和表达分析,但未进行生物学功能验证[7]。PI 基因是B 类花发育基因之一,参与花瓣和雄蕊的发育调控,是研究板栗苞片发育调控机制的理想基因之一。笔者以板栗(Castanea mollissima)品种燕山红栗为试验材料,通过对板栗B类花器官发育基因CmPI的同源克隆、进化分析、时空表达分析和功能验证,为解析板栗苞片性二态发育的分子调控机制奠定基础。
1 材料和方法
1.1 试验材料
以板栗品种燕山红栗为材料,分别取雄花、雌花、胚、叶芽、花芽、茎尖及叶片,雄花从5月11日始,每隔7 d取样1次,共取样5次,样品经液氮速冻后置于-80 ℃冰箱保存。
课题组实验室保存的Columbia 野生型拟南芥(Arabidopsis thaliana)用于目的基因的异源过表达验证,本氏烟草(Nicotiana benthamiana)用于亚细胞定位融合载体的瞬时转化。
1.2 试验方法
1.2.1 总RNA 提取与反转录 取液氮速冻后存于-80 ℃冰箱的100 mg 植物组织进行RNA 提取。按照TaKaRa 公司植物RNA 提取试剂盒(TaKaRa MiniBEST Universal RNA Extraction Kit,9769)说明书提取总RNA。提取完成后检测总RNA 的浓度和纯度,再用1.5%琼脂糖凝胶电泳进一步检测总RNA的完整性。利用TaKaRa 公司的反转录试剂盒(PrimeScript ™RT reagent Kit with gDNA Eraser,RR047Q),按照其说明书合成cDNA 第一链,反转录产物存放于-20 ℃冰箱。
1.2.2 板栗PI 基因克隆 根据板栗基因组测序数据(http://chestnutgenome.cn)利用DNAMAN 软件设计板栗CmPI 编码区序列(CDS)全长引物(表1)。以反转录产物为模板进行CmPI 基因的CDS 扩增。PCR 产物经1.5%琼脂糖凝胶电泳检测后,利用TAKARA 胶回收试剂盒(TaKaRa MiniBEST DNA Fragment Purification Kit Ver.4.0,9761)回收目的条带。将回收产物连接pBM21(pBM21 Topsmart Cloning Kit)载体上,转化大肠杆菌DH5α,在含氨苄霉素的LB 培养基上,放置37 ℃恒温培养16 h,送至生工生物工程(上海)股份有限公司测序。
表1 PCR 引物信息
Table 1 Primers used for gene cloning
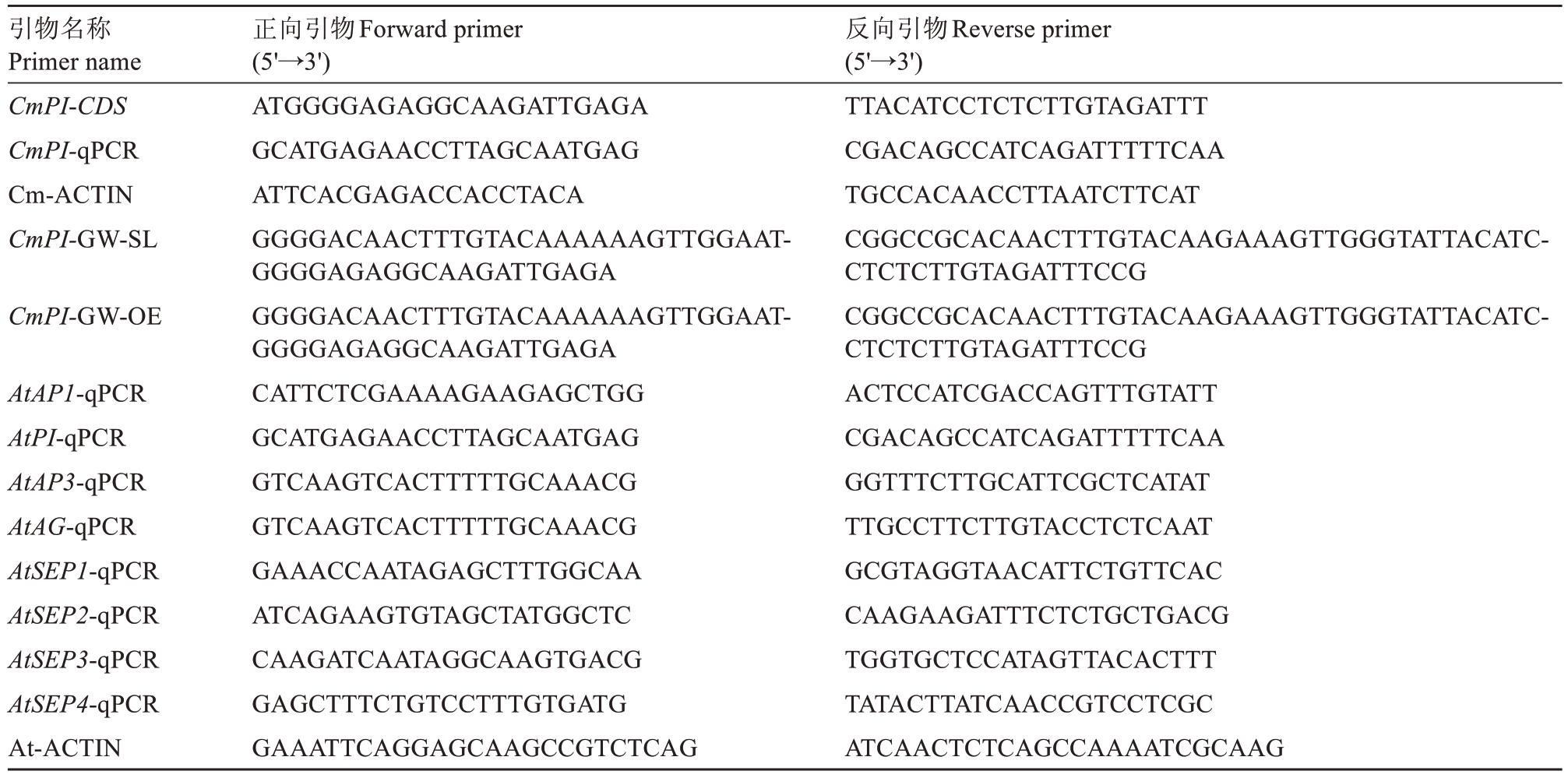
引物名称Primer name CmPI-CDS CmPI-qPCR Cm-ACTIN CmPI-GW-SL CmPI-GW-OE AtAP1-qPCR AtPI-qPCR AtAP3-qPCR AtAG-qPCR AtSEP1-qPCR AtSEP2-qPCR AtSEP3-qPCR AtSEP4-qPCR At-ACTIN正向引物Forward primer(5'→3')ATGGGGAGAGGCAAGATTGAGA GCATGAGAACCTTAGCAATGAG ATTCACGAGACCACCTACA GGGGACAACTTTGTACAAAAAAGTTGGAATGGGGAGAGGCAAGATTGAGA GGGGACAACTTTGTACAAAAAAGTTGGAATGGGGAGAGGCAAGATTGAGA CATTCTCGAAAAGAAGAGCTGG GCATGAGAACCTTAGCAATGAG GTCAAGTCACTTTTTGCAAACG GTCAAGTCACTTTTTGCAAACG GAAACCAATAGAGCTTTGGCAA ATCAGAAGTGTAGCTATGGCTC CAAGATCAATAGGCAAGTGACG GAGCTTTCTGTCCTTTGTGATG GAAATTCAGGAGCAAGCCGTCTCAG反向引物Reverse primer(5'→3')TTACATCCTCTCTTGTAGATTT CGACAGCCATCAGATTTTTCAA TGCCACAACCTTAATCTTCAT CGGCCGCACAACTTTGTACAAGAAAGTTGGGTATTACATCCTCTCTTGTAGATTTCCG CGGCCGCACAACTTTGTACAAGAAAGTTGGGTATTACATCCTCTCTTGTAGATTTCCG ACTCCATCGACCAGTTTGTATT CGACAGCCATCAGATTTTTCAA GGTTTCTTGCATTCGCTCATAT TTGCCTTCTTGTACCTCTCAAT GCGTAGGTAACATTCTGTTCAC CAAGAAGATTTCTCTGCTGACG TGGTGCTCCATAGTTACACTTT TATACTTATCAACCGTCCTCGC ATCAACTCTCAGCCAAAATCGCAAG
