石榴(Punica granatum L.)属于千屈菜科(Lythraceae)石榴属(Punica)植物,在中国有2000 余年栽培历史,味好、色美、富含营养和生物活性物质,备受消费者喜爱。石榴产业已发展为中国云南、四川和河南等地脱贫攻坚和乡村振兴的支柱产业[1-3]。颜色是影响石榴果实品质和观赏价值的重要性状[4]。花色苷是一种广泛存在于植物体中的天然水溶性色素,决定了石榴叶片、花器官[5]、果实及果汁色泽呈白色到深红色甚至紫黑色等颜色特征[6-8];同时花色苷也是一类被熟知的抗氧化活性物质,其含量是石榴果实品质及工业加工价值的重要指示性状[5,9],因此,花色苷合成和颜色形成研究对石榴品种改良和新品种培育意义重大。
石榴花色苷主要有飞燕草-3-葡萄糖苷、飞燕草-3,5-葡萄糖苷和矢车菊-3-葡萄糖苷等[10-11],其形成与积累因组织器官、品种和地源等不同存在一定变异。总体上,泰山红[5,7-8]、突尼斯软籽[1,8]和墨石榴[7]等红色、深红色系石榴品种果实花色苷含量随发育成熟期呈上升趋势,相比之下,三白和泰山三白甜等浅或白色系石榴品种果实花色苷含量较低,而水晶甜果皮中不含花青苷[12-15]。榴花红色石榴盛花期花瓣颜色较榴花粉色和榴花白色红艳,其花色苷含量及其合成相关酶类活性均高于后者花瓣[16]。突尼斯软籽石榴是中国流行栽培、品质优良的软籽石榴品种,其果实色泽、花色苷含量在云贵高原产区均优于河南等北方产区[6,17-18]。豫大籽和突尼斯软籽石榴杂种后代幼叶颜色表现绿色、黄绿、红和深红性状分离,这与其花色苷、叶绿素含量及其比值有关[19-20]。这些研究基础为石榴色泽形成的分子机制研究提供了重要支撑。
植物花色苷通常经苯丙氨酸解氨酶(PAL)、查尔酮合成酶(CHS)、二氢黄酮醇3’-羟化酶(F3’H)、二氢黄酮醇4-还原酶(DFR)和花色素苷合酶(ANS)等一系列苯丙烷代谢途径相关酶作用,再由糖基转移酶(UGT)等催化与不同单糖结合形成,该生物过程主要受MYB、bHLH、WD40 等转录因子调控[5,21]。随着石榴基因组信息的公布,相继克隆了PgMYB[11]、PgbZIP[11]、PgDFR[14]、PgANS[14]、PgUGT[22]和PgCHS 等与石榴花色苷合成及红色形成相关的基因,并分析了其序列与表达情况[15,23-25]。矢车菊素是石榴中主要的花色苷种类之一,F3’H 是合成矢车菊素的重要限速酶。Yuan 等[26]及Harel-beja 等[15]通过多组学分析发现石榴果皮着色过程中PgF3’H 基因表达上调,但人们对石榴PgF3’H 基因家族成员的数目、大小、结构和系统发育等基本信息尚不了解,它们在石榴籽粒着色过程中的表达情况及其功能并不清楚。因此,笔者在本研究中对石榴PgF3’H 基因家族成员进行了全基因组鉴定和分析,介绍了其长度、大小、结构、系统发育,分析了其顺式作用元件情况,并通过已有转录组数据分析了石榴PgF3’H基因家族成员在石榴籽粒着色中的可能作用。
F3’H 属于P450 细胞色素单加氧酶,主要负责在类黄酮分子B 环上C 位基团上加第二个羟基[15,27-28]。Nitarska等[29]通过DNA基因编辑技术获得一品红(Euphorbia pulcherrima Willd. et Kl.)Christmas EveEpF3’H敲除株系,其矢车菊素含量下降,红色变浅,验证了该基因在矢车菊花色苷合成中的重要作用。在葡萄(Vitis vinifera L.)中鉴定出1 个VvF3’H基因,VvF3’H1表达量在红色浆果中持续显著高于绿色浆果[28,30]。在苹果(Malus domestica)中鉴定出2个MdF3’H基因[31]。草莓(Fragaria×ananassa Duch.)FaF3’H 受FaMYB5 正调控参与果实红色色泽形成[32]。由此可见,F3’H广泛参与了植物花、种皮和果实着色。然而,截止到目前石榴PgF3’H基因家族成员调控花色苷合成的功能并不明确,因此,笔者在本研究中进一步克隆了石榴PgF3’H 基因家族成员中PgF3’H3 基因,并初步验证了其调控花色苷合成的功能。研究结果有助于加深对石榴色泽形成分子机制的理解,也为石榴品种色泽改良提供了重要理论依据。
1 材料和方法
1.1 试验材料
采集河南省郑州市毛庄果园的突尼斯软籽自交F1代深红色株系叶片,用于克隆PgF3’H3 基因。用于基因功能验证的拟南芥(Arabidopsis thaliana,ecotype Columbia)分别为野生型植株(WT)和PgF3’H3过表达植株(OE),均种植于实验室植物培养间,培养条件为温度26 ℃、16 h/8 h 光照/黑暗交替、湿度75%~85%。采集健康、发育相对一致的种植于河南省荥阳市的石榴品种突尼斯软籽(TS,籽粒呈浅红色)和美丽酸(MLS,籽粒呈深红色)的坐果期、幼果期、转色期、膨大期和成熟期果实籽粒,以及种植于四川省攀枝花市石榴品种紫美(ZM,籽粒呈紫红色)幼果期、转色期和成熟期果实籽粒(图1),用于PgF3’H基因家族成员表达分析。

图1 不同发育期石榴果实及籽粒表型
Fig.1 Fruit and aril performances of pomegranate at different development stages
1.2 试验方法