1.2.3 生物信息学分析 利用EXPACY网站(http://www.expacy.org/)预测其编码蛋白的等电点和分子质量大小及其亲/疏水性;在NCBI数据库(http://blast.ncbi.nlm.nih.gov/)中通过Conserved Domains进行CmPI 基因的保守域预测。从NCBI数据库下载大豆(Glycine max)、黄瓜(Cucumis sativus)、葡萄(Vitis vinifera)、白桦(Betula platyphylla)、向日葵(Helianthus annuus)、鹅掌楸(Liriodendron chi‐nense)、红皮柳(Salix purpurea)、睡莲(Nymphaea te‐tragona)、藜麦(Chenopodium quinoa)、拟南芥(Ara‐bidopsis thaliana)的PI氨基酸序列并与CmPI 借助MEGAX 软件,用邻接法(Neighbor-Joining)构建进化树,校验参数为bootsrap=1000。
1.2.4 CmPI 在板栗中的时空表达模式分析 以不同组织和不同发育时期雄花RNA 反转录产物为模板,进行实时荧光定量PCR。利用DNAMAN 设计特异引物。以CmActin基因为内参基因(表1)[8]。具体步骤参照TaKaRa 试剂盒(TB Green® Premix Ex Taq™Ⅱ,RR820A)说明书。反应程序为:95 ℃预变性30 s,95 ℃变性5 s,60 ℃延伸30 s,40个循环。试验设置3 个重复,并采用2-∆∆Ct法计算基因的相对表达量。通过GraphPad Prism8 软件对基因的表达量进行差异分析。
1.2.5 载体构建 根据试剂盒(Life technologies,Carlsbad,CA,USA)说明书分别设计亚细胞定位载体和过表达载体引物(表1)。利用PCR 扩增得到含有载体接头的CmPI 基因,并将其克隆至pEGOEP35S 中间载体上。将中间载体上不含终止密码子CmPI-GFP 基因和完整CmPI-GFP 基因分别重组至pVS1 RepA 和pVS1 StaA 载体上,得到含35S::CmPI-GFP 亚细胞定位载体和35S::CmPI 过表达载体。
1.2.6 亚细胞定位 利用冻融法,将35S::CmPIGFP亚细胞定位载体质粒转化到农杆菌中。参照杨国栋[9]的方法侵染本氏烟草叶片,将pEGOEP35S-HGFP 空载作为对照。烟草侵染2~3 d 后,用激光共聚焦显微镜(Leica TCS SPII)观察叶片细胞中荧光的分布情况。
1.2.7 板栗CmPI 过表达转基因拟南芥的获取及表型分析 将含有目的基因的过表达载体通过冻融法转入GV31001 农杆菌感受态细胞。参照Clough等[10]报道的花粉管侵染法转化拟南芥。用50 mg·L-1的潮霉素筛选抗性植株。将长出真叶的拟南芥幼苗移栽到花盆中置于光照培养箱培养,培养条件为光照16 h(24 ℃),黑暗8 h(20 ℃)。选择表型变化明显的5 个株系继续培养至T2 代。挑选目的基因表达量较高的3 个转基因株系及野生型株系分别移栽10 株至气候箱,对其进行定期观察,待进入盛花期后,在顶部花序中,每株随机选取5 朵处于第六阶段的拟南芥花,分别测量其花瓣与萼片的长度,对测量结果利用GraphPad Prism8软件进行分析[11]。
1.2.8 CmPI 过表达拟南芥中ABCE 类花发育基因表达分析 取5 株CmPI-OE 拟南芥,以CmPI-OE 拟南芥花RNA 反转录产物为模板,进行实时荧光定量PCR。花发育基因AtAP1、AtAP3、AtPI、AtAG、At‐SEP1、AtSEP2、AtSEP3、AtSEP4及内参基因AtACTIN引物参考https://biodb.swu.edu.cn/qprimerdb/(表1)。具体步骤参照TaKaRa 试剂盒(TB Green® Premix Ex Taq™I,RR820A)说明书进行。反应程序为:95 ℃预变性30 s;95 ℃变性5 s,60 ℃延伸30 s,40个循环。试验设置3 个重复,并采用2-∆∆Ct法计算基因的相对表达量。通过GraphPad Prism8 软件对基因的表达量进行差异分析。
2 结果与分析
2.1 板栗CmPI基因克隆及生物信息学分析
以板栗雄花的cDNA为模板,通过RT-PCR克隆板栗CmPI(图1-A)。克隆到的板栗CmPI 编码区序列长度为630 bp,编码209 个氨基酸。NCBI分析表明,CmPI 蛋白理论分子质量为82 015.9 Da,等电点为5.1,为亲水性蛋白。

图1 板栗CmPI 基因克隆及序列分析
Fig.1 Cloning and sequence analysis of CmPI gene in chestnut
A.CmPI 同源克隆PCR 产物琼脂糖电泳图;B.板栗CmPI保守结构域示意图;C.不同物种PI氨基酸序列比对。
A.Agarose electrophoresis of PCR product of CmPI homologous cloning;B.Schematic diagram of CmPIconserved domain of Castanea mollissi‐ma;C.Amino acid sequence alignment of PIin different species.