1.2.1 石榴PgF3’H基因家族成员鉴定 从NCBI数据库(https://www.ncbi.nlm.nih.gov/assembly/GCF_007655135.1)中检索并下载突尼斯软籽石榴基因组数据(PRJNA355913)。从Ensemble Plants 数据库(http://plants.ensembl.org/index.html)获得水稻(Oryzasativa)OsF3’H 序列,通过BLASTP 工具进行双向blast 比对筛选获得PgF3’H 候选基因。利用NCBI CDD 数据库(Conserved Domain Database;http://www.ncbi.nlm.nih.gov//Structure/bwrpsb/bwrpsb.cgi)对候选PgF3’H基因进行保守结构域分析,最终确定石榴PgF3’H 基因家族的成员。利用石榴基因组注释文件(https://www.ncbi.nlm.nih.gov/)分析PgF3’H基因家族成员在染色体上的位置,并依次对各成员进行重命名,利用TBtools工具进行可视化作图。使用TBtools工具分析石榴PgF3’H基因家族编码蛋白的理化性质,包括分子质量、等电点、氨基酸数量、不稳定系数和亲水指数。利用Cell-PLoc 2.0(http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/)对PgF3’H基因家族编码蛋白亚细胞定位进行预测分析。利用SOPMA(https://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html)和SWISS-MODEL(https://swissmodel.expasy.org/)对PgF3’H 基因家族成员编码蛋白质的二级结构和三级结构进行预测。
1.2.2 石榴PgF3’H 基因的系统发育、分类、保守基序和基因结构分析 使用MEGA11 软件,根据最大似然方法(maximum likelihood,ML),Bootstrap 检验重复次数为1000 次,其他参数设定为默认,对拟南芥、水稻等单子叶植物和葡萄、苹果等双子叶植物中共47个F3’H 全长蛋白序列构建系统发育树。利用MEME(https://meme-suite.org/meme/tools/meme)在线数据库分析PgF3’H 蛋白保守基序,使用TBtools将外显子-内含子结构进行可视化。
1.2.3 石榴PgF3’H 基因启动子顺式作用元件分析 使用TBtools 提取石榴PgF3’H 基因上游2000 bp的启动子区序列信息;使用PlantCARE(http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)在线工具预测分析PgF3’H 基因启动子区顺式元件,然后使用TBtools工具将结果可视化。
1.2.4 石榴PgF3’H 家族基因表达分析 提取不同发育期突尼斯软籽、美丽酸和紫美石榴果实籽粒RNA,设置3次重复,转录组测序委托武汉迈特维尔生物科技有限公司完成,从转录组数据中提取PgF3’H 基因家族成员在不同品种不同发育期籽粒样品中的表达信息,使用TBtools生成热图。
1.2.5 石榴PgF3’H3 基因克隆及序列分析 利用RNA 提取试剂盒(上海生工B518631-0100)提取突尼斯软籽石榴自交F1 代深红色株系叶片总RNA,利用RNA 反转录试剂盒(南京诺唯赞R233-01)获得cDNA。参考石榴基因组PRJNA355913,使用Primer5.0 设计PgF3’H 基因引物(F:ATGGACTCTCTCCCCTTCTTCCTGGC;R:TCAATAAACATGTCGCGGCAGCCTC),从上述cDNA 中克隆PgF3’H3 基因。利用DNAMAN 软件和NCBIBlast 对PgF3’H3 和草莓(Fragaria × ananassa)、樱桃(Prunus avium)、苹果(Malus domestica)和葡萄(Vitis vinifera)F3’H 进行多序列比对分析,使用MEGA11.0软件构建系统发育树。
1.2.6 石榴PgF3’H3基因功能验证 构建PgF3’H3基因过表达载体pSAK277:PgF3’H3,将重组质粒pSAK277:PgF3’H3 转化入农杆菌EHA105 中。然后,利用农杆菌侵染转化方法[11]转化野生型-哥伦比亚拟南芥(WT),经50 mg·L-1 Kan 筛选获得转化株系,提取转化株系和WT株系叶片总DNA,通过PCR鉴定后获得稳定遗传的拟南芥过表达PgF3’H3株系(OE)。参考万然等[20]和胡悦[33]的方法,采用pH示差法检测OE 和WT(14 d)子叶中花色苷含量,采用石油醚方法检测其类胡萝卜素含量,采用分光光度计法检测其叶绿素含量;每个样品重复检测3次。
1.2.7 数据处理及分析 用Microsoft Excel 2016对数据进行整理,用SPSS 19.0软件对数据进行方差分析和多重比较(Duncan’s test,显著水平p=0.05)。
2 结果与分析
2.1 石榴PgF3’H基因家族成员鉴定
从突尼斯软籽石榴基因组中共鉴别到3 个PgF3’H 基因家族成员,根据它们在染色体上的位置,按照顺序依次将其重命名为PgF3’H1、PgF3’H2、PgF3’H3,它们分别位于LG0、LG0 和LG6 上,序列长度分别为2055、3933、2606 bp(表1)。这3 个PgF3’H 基因编码蛋白所含氨基酸数目分别为520、580 和508 个,其相对分子质量分别为58.3、66.2 和56.4 kDa,它们均为碱性、亲水性蛋白,另外,PgF3’H1、PgF3’H2 蛋白质稳定性较PgF3’H3更好。经亚细胞定位预测分析,发现PgF3’H1、PgF3’H2和PgF3’H3蛋白均可能定位于内质网发挥作用(表1)。蛋白结构预测分析表明,PgF3’H蛋白以α-螺旋和无规则卷曲为主,占比80%以上,其次为延伸链和β-折叠(表2,图2)。以上这些结果为进一步研究PgF3’H蛋白的纯化、活性和功能提供了理论依据。
表1 PgF3’H 基因家族成员信息
Table 1 The information of PgF3’H gene family members
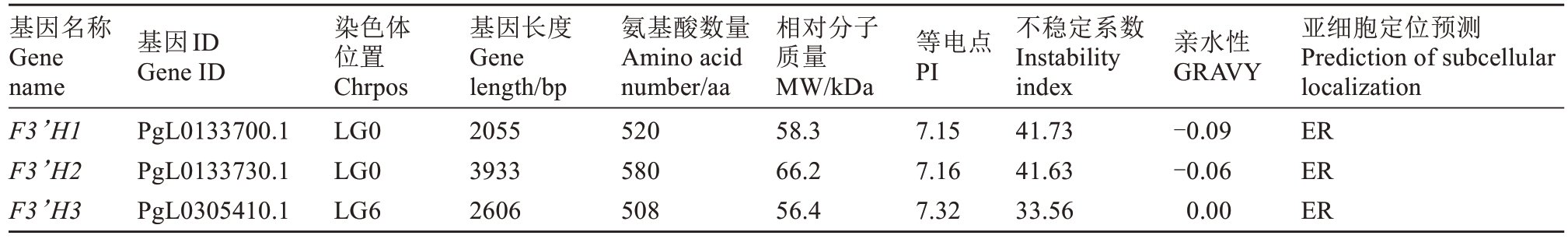
注:ER,内质网。
Note:ER,Endoplasmic reticulumn.
基因名称Gene name F3’H1 F3’H2 F3’H3亚细胞定位预测Prediction of subcellular localization ER ER ER基因ID Gene ID等电点PI亲水性GRAVY PgL0133700.1 PgL0133730.1 PgL0305410.1染色体位置Chrpos LG0 LG0 LG6基因长度Gene length/bp 2055 3933 2606氨基酸数量Amino acid number/aa 520 580 508相对分子质量MW/kDa 58.3 66.2 56.4 7.15 7.16 7.32不稳定系数Instability index 41.73 41.63 33.56-0.09-0.06 0.00
表2 PgF3’H 蛋白的二级结构预测
Table 2 Predicition of secondary structure of PgF3’Hs
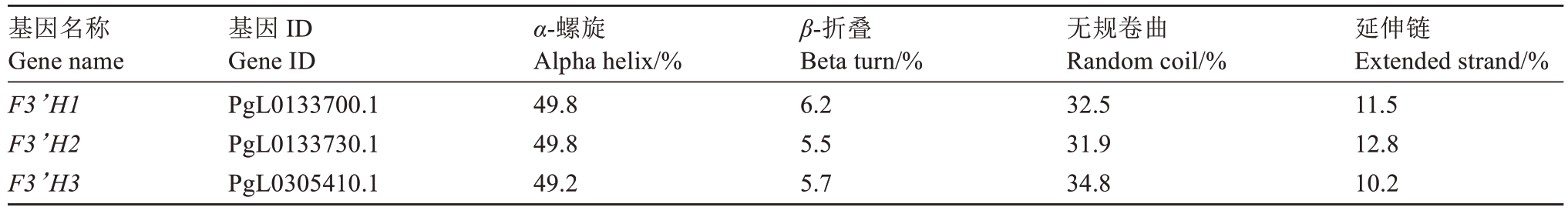
基因名称Gene name F3’H1 F3’H2 F3’H3基因ID Gene ID PgL0133700.1 PgL0133730.1 PgL0305410.1 α-螺旋Alpha helix/%49.8 49.8 49.2 β-折叠Beta turn/%6.2 5.5 5.7无规卷曲Random coil/%32.5 31.9 34.8延伸链Extended strand/%11.5 12.8 10.2
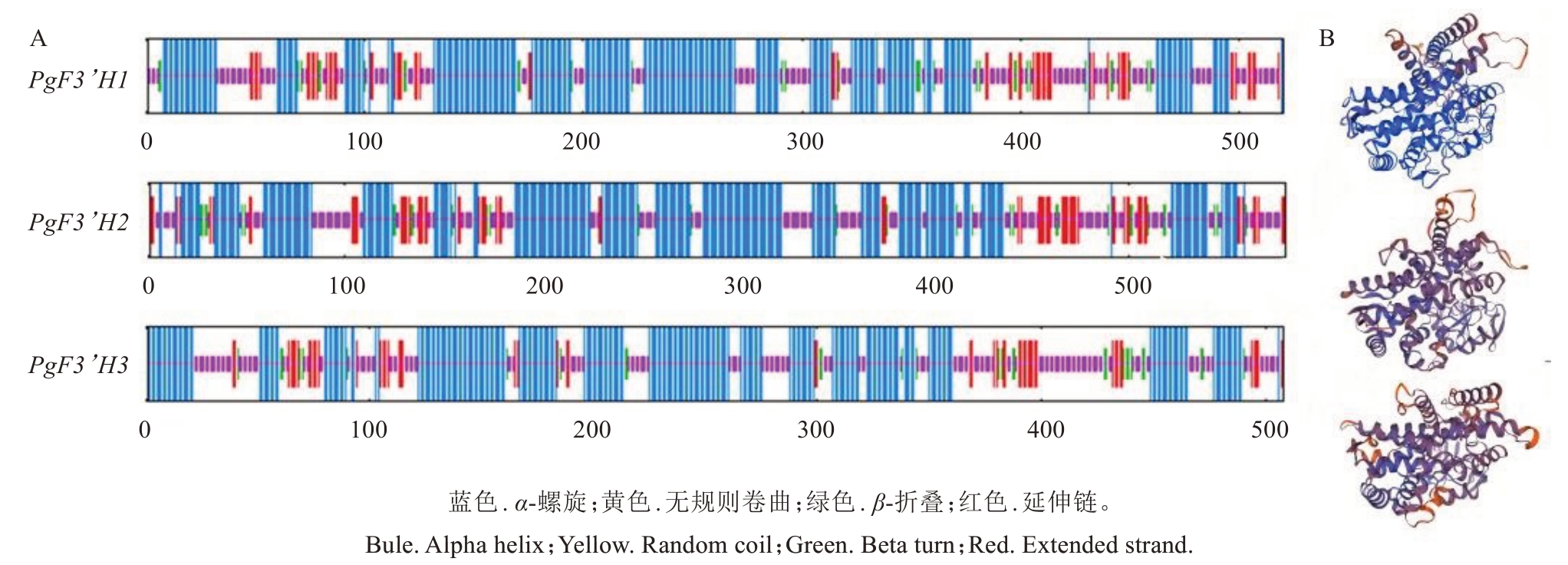
图2 PgF3’Hs 二级结构(A)与三级结构(B)
Fig.2 Secondary(A)and tertiary(B)structures of PgF3’Hs
2.2 石榴PgF3’H基因家族成员系统发育分析
为了揭示石榴PgF3’Hs与其他植物F3’Hs的进化关系,将节节麦和水稻2 种单子叶植物以及拟南芥、葡萄、苹果、桃等31 种双子叶植物共47 个F3’H共同用于构建系统发育树。系统发育分析表明,GmF3’H 蛋白位于系统进化树末端单独一分支上,形成单独的一个亚族,即Group Ⅴ;剩余39 个植物F3’H 蛋白被划分为4 个亚族,即Group Ⅰ、Ⅱ、Ⅲ、Ⅳ(图3)。由进化树可推知,在植物F3’H 蛋白进化树末端分支上,存在11 对同源基因,其中10 对属于直系同源基因,5 对属于旁系同源基因;旁系同源基因包括OsF3’H4/OsF3’H5、AtaF3’H2/AtaF3’H3、PgF3’H1/PgF3’H2、OsF3’H2/OsF3’H3和IbF3’H1/OsF3’H2 等,它们起源于物种特异的基因复制扩增;直系同源基因包括OsF3’H1/AtaF3’H1、TcF3’H/GbF3’H、VaF3’H/VvF3’H、AtF3’H/PgF3’H3、MdF3’H2/PpyF3’H1 和IbF3’H1/OsF3’H2 等等,它们起源于物种分化事件(图3)。此外,F3’H 基因数量在两个及以上的物种有水稻、石榴、苹果、番薯和节节麦等7个,其中只有水稻和石榴F3’H 基因聚类到不同的亚族,PgF3’H1/PgF3’H2 与FoF3’H、OsF3’H2/OsF3’H3同源,聚到Group Ⅲ,而PgF3’H3与AtF3’H 同源,聚到Group Ⅱ;其余同一物种内的F3’H基因均在同一亚族(图3)。
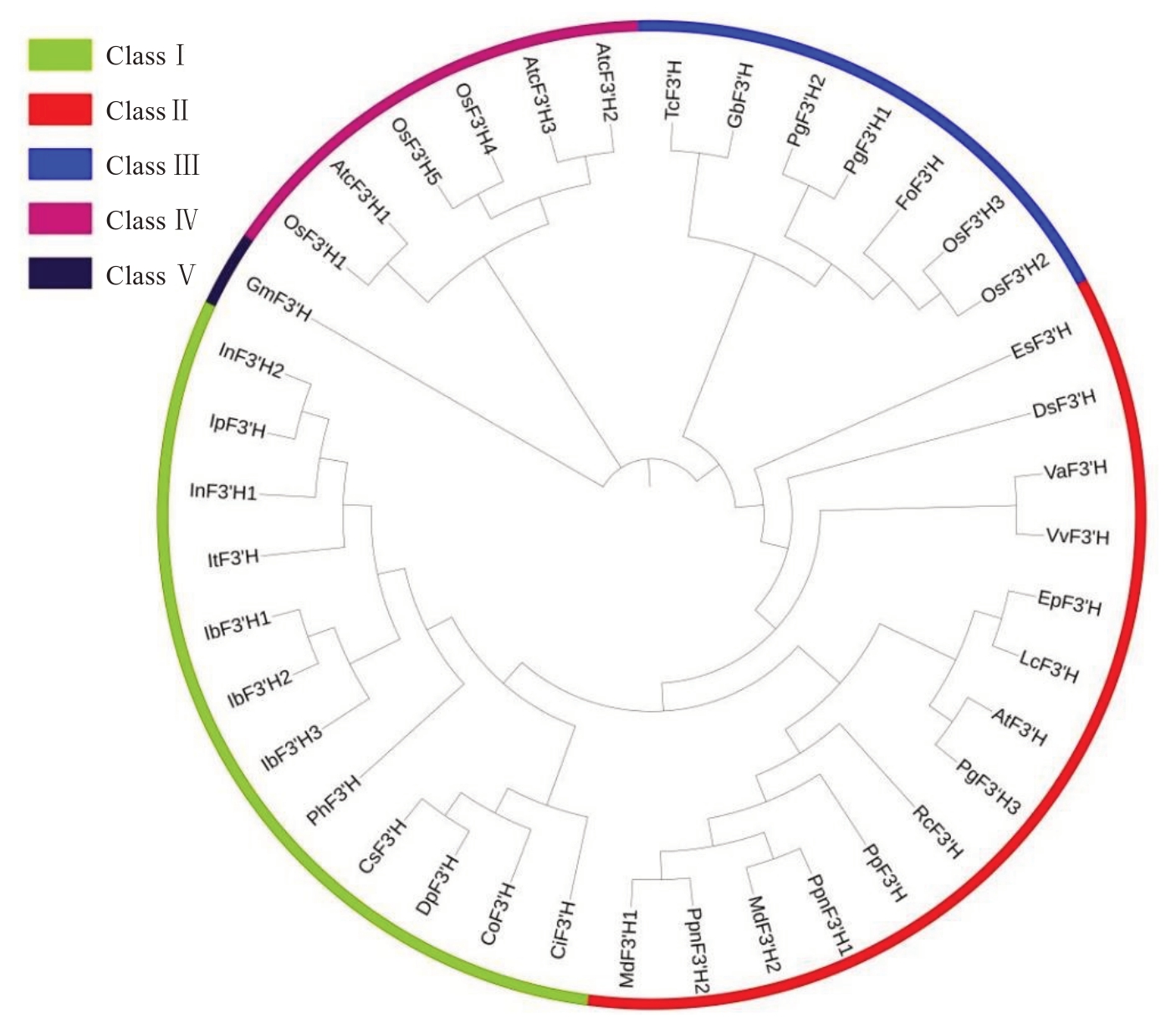
图3 植物F3’H 基因家族的系统发育分析
Fig.3 Phylogenetic analyseof plant F3’H family members
2.3 PgF3’H基因的保守基序和基因结构分析