在NCBI数据库中通过Conserved Domains 进行基因的保守域预测,结果(图1-B)表明,PI蛋白在2~75 aa 处含有高度保守的MADS 域(MADS-box),在84~165 aa 处含有K 域(K-box),为MADS 的Ⅱ型亚家族成员。
在NCBI库中用Blast 对CmPI蛋白序列进行相似性比对分析(图1-C)。利用MEGAX 构建板栗与大豆、黄瓜、葡萄、白桦、向日葵、鹅掌楸、紫柳、睡莲、藜麦、拟南芥PI同源蛋白的系统进化树(图2)。结果表明,板栗与白桦PI蛋白亲缘关系最近,大豆、黄瓜和葡萄次之。
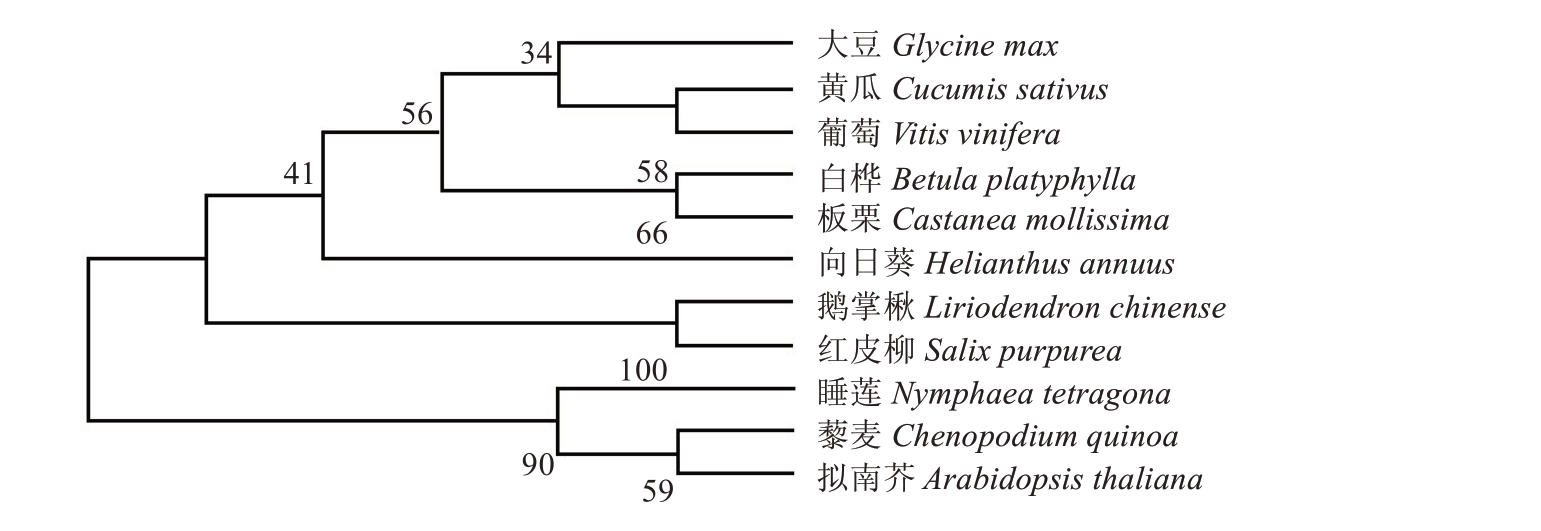
图2 使用邻接法(NJ)构建的11 个物种的PI 氨基酸序列系统树
Fig.2 PI amino acid sequence tree of 11 species constructed by neighbor-joining method(NJ)
2.2 CmPI在板栗中的时空表达模式分析
以板栗雄花、雌花、叶片、叶芽、花芽、胚、茎尖7个不同植物组织为材料,进行实时荧光定量PCR 分析,结果(图3)表明,CmPI 主要在雄花中表达。以板栗不同时期雄花为材料,进行实时荧光定量PCR分析,结果(图4)表明,CmPI 基因的表达水平在雄花发育过程中逐渐升高,并在5 月31 日(盛花期)达到最大值,然后开始降低。
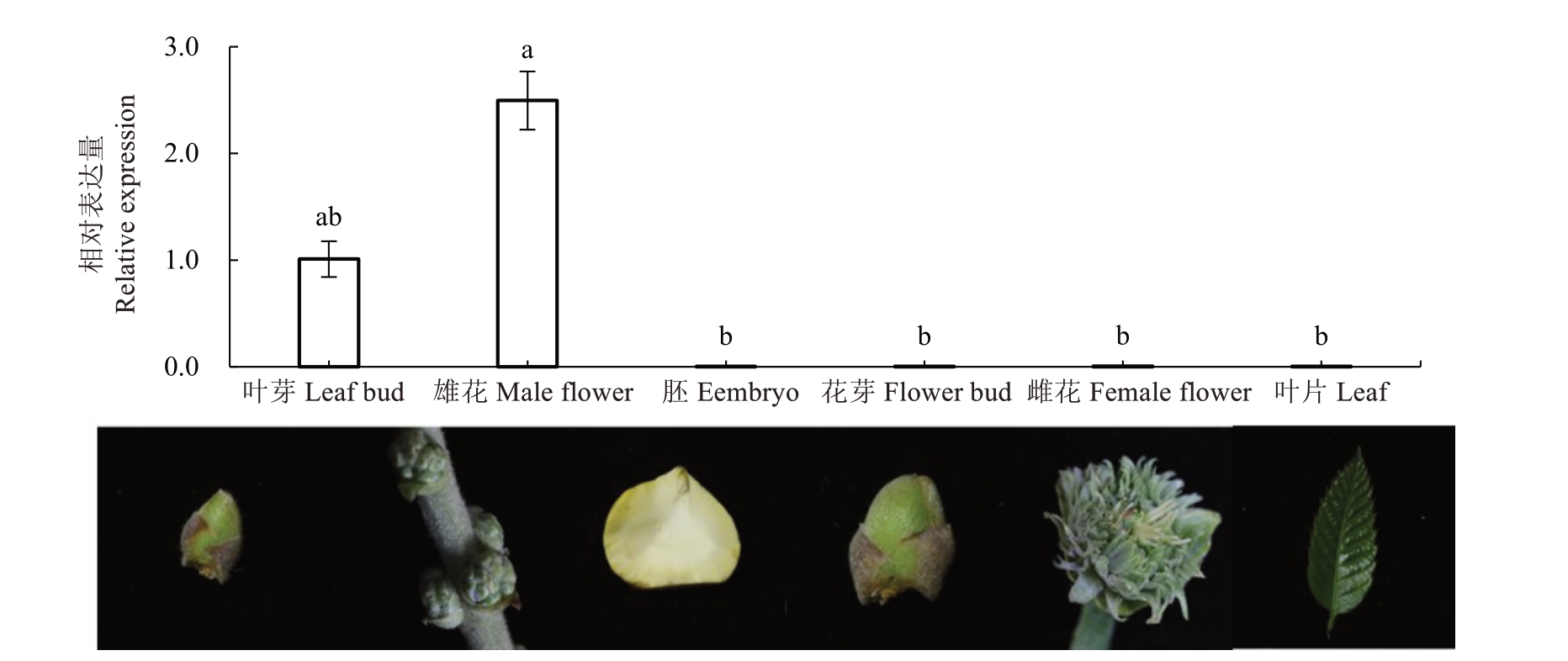
图3 CmPI 在板栗不同组织中的表达
Fig.3 The expression analysis of CmPI in different tissues of chestnut
用LSD 法分析不同组间的差异显著性。数值为平均值±标准误,n=3。不同小写字母表示差异显著(p<0.05)。下同。
Least significant difference (LSD) tests were used to determine significant differences between groups.Data are mean ± SE,n=3.Different small letters indicate significant difference(p<0.05).The same below.