利用MEME在线工具对PgF3’H基因的保守基序(motif)及保守结构域进行了分析(图4)。结果表明,PgF3’H 基因结构保守,除了PgF3’H3 没有motif 7和motif 8以外,PgF3’H1、PgF3’H2和PgF3’H3基因均含有motif 1~10,一般一个基因中一种motif都只有1 个;它们从5’端到3’端均依次分布motif 10、motif 5、motif 2、motif 7(除PgF3’H3)、motif 6、motif 8(除PgF3’H3)、motif 4、motif 1、motif 3和motif 9;根据基序编码氨基酸序列显示出3 个PgF3’H蛋白间的保守性与变异性,每个堆叠高度越高代表此处序列越高保守,每个氨基酸符号的高度代表它在此位点出现的频率(图4-A、C)。此外,PgF3’H基因均含有典型的CYP75B 结构域,并具有4~5 个内含子(图4-B)。这些关于基因结构分析的结果表明了PgF3’H基因的保守性。
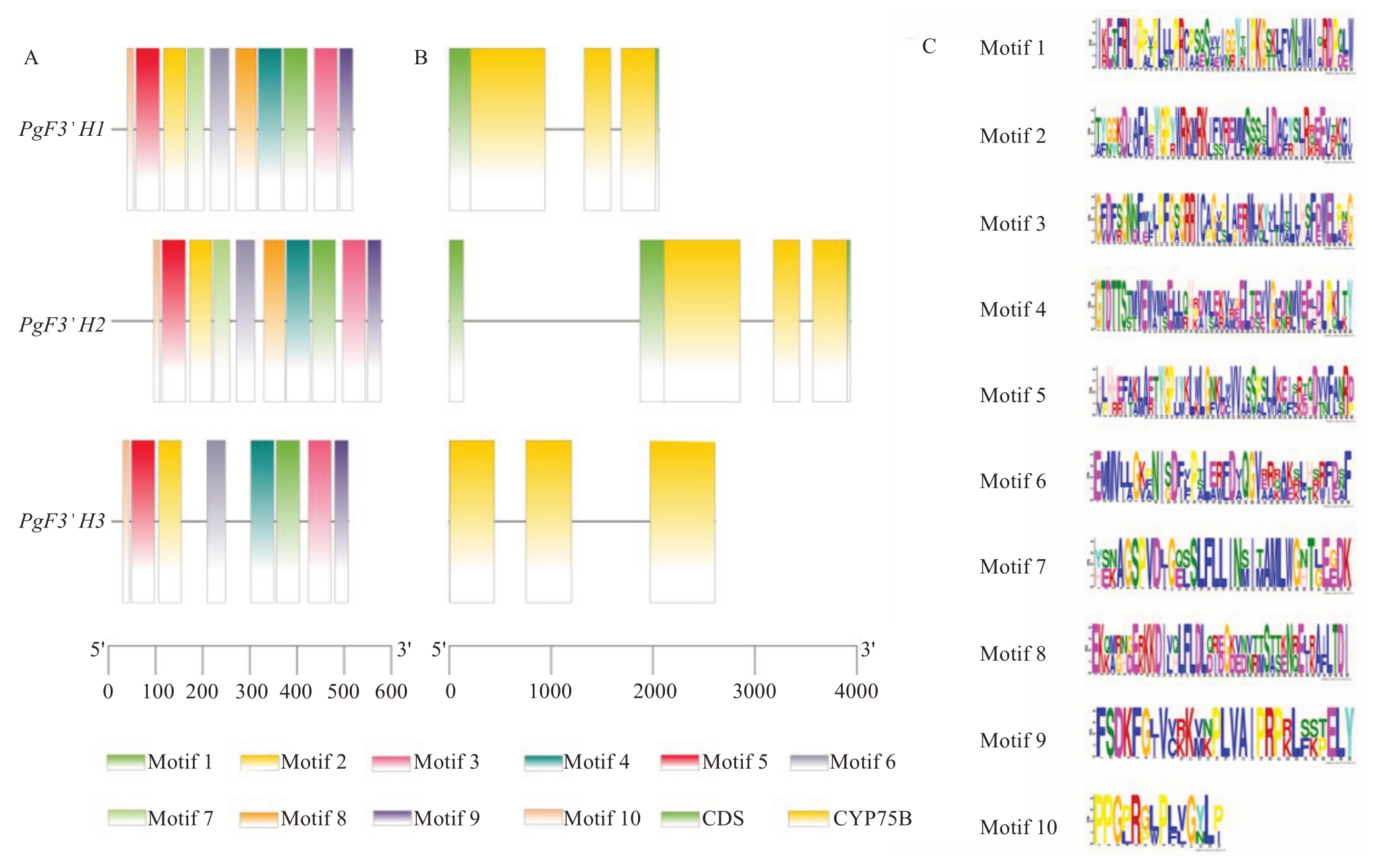
图4 PgF3’H 基因保守基序(A)、基因结构(B)和motif 序列信息(C)
Fig.4 Conserved motifs(A),gene structures(B)and motif sequences(C)of PgF3’H genes
2.4 石榴PgF3’H基因启动子顺式作用元件(cis-elements)分析
花色苷合成是植物生长发育和抗逆反应的重要生物学过程。通过顺式作用元件分析,发现PgF3’H基因家族成员的启动子区均分布有生长发育相关元件、光响应元件、MYB相关元件、激素响应元件和应激反应元件(图5)。PgF3’H1基因启动子区含有16个不同的顺式作用元件,其中有6种激素响应元件各1个以及1 个MYB 转录因子结合位点CCAAT-box。PgF3’H2基因启动子区具有最多的顺式作用元件,共29 个,分别包括9 个生长发育相关元件(其中有4 个TGACG-motif和4个CGTCA-motif);8个光响应元件(其中有2个G-box和2个TCT-box);9个激素响应元件[其中有3个ABRE(脱落酸ABA)和4个TGA-element(生长素AUX)];以及1个MYB转录因子结合位点(MBS)和2 个应激反应元件(图5)。PgF3’H3 基因启动子区分别含有9个光响应元件(其中有5个Gbox);8 个激素响应元件(其中有6 个ABRE);4 个生长发育相关元件(其中2 个TGACG-motif 和2 个CGTCA-motif);1个MYB转录因子结合位点(MBS)和2个应激反应元件(ARE)(图5)。
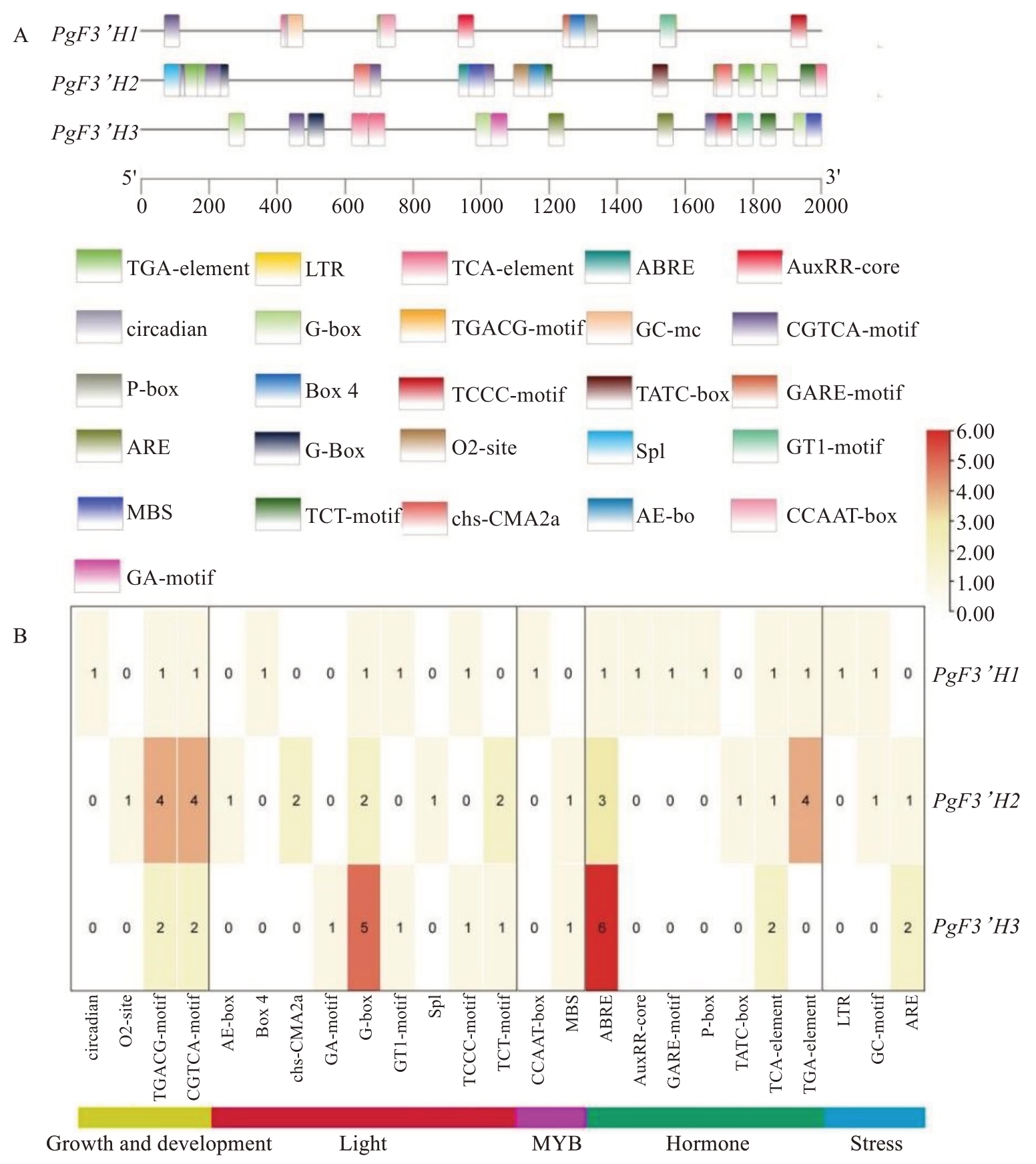
图5 PgF3’H 基因启动子区顺式作用元件
Fig.5 Cis-elements in promoters of PgF3’H genes
2.5 PgF3’H 基因在不同颜色石榴籽粒着色过程中的表达分析
为了进一步了解PgF3’H 基因在石榴果实籽粒着色及花色苷积累中的作用,从已有的突尼斯软籽、美丽酸和紫美在不同发育期的籽粒样品转录组数据中,获得PgF3’H1、PgF3’H2和PgF3’H3的表达情况(图6)。结果表明,在这3 个石榴品种中,PgF3’H1基因的表达量始终处于较低水平,说明该基因可能与石榴籽粒着色关系不大;相比而言,PgF3’H2 和PgF3’H3 具有更高的表达水平;此外,随果实发育和籽粒着色,PgF3’H2 和PgF3’H3 均在不同石榴品种中表现出相似的表达趋势,这说明它们与石榴籽粒着色有重要关系。

图6 PgF3’Hs 在不同品种石榴籽粒中的表达量
Fig.6 Expression levels of PgF3’H genes in pomegranate arils of different shades of red color at the indicated fruit development stages
PgF3’H2 在这3 个石榴品种籽粒中均呈显著上调表达趋势,其表达水平在幼果期之前极低且无显著变化;此后开始快速大幅上升,至转色期分别显著增加了11 616.68%、9 381.51%和10 942.06%;然后,其表达水平继续显著上调表达,至成熟期又分别增加了254.98%、602.27%和198.76%(图6)。PgF3’H3 在坐果期突尼斯软籽、美丽酸和紫美籽粒中即已转录表达,至转色期其表达水平分别提高了271.41% 38.70%和793.05%;不过,PgF3’H3 在突尼斯软籽二次膨大期籽粒中未继续上调表达,到成熟期反而下降了38.84%;在美丽酸中却持续小幅上调表达,至成熟期其表达水平又提高了48.76%;另外,PgF3’H3 在紫美籽粒中至转色期其表达水平提高了793.05%,至成熟期又下降了38.84%。此外,至成熟期时,在这3个石榴品种籽粒中,PgF3’H2 的表达量均显著高于PgF3’H3。比较分析以上研究结果说明,PgF3’H3 可能主要作用于石榴籽粒早期花色苷合成和颜色形成,而PgF3’H2 可能是石榴成熟籽粒花色苷合成和呈色的主要作用基因。
38.70%和793.05%;不过,PgF3’H3 在突尼斯软籽二次膨大期籽粒中未继续上调表达,到成熟期反而下降了38.84%;在美丽酸中却持续小幅上调表达,至成熟期其表达水平又提高了48.76%;另外,PgF3’H3 在紫美籽粒中至转色期其表达水平提高了793.05%,至成熟期又下降了38.84%。此外,至成熟期时,在这3个石榴品种籽粒中,PgF3’H2 的表达量均显著高于PgF3’H3。比较分析以上研究结果说明,PgF3’H3 可能主要作用于石榴籽粒早期花色苷合成和颜色形成,而PgF3’H2 可能是石榴成熟籽粒花色苷合成和呈色的主要作用基因。
2.6 PgF3’H3基因克隆及功能验证
笔者课题组前期研究已发现,突尼斯软籽石榴自交1代植株幼叶红色深浅与其花色苷含量呈正相关[20],PgF3’H3 表达量随着幼叶颜色由红转绿而逐渐下降[20]。故PgF3’H3 可能与石榴器官发育前期着色有关。因此克隆了PgF3’H3基因,并对PgF3’H3、VvF3’H、FaF3’H、CaF3’H和MdF3’H等不同园艺作物中F3’H蛋白进行了多序列比对分析,结果发现PgF3’H3 与VvF3’H和MdF3’H相似度较高(75%左右)(图7-A)。

图7 PgF3’H3 序列分析及其功能研究
Fig. 7 Sequence analysis and function of PgF3’H3
为了探明PgF3’H3 在调控石榴花色苷合成方面的功能,通过农杆菌介导的遗传转化技术,成功获得PgF3’H3 过表达拟南芥植株,该基因被稳定整合到拟南芥基因组中,表达量显著高于野生型(图7-B~D)。根据表型观察,发现野生型植株幼苗子叶呈现绿色,PgF3’H3 过表达拟南芥植株幼苗子叶呈现红色(图7-B)。通过检测花色苷、类胡萝卜素和叶绿素含量,发现PgF3’H3 过表达拟南芥子叶中花色苷含量为0.308 3 mg·g-1,然而在野生型子叶中未检测到花色苷,此外,其类胡萝卜素和叶绿素含量显著低于野生型(图7-E)。
3 讨 论
“榴枝婀娜榴实繁,榴膜轻明榴子鲜”,石榴兼具营养、保健、观赏、生态、文化及文明等多重价值,备受国内外关注和喜爱。石榴花色苷是其色泽性状构成的重要物质基础,也是其果实营养保健价值的重要物质来源[4,19]。国内外学者围绕石榴不同品种[16]、发育期[1]、组织及地源间花色苷种类、含量情况进行了诸多研究[5,19],尽管如此,石榴花色苷合成相关基因的研究仍有待加强。F3’H 参与决定了植物叶色[30]、花色[29]、种皮和果实着色等性状[34-35]。因此,笔者在本研究中比较详细地介绍了石榴PgF3’H 基因家族成员的序列、大小、编码蛋白结构预测、基因结构和顺式作用元件等信息,并分析了PgF3’H 基因家族成员在石榴籽粒花色苷合成中的作用,为石榴色泽形成研究和品种改良提供科学依据。