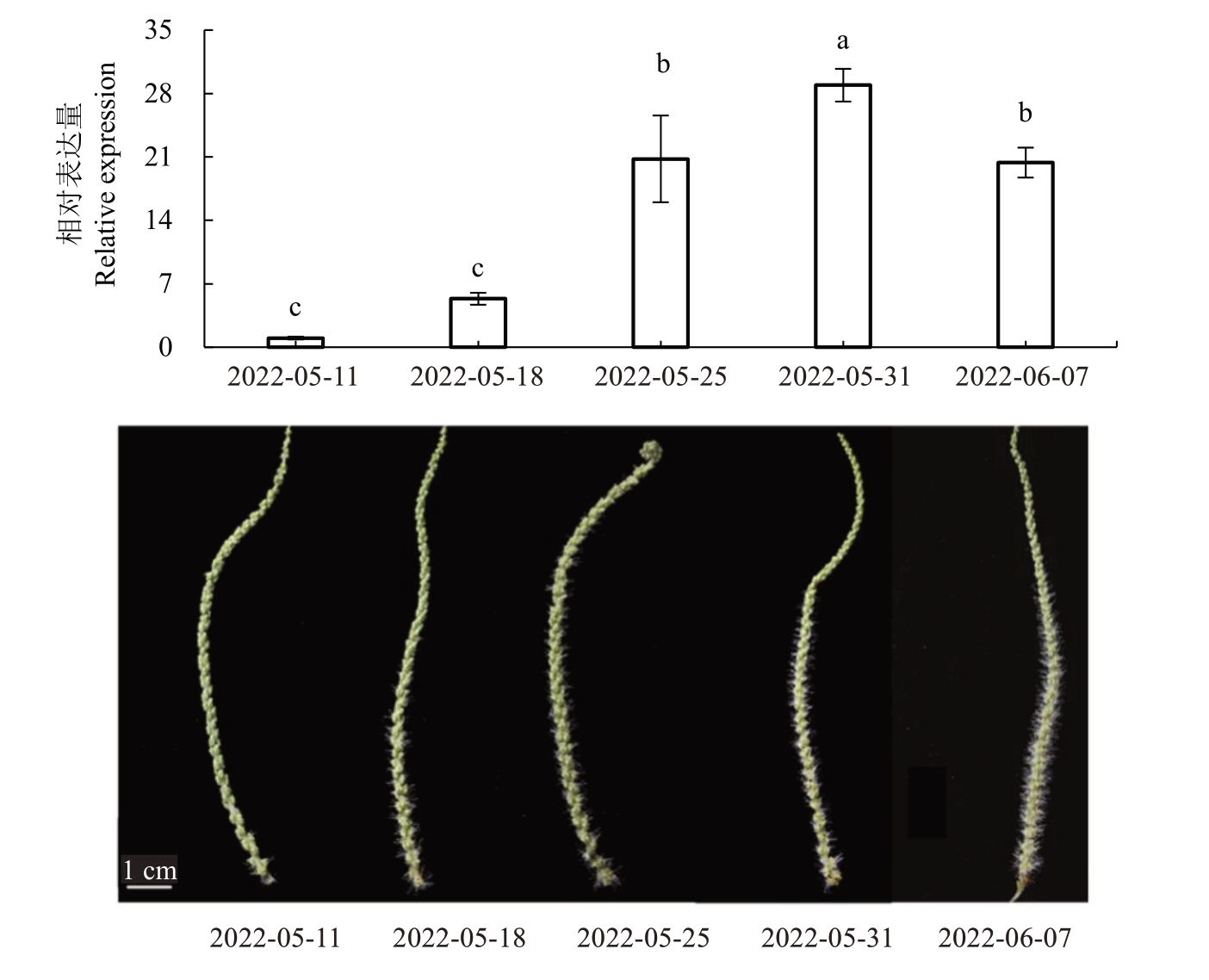
图4 CmPI 在不同发育时期板栗雄花中的表达
Fig.4 The expression analysis of CmPI in different development stage male flower of chestnut
2.3 亚细胞定位分析
将构建好的pEGOEP35S::CmPI-GFP 载体和pEGOEP35S::GFP 空载体通过农杆菌介导法侵染本氏烟草叶片,进行瞬时表达,观察GFP 荧光信号。结果如图5所示,空载体的荧光信号分布在细胞膜、细胞质和细胞核上,而pEGOEP35S::CmPI-GFP 的荧光信号只分布在细胞核上,表明CmPI 基因定位于细胞核。
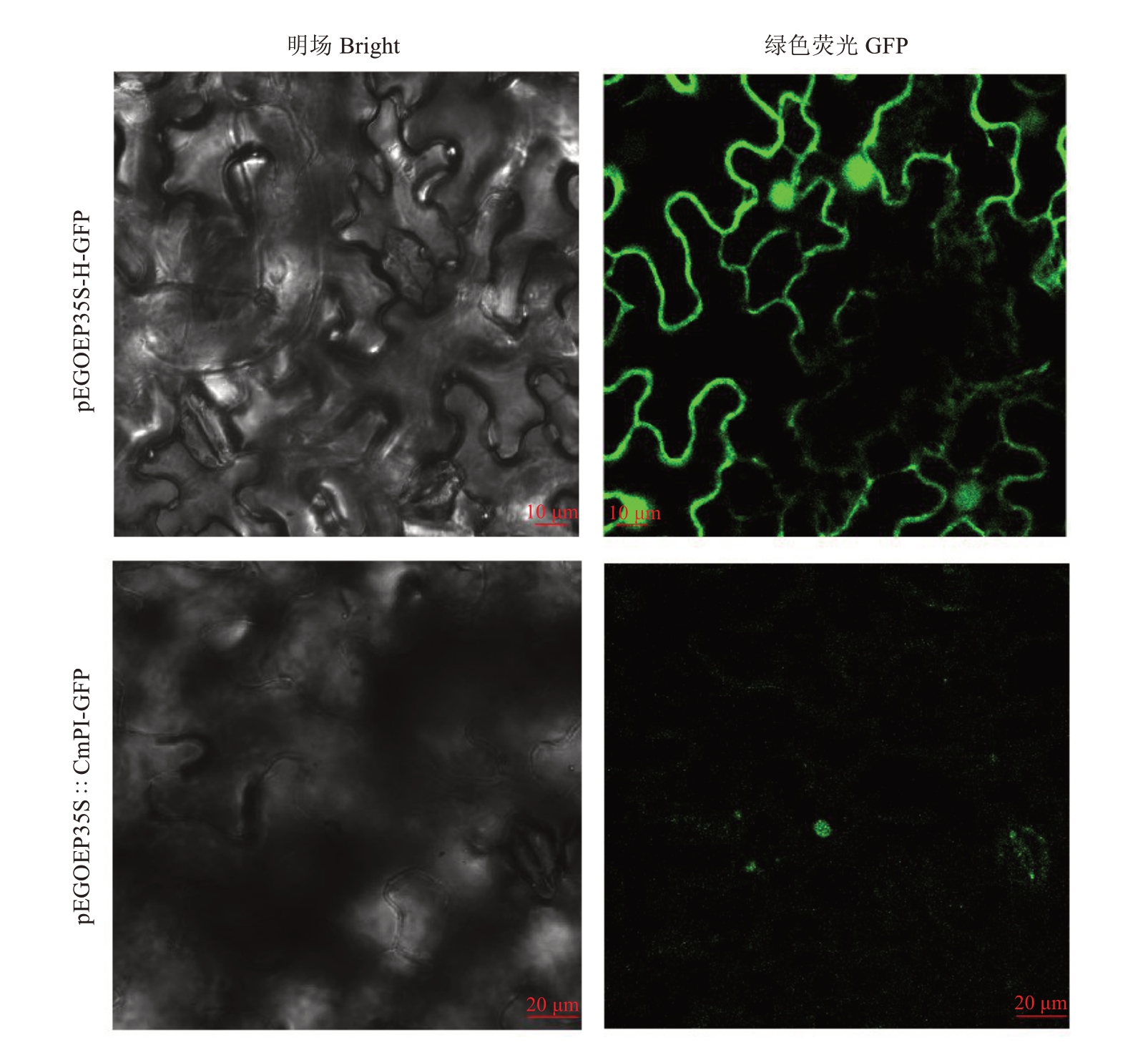
图5 CmPI 亚细胞定位
Fig.5 Subcellular localization of CmPI
2.4 过表达CmPI影响花器官发育及相关基因的表达
为进一步研究CmPI 的生物学功能,笔者构建了CmPI 过表达载体,并转入拟南芥获取了转基因植株。通过表型观察发现,CmPI过表达拟南芥植株花发育早期,萼片变窄,萼片间出现缝隙,无法包裹内部花器官结构。CmPI 过表达拟南芥植株花开放后,萼片和花瓣呈离散状(图6-A~B)。测量结果表明,CmPI转基因植株萼片宽度为0.59 mm,显著低于野生型拟南芥(图6-C);萼片长度为2.49 mm,显著高于野生型拟南芥(图6-D)。而CmPI转基因植株花瓣长度与野生型拟南芥相比,无显著差异(图6-E)。对转基因株系和野生型拟南芥调控萼片和花瓣的相关A、B、C、E 基因进行表达分析,结果表明,C 类基因AtAG在CmPI过表达拟南芥中的表达量极显著高于野生型,而A 类基因AtAP1、B 类基因AtAP3/PI、E 类基因AtSEP1/AtSEP2/AtSEP3/AtSEP4在CmPI过表达拟南芥中的表达量显著低于野生型(图7)。
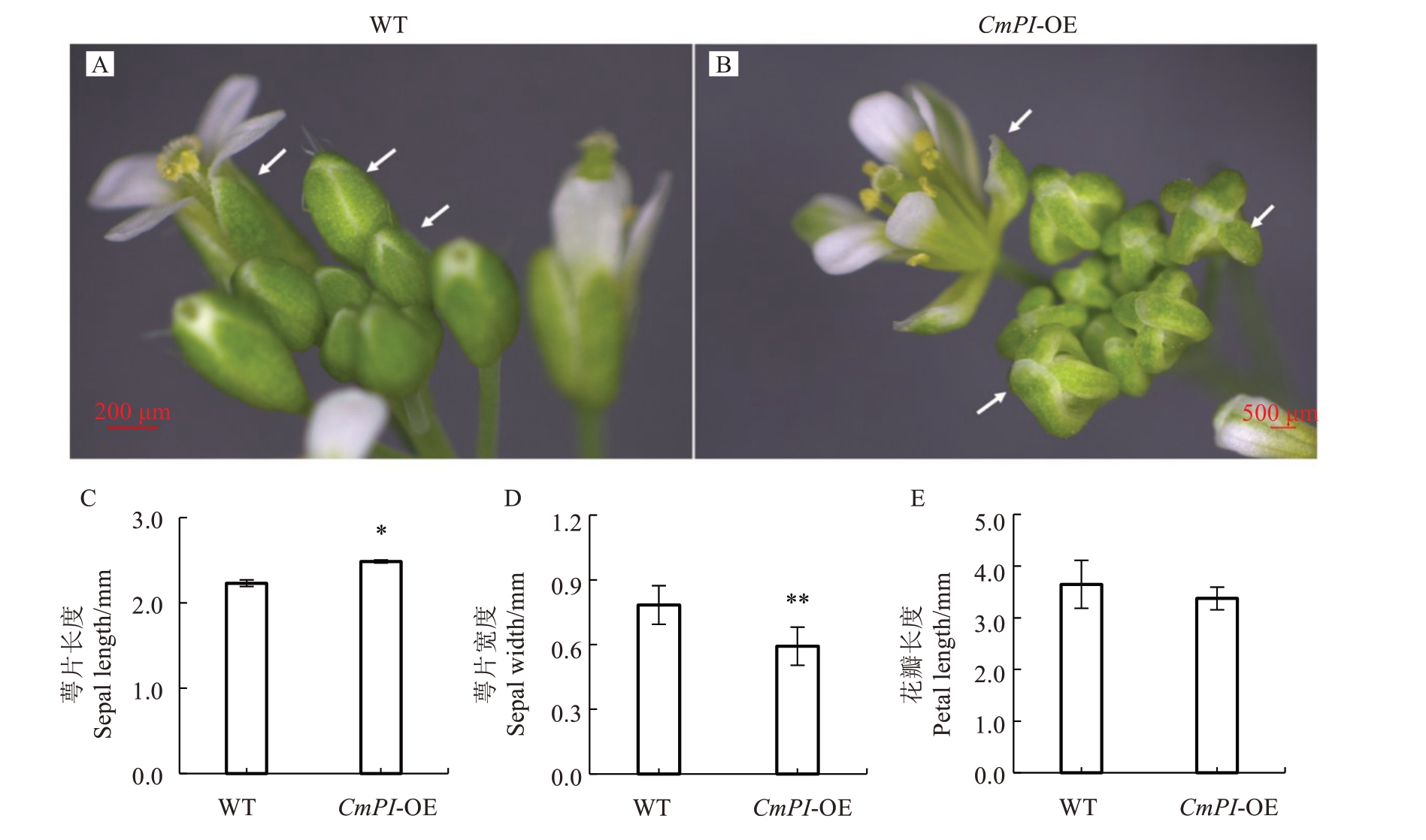
图6 过表达CmPI 对拟南芥花器官发育的影响
Fig.6 The effects of CmPI overexpression on flower development in Arabidopsis thaliana
A.野生型拟南芥花表型;B.CmPI 过表达拟南芥花发育表型;C~E.依次为过表达CmPI 对萼片长度、萼片宽度及花瓣长度的影响。WT 代表野生型拟南芥,CmPI-OE 代表过表达转基因拟南芥。白色箭头示萼片。“*”代表p 在<0.05 水平显著差异;“**”代表p 在<0.01 水平极显著差异。下同。