根据已有报道,水稻(5 个)和节节麦(3 个)等单子叶植物以及拟南芥(1 个)、欧洲葡萄(1 个)[31]和苹果(2 个)[32]等双子叶植物中F3’H 基因数量均较少,也有研究从12种单子叶植物中共鉴定了44个F3’H基因[34]。类似地,笔者在本研究中根据突尼斯软籽石榴基因组信息,共鉴定到3 个PgF3’H 基因,它们均具有典型的CYP75B结构域,它是决定F3’H控制植物器官花色苷合成和呈色的功能结构域[36]。进而对来自不同物种的47 个PgF3’H 构建了系统发育树,结果发现PgF3’H3 和AtF3’H 具有较近的亲缘关系,属于Group Ⅱ,而PgF3’H1和PgF3’H2同源,与OsF3’H2/3具有较近的亲缘关系,属于Group Ⅲ,这说明它们可能具有相似的功能。研究结果还证明了PgF3’H1 和PgF3’H2 具有比较保守的基序和结构域(CYP75B),并且这些保守基序的排列顺序也相当保守。这与已有研究发现“不同单子叶和双子叶植物中F3’H 基因保守基序的排列顺序及其CYP75B 结构域相当保守”的论点一致[26,33]。然而,相比旁系同源基因对PgF3’H1/PgF3’H2,PgF3’H3缺失了motif 7,这可能是它们分属于不同亚族的原因,它们在具体生物学功能上可能有不同分工。
笔者在本研究中进一步分析了石榴PgF3’Hs上游2000 bp启动子序列内的顺式作用元件,以了解其可能的调控机制,结果发现PgF3’H2 和PgF3’H3启动子上具有比PgF3’H1 数量更多的顺式作用元件,而且PgF3’H2 启动子上有8 个生长发育相关及4 个AUX 相关的作用元件,而PgF3’H3 有5 个Gbox 光响应及6 个ABA 相关的作用元件。植物花色苷合成受AUX 和ABA 显著调控,苹果中生长素响应因子MdARF 结合MdDFR 启动子抑制其表达从而负调控花色苷合成[37];ABA 处理提高了夏黑葡萄锦葵素、矮牵牛素水平[38];在草莓中,ABA 处理显著促进了果实红色素及花色苷积累,而AUX处理则相反[39]。以上分析暗示着PgF3’H2 和PgF3’H3 的表达与激素和光有关,相比PgF3’H1,是作用于石榴籽粒花色苷合成更重要的基因。还发现与PgF3’H2和PgF3’H3 相比,PgF3’H1 在石榴籽粒发育过程中表达量极低,这也说明了PgF3’H2和PgF3’H3的重要性。另外,PgF3’H2 和PgF3’H3 启动子上均具有MYB 结合位点。已有研究表明PgMYB1 能够直接结合下游PgGSTF6 启动子区,从而调控石榴花色苷合成[40];草莓FaMYB5 与FaEGL3、FaLWD1/FaLWD1-like 互作形成MYB-bHLH-WD40 复合体(MBW)直接激活FaF3’H 转录表达,从而赋予草莓果皮和果肉红色色泽[27];此外,龙眼和猕猴桃等园艺作物果皮着色机制中也存在类似的作用模式[41-42]。由此笔者课题组推测PgF3’H2 或PgF3’H3 基因表达受MYB 转录调控,通过影响花色苷水平和种类,在石榴籽粒颜色形成中发挥重要作用。因此,进一步解析PgF3’Hs 基因功能,深入研究其调控机制对阐明石榴籽粒颜色形成的作用非常重要。
F3’H 基因调控植物组织器官花色苷合成及颜色形成在不同物种中均有报道[27,30,34]。笔者在本研究中进而分析了石榴PgF3’H 家族成员随果实籽粒着色的表达情况,发现PgF3’H3 可能主要作用于石榴籽粒早期花色苷合成,而PgF3’H2 是石榴成熟籽粒呈色的主要作用基因。笔者实验室前期利用突尼斯软籽石榴呈颜色分离的自交后代材料为研究对象,从中克隆到一个和石榴幼叶呈红色相关的PgF3’H3 基因[20,33],PgF3’H3[20,33]与园艺作物樱桃、葡萄[30]和苹果[31]中的F3’H 相似度较高,而据报道它们均与果实着红色有关[27-28,43],因此,PgF3’H3 可能是石榴籽粒花色苷合成的重要作用基因。景宗慧等[27]通过同源遗传转化试验证明了过表达草莓(Fragaria×ananassa)FaF3’H 后其果肉由浅红色变成深红色,矢车菊素-3-O葡萄糖苷含量显著增加,而天竺葵色素-O-葡萄糖苷含量显著下降。笔者在本研究中通过异源转化拟南芥试验,发现过表达PgF3’H3株系的幼苗子叶呈红色而野生型呈绿色,并且其花色苷含量显著高于野生型而类胡萝卜素和叶绿素含量显著低于野生型。以上研究结果表明了PgF3’H3具有调控花色苷积累和红色形成的功能,并在石榴籽粒颜色形成过程中发挥重要作用。