A.The wild type Arabidopsis flower phenotype;B.The flower development phenotype of Arabidopsis overexpression of CmPI;C-E.The effect of overexpression of CmPIon sepal length, sepal width and petal length respectively.WT stands for wild-type Arabidopsis thaliana,and CmPI-OE stands for overexpressing transgenic Arabidopsis.The white arrow shows sepals.“*”represents significant difference at p<0.05;“**”represents extremely significant difference at p<0.01.The same below.
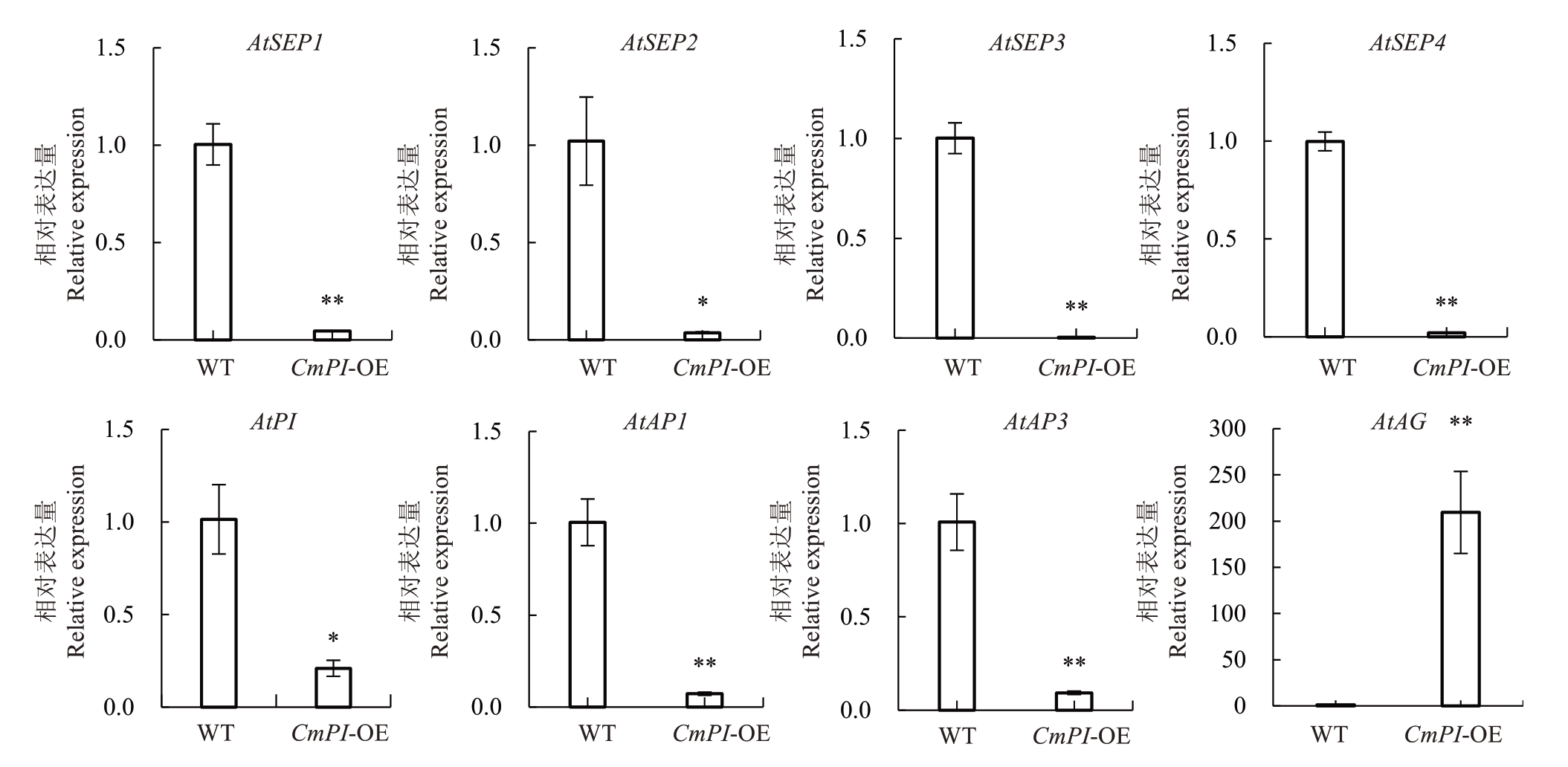
图7 CmPI 过表达对拟南芥花器官发育基因表达的影响
Fig.7 The effects of CmPI overexpression on the expression of flower development related genes in Arabidopsis thaliana
3 讨 论
MADS-box 基因调控花果发育的功能备受关注。PI 属于ABCDE 花发育模型中的B 类MADSbox 功能基因,在花发育过程中参与花瓣和雄蕊的发育[12]。笔者克隆的板栗花发育B 类基因CmPI 有高度保守的MADS域(MADS-box)和K 域(K-box),属MADS 的Ⅱ型亚家族转录因子[13-14]。板栗、白桦、向日葵、葡萄等8 个物种的PI 同源基因编码134~186 个氨基酸,相似度58.7%。该结果表明PI 在不同物种中蛋白结构高度保守,可能具有相似的生物学功能。CmPI定位于细胞核,符合其转录因子的定位特征。值得注意的是,本研究中的进化分析结果表明,板栗与白桦的PI 基因关系最近,这可能与两者均是柔荑花序木本植物有关。该结果暗示柔荑花序植物的PI 基因在进化中发生了区别于其他类型花序的变异。
基因的时空表达模式是其发挥生物学功能的关键因素之一。在笔者的研究中,CmPI主要在雄花中表达,茎尖、叶片、根等组织中几乎不表达,且CmPI在雄花中的表达量随雄花发育进程不断升高。这与Liu 等[7]对CmPI 在板栗不同组织中的表达分析结果一致。该结果暗示CmPI 是参与板栗雄花发育的关键基因。在拟南芥[15]、辣椒(Capsicum annuum)[16]、烟草[17]、摩洛哥坚果树(Argania spinosa)[7]、蒲公英(Taraxacum mongolicum)[18]等完全花植物中,PI 主要在第二轮和第三轮花器官中表达。但板栗花为不完全花,其花的第一轮和第二轮器官被合生苞片取代。雄花主要由苞片和雄蕊组成,雌花则由苞片和雌蕊组成。CmPI 在板栗苞片、雌蕊、雄蕊中的表达模式需要进一步研究,以揭示其时空表达模式在调控花器官发育中的作用。
为进一步验证CmPI的生物学功能,笔者将CmPI过表达载体转入野生型拟南芥,结果表明,CmPI 过表达转基因拟南芥的萼片发育为花瓣状组织。夏堇(Torenia fournieri)TfPI 基因在拟南芥中过表达导致萼片转变为花瓣组织[19]。将蝴蝶兰(Phalaenopsis aphrodite Rchb.)PI-like 基因PeMADS6 在拟南芥中过表达会导致萼片膨大、伸长,并在近轴侧沿边缘呈现花瓣状结构[20]。在麝香百合(Lilium longiflorum Thumb.)中将LMADS8/9 的异位表达可以挽救拟南芥pi-1 突变体第二轮花瓣的发育,并使部分萼片转化为花瓣状组织[21-22]。上述研究表明不同物种PI 基因具有保守的抑制萼片发育,而促进花瓣发育的生物学功能。De Craene[5]认为苞片与萼片和花瓣由同一器官进化而来,而苞片在板栗雌花和雄花中进一步进化为形态差异的器官。CmPI 在板栗雌花中的表达量显著低于雄花,暗示了其能够抑制苞片发育为壳斗。
PI通过与其他MADS-box蛋白形成二聚体来调控花器官发育[23-24]。AP1 可激活AP3 和PI[23]。PI 则直接与AP1 的启动子结合,进行负反馈调节[25]。在笔者的研究中,CmPI过表达拟南芥株系中A、B 和E三类基因的表达量均显著低于野生型拟南芥。这可能是因为CmPI 与拟南芥PI 具有相似的结构域,均能结合AP1 的启动子,抑制其转录激活[26]。而AP3和PI 的转录激活依赖于AP1,进而表达受到抑制。值得注意的是,CmPI 过表达拟南芥中C 类基因AG的表达水平显著高于野生型拟南芥。AG 基因主要调控雄蕊和心皮的发育,但本研究中转基因株系的雄蕊和心皮形态均未发生异常。花器官的发育调控网络复杂,是多基因与多因素共同作用的结果。板栗CmPI 在花发育中的生物学功能及其分子机制尚需进一步研究。
4 结 论
板栗与其他物种相同,均具有结构高度保守的B 类开花基因CmPI。CmPI 为核定位基因,主要在雄花中表达。CmPI 在野生型拟南芥中过表达可促进萼片转变为花瓣组织,可能是板栗苞片进一步发育的抑制基因。