此外,笔者在本研究中还发现PgF3’H 基因的表达水平及表达趋势在不同品种石榴籽粒中比较相似,但其成熟籽粒的红色差别比较大,推测除了PgF3’H 基因外,与花色苷合成相关的苯丙烷代谢途径中其他支路基因应该也发挥了作用。植物颜色形成是多级调控多基因参与的一个复杂网络作用的结果,包括参与早期柚皮素前体物质合成的PAL、CHS 和CHI,负责合成中间代谢物二氢黄酮醇、二氢杨梅黄酮和二氢栎皮酮的F3H、F3’H和F3’5’H,后期负责合成呈蓝紫色的锦葵素、呈砖红色的花葵素和呈紫红色的芍药素的DFR、ANS、UFGT 等,以及负责转运至液泡的GST、MATE 及ABC transport等[20-22]。因此,需要通过基因同源转化及基因编辑技术积极探索PgF3’H2 和PgF3’H3 基因对石榴籽粒花色苷组分的影响,通过与其他相关基因比较,分析它们对石榴籽粒花色苷积累及颜色形成的贡献。
4 结 论
笔者在本研究中对石榴PgF3’H 基因家族成员进行了全基因组鉴定和分析,提供了石榴PgF3’H基因家族3个成员包括染色体位置、基因大小、蛋白结构、系统发育、基因结构、保守结构域和启动子区顺式作用元件等信息。利用已有转录组数据,分析了石榴PgF3’H 基因家族3 个成员在不同品种石榴籽粒中随发育期的表达情况,明确了PgF3’H2 和PgF3’H3 可能是石榴籽粒着色和花色苷合成的重要作用基因。结合前期研究结果,通过遗传转化拟南芥初步验证了PgF3’H3 具有调控花色苷合成和红色形成的功能。PgF3’H2 和PgF3’H3 可能受光和激素影响,通过MYB调控石榴籽粒着色的功能和作用机制有待深入研究。
[1] 胡青霞,冯梦晨,陈延惠,郭强,简在海,史江莉,万然,郑先波.突尼斯软籽石榴果实生长发育及其品质形成规律研究[J].果树学报,2022,39(3):426-438.HU Qingxia,FENG Mengchen,CHEN Yanhui,GUO Qiang,JIAN Zaihai,SHI Jiangli,WAN Ran,ZHENG Xianbo. Growth and development of Tunisia soft seed pomegranate and its quality formation regulation[J].Journal of Fruit Science,2022,39(3):426-438.
[2] 陈延惠,史江莉,万然,简在海,胡青霞.中国软籽石榴产业发展现状与发展建议[J].落叶果树,2020,52(3):1-4.CHEN Yanhui,SHI Jiangli,WAN Ran,JIAN Zaihai,HU Qingxia.Development status and suggestions of soft seed pomegranate industry in China[J].Deciduous Fruits,2020,52(3):1-4.
[3] BOROCHOV-NEORI H,JUDEINSTEIN S,HARARI M,BARYA’AKOV I,PATIL B S,LURIE S,HOLLAND D.Climate effects on anthocyanin accumulation and composition in the pomegranate(Punica granatum L.)fruit arils[J].Journal of Agricultural and Food Chemistry,2011,59(10):5325-5334.