[1] HU T Q,LIX H,DU L R,MANUELA D,XU M L.Leafy and apetala1 down-regulate zinc finger protein 1 and 8 to release their repression on class B and C floral homeotic genes[J].Proceedings of the National Academy of Sciences of the United States of America,2023,120(22):e2221181120.
[2] 马小杰,闫先喜,张宪省.高等植物花发育的基因调控[J].山东农业大学学报,1995,26(2):263-272.MA Xiaojie,YAN Xianxi,ZHANG Xiansheng.Gene-regulated flower development in higher plants[J].Journal of Shandong Agricultural University,1995,26(2):263-272.
[3] 冯献忠,杨素欣,郭蔼光.高等植物花发育的研究进展[J].西北农业大学学报,1998,26(3):94-99.FENG Xianzhong,YANG Suxin,GUO Aiguang.Progress of flower development in higher plants[J].Acta Universitatis Agriculturalis Boreali Occidentalis,1998,26(3):94-99.
[4] PELAZ S,DITTA G S,BAUMANN E,WISMAN E,YANOFSKY M F.B and C floral organ identity functions require SE‐PALLATA MADS-box genes[J].Nature,2000,405(6783):200-203.
[5] DE CRAENE L P R.Are petals sterile stamens or bracts?The origin and evolution of petals in the core eudicots[J].Annals of Botany,2007,100(3):621-630.
[6] ZHAO Y H,ZHANG X M,LID Z.Development of the petaloid bracts of a paleoherb species,Saururus chinensis[J].PLoS One,2021,16(9):e0255679.
[7] LIU Y,CHEN G S,GAO Y R,FANG K F,ZHANG Q,CAO Q Q,QIN L,XING Y,SU S C.Identification and characterization of MADS-box genes involved in floral organ development in Chinese chestnut (Castanea mollissima Blume)[J].Horticultural Science and Technology,2021,39(4):482-496.
[8] 程运河,程丽莉,胡广隆,杨前宇,韩笑,兰彦平.板栗CmFT基因的克隆及功能分析[J].果树学报,2022,39(6):935-944.CHENG Yunhe,CHENG Lili,HU Guanglong,YANG Qianyu,HAN Xiao,LAN Yanping.Cloning and functional analysis of CmFT in Chinese chestnut[J].Journal of Fruit Science,2022,39(6):935-944.
[9] 杨国栋.棉花耐盐基因GhNHX1 启动子的克隆及功能分析[D].泰安:山东农业大学,2007.YANG Guodong.Isolation and characterization of a salt inducible GhNHX1 promoter from Gossypium hirsutum[D].Tai’an:Shandong Agricultural University,2007.
[10] CLOUGH S J,BENTAF.Floral dip:Asimplified method forAgro‐bacterium-mediated transformation of Arabidopsis thaliana[J].The Plant Journal,1998,16(6):735-743.
[11] SMYTH D R,BOWMAN J L,MEYEROWITZ E M.Early flower development in Arabidopsis[J].The Plant Cell,1990,2(8):755-767.
[12] 徐启江,丁一,黄赫,王勇,崔成日,梁毅.洋葱花发育B 类MADS-box 基因AcDEF 的克隆与表达分析[J].园艺学报,2013,40(6):1090-1100.XU Qijiang,DING Yi,HUANG He,WANG Yong,CUIChengri,LIANG Yi.Cloning and expression analysis of B class MADSbox gene AcDEF associated with floral development in onion[J].Acta Horticulturae Sinica,2013,40(6):1090-1100.
[13] KIM S,YOO M J,ALBERT V A,FARRIS J S,SOLTIS P S,SOLTIS D E.Phylogeny and diversification of B- function MADS-box genes in angiosperms:Evolutionary and functional implications of a 260-million-year-old duplication[J].American Journal of Botany,2004,91(12):2102-2118.
[14] 陈磊,翟笑雨,徐启江.B 类MADS-Box 基因的系统发育与功能演化[J].植物生理学报,2015,51(9):1359-1372.CHEN Lei,ZHAIXiaoyu,XU Qijiang.The phylogeny and functional evolution of B-class MADS-Box genes[J].Plant Physiology Journal,2015,51(9):1359-1372.
[15] GOTO K,MEYEROWITZ E M.Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA[J].Genes & Development,1994,8(13):1548-1560.
[16] 廖铭宇,肖佳林,李丽缘,宋钰,黄湖荣,杨博智.辣椒雄性不育系和保持系CaMADS6 基因克隆及表达分析[J].华北农学报,2023,38(1):46-52.LIAO Mingyu,XIAO Jialin,LILiyuan,SONG Yu,HUANG Hurong,YANG Bozhi.Cloning and expression analysis of Ca‐MADS6 in cytoplasmic male sterile line and maintainer line of pepper[J].Acta Agriculturae Boreali-Sinica,2023,38(1):46-52.
[17] HANSEN G,ESTRUCH J J,SOMMER H,SPENA A. NTGLO:A tobacco homologue of the GLOBOSA floral homeotic gene of Antirrhinum majus:cDNA sequence and expression pattern[J].Molecular&General Genetics,1993,239(1/2):310-312.
[18] VIJVERBERG K,WELTEN M,KRAAIJ M,VAN HEUVEN B J,SMETS E,GRAVENDEEL B.Sepal identity of the pappus and floral organ development in the common dandelion(Taraxa‐cum officinale;Asteraceae)[J].Plants,2021,10(8):1682.
[19] SASAKIK,YAMAGUCHIH,NAKAYAMA M,AIDA R,OHTSUBO N.Co-modification of class B genes TfDEF and TfGLO in Torenia fournieri Lind.alters both flower morphology and inflorescence architecture[J].Plant Molecular Biology,2014,86(3):319-334.
[20] TSAIW C,LEE P F,CHEN H I,HSIAO Y Y,WEIW J,PAN Z J,CHUANG M H,KUOH C S,CHEN W H,CHEN H H.PeMADS6,a GLOBOSA/PISTILLATA-like gene in Phalaenop‐sis equestris involved in petaloid formation,and correlated with flower longevity and ovary development[J].Plant & Cell Physiology,2005,46(7):1125-1139.
[21] CHEN M K,HSIEH W P,YANG C H.Functional analysis reveals the possible role of the C-terminal sequences and PImotif in the function of lily (Lilium longiflorum)PISTILLATA(PI)orthologues[J].Journal of Experimental Botany,2012,63(2):941-961.
[22] MAO W T,HSU H F,HSU W H,LIJ Y,LEE Y I,YANG C H.The C-terminal sequence and PImotif of the orchid (Oncidium Gower Ramsey) PISTILLATA(PI) ortholog determine its ability to bind AP3 orthologs and enter the nucleus to regulate downstream genes controlling petal and stamen formation[J].Plant &Cell Physiology,2015,56(11):2079-2099.
[23] NG M,YANOFSKY M F.Activation of the Arabidopsis B class homeotic genes by APETALA1[J].The Plant Cell,2001,13(4):739-753.
[24] 杜朝金,张汉尧,罗心平,宋云连,毕珏,王跃全,张惠云.基因调控植物花器官发育的研究进展[J].植物遗传资源学报,2024,25(2):151-161.DU Chaojin,ZHANG Hanyao,LUO Xinping,SONG Yunlian,BIJue,WANG Yuequan,ZHANG Huiyun.Progress in gene regulation of plant floral organ development[J].Journal of Plant Genetic Resources,2024,25(2):151-161.
[25] SUNDSTRÖM J F,NAKAYAMA N,GLIMELIUS K,IRISH V F.Direct regulation of the floral homeotic APETALA1 gene by APETALA3 and PISTILLATA in Arabidopsis[J].The Plant Journal,2006,46(4):593-600.
[26] WANG S,HUANG H J,HAN R,LIU C Y,QIU Z N,LIU G F,CHEN S,JIANG J.Negative feedback loop between BpAP1 and BpPI/BpDEF heterodimer in Betula platyphylla × B.pendula[J].Plant Science,2019,289:110280.