[4] 曹尚银,牛娟,曹达,李好先,薛辉,陈利娜,张富红,赵弟广.石榴果实成熟期果皮色泽差异蛋白质组比较分析[C]//曹尚银,李好先. 中国石榴研究进展(二). 北京:中国林业出版社,2014:141-149.CAO Shangyin,NIU Juan,Cao Da,LI Haoxian,XUE Hui,CHEN Lina,ZHANG Fuhong,ZHAO Diguang.Comparative proteomics analysis of fruit skin color in pomegranate (Punica granatum L.)[C]//The Research Progress of Pomegranate (Ⅱ).Beijing:China Forestry Publishing House,2014:141-149.
[5] ZHANG X H,ZHAO Y J,REN Y,WANG Y Y,YUAN Z H.Fruit breeding in regard to color and seed hardness:a genomic view from pomegranate[J].Agronomy,2020,10(7):991.
[6] 招雪晴,苑兆和.2 个石榴品种果皮花色苷合成相关基因表达分析[J].西北植物学报,2018,38(5):823-829.ZHAO Xueqing,YUAN Zhaohe. Expression profiles of anthocyanin biosynthetic genes in two cultivars of Punica granatum L.[J]. Acta Botanica Boreali-Occidentalia Sinica,2018,38(5):823-829.
[7] 冯立娟,尹燕雷,苑兆和,招雪晴,陶吉寒,侯乐峰.不同发育期石榴果实果汁中花青苷含量及品质指标的变化[J].中国农学通报,2010,26(3):179-183.FENG Lijuan,YIN Yanlei,YUAN Zhaohe,ZHAO Xueqing,TAO Jihan,HOU Lefeng. Change of anthocyanin content and quality index in pomegranate fruits during different developmental period[J].Chinese Agricultural Science Bulletin,2010,26(3):179-183.
[8] YUAN L,NIU H H,YUN Y R,TIAN J,LAO F,LIAO X J,GAO Z Q,REN D B,ZHOU L Y.Analysis of coloration characteristics of Tunisian soft-seed pomegranate arils based on transcriptome and metabolome[J]. Food Chemistry,2022,370:131270.
[9] ALAM M A,ISLAM P,SUBHAN N,RAHMAN M M,KHAN F,BURROWS G E,NAHAR L,SARKER S D. Potential health benefits of anthocyanins in oxidative stress related disorders[J].Phytochemistry Reviews,2021,20(4):705-749.
[10] 胡悦,胡青霞,谭彬,陈延惠,简在海,史江莉,万然.红皮石榴着色机制与花青苷合成研究进展[J].分子植物育种,2019,17(8):2692-2700.HU Yue,HU Qingxia,TAN Bin,CHEN Yanhui,JIAN Zaihai,SHI Jiangli,WAN Ran. Research progress on coloring mechanism and anthocyanin biosynthesis of red- skinned pomegranate[J].Molecular Plant Breeding,2019,17(8):2692-2700.
[11] 王沙.石榴花瓣花青苷生理指标及其生物合成相关基因表达分析[D].南京:南京林业大学,2022.WANG Sha.Analysis of physiological indicators and biosynthesis-related gene expression of anthocyanins in pomegranate petals[D].Nanjing:Nanjing Forestry University,2022.
[12] LI Q L,TAN W,ZHAO L,LUO H,ZHOU Z D,ZHANG Y W,BI R X,ZHAO L N. A comprehensive evaluation of 45 pomegranate (Punica granatum L.) cultivars based on principal component analysis and cluster analysis[J]. International Journal of Fruit Science,2023,23(1):135-150.
[13] SHAHKOOMAHALLY S,KHADIVI A,BRECHT J K,SARKHOSH A. Chemical and physical attributes of fruit juice and peel of pomegranate genotypes grown in Florida,USA[J]. Food Chemistry,2021,342:128302.
[14] ATTANAYAKE R,RAJAPAKSHA R,WEERAKKODY P,BANDARANAYAKE P C G. The effect of maturity status on biochemical composition,antioxidant activity,and anthocyanin biosynthesis gene expression in a pomegranate (Punica granatum L.) cultivar with red flowers,yellow peel,and pinkish arils[J]. Journal of Plant Growth Regulation,2019,38(3):992-1006.
[15] HAREL-BEJA R,TIAN L,FREILICH S,HABASHI R,BOROCHOV-NEORI H,LAHAV T,TRAININ T,DORON-FAIGENBOIM A,OPHIR R,BAR-YA’AKOV I,AMIR R,HOLLAND D. Gene expression and metabolite profiling analyses of developing pomegranate fruit peel reveal interactions between anthocyanin and punicalagin production[J]. Tree Genetics & Genomes,2019,15(2):22.
[16] 王沙,张心慧,赵玉洁,李变变,招雪晴,沈雨,董建梅,苑兆和.石榴花青苷合成相关基因PgMYB111 的克隆与功能分析[J].园艺学报,2022,49(9):1883-1894.WANG Sha,ZHANG Xinhui,ZHAO Yujie,LI Bianbian,ZHAO Xueqing,SHEN Yu,DONG Jianmei,YUAN Zhaohe.Cloning and functional analysis of PgMYB111 related to anthocyanin synthesis in pomegranate[J]. Acta Horticulturae Sinica,2022,49(9):1883-1894.
[17] 史江莉,仝瑞冉,王森,姚佳男,韦爱娜,王飒,王苗苗,万然,焦健,张春玲,张珍,郑先波.低温气调贮藏对‘突尼斯软籽’石榴果实品质的影响[J]. 河南农业大学学报,2022,56(5):779-787.SHI Jiangli,TONG Ruiran,WANG Sen,YAO Jianan,WEI Aina,WANG Sa,WANG Miaomiao,WAN Ran,JIAO Jian,ZHANG Chunling,ZHANG Zhen,ZHENG Xianbo. Effect of cold controlled atmosphere storage on fruit quality of‘Tunisia’soft-seed pomegranate[J]. Journal of Henan Agricultural University,2022,56(5):779-787.
[18] 袁蕾.基于代谢组和转录组学分析突尼斯软籽石榴品质调控机制[D].昆明:昆明理工大学,2022.YUAN Lei. Analysis of the quality control mechanism of Tunisian Soft Seed pomegranate based on metabolomics and transcriptomics[D]. Kunming:Kunming University of Science and Technology,2022.
[19] GUO X L,LI H T,LI D W,ZHANG L T,WU Y J,CHEN Y H.Determination and comparison of pigment contents in leaves of Punica granatum L. hybrids with different leaf colors[J]. Agricultural Science&Technology,2013,14(2):243-247.
[20] 万然,史江莉,胡悦,胡青霞,简在海,陈延惠.‘突尼斯软籽’石榴自交一代颜色性状分离及形成基础研究[C]//曹尚银,李好先. 中国石榴研究进展(三). 北京:中国林业出版社,2018:225-236.WAN Ran,SHI Jiangli,HU Yue,HU Qingxia,JIAN Zaihai,CHEN Yanhui. Reseaches on color segregation and formation in generation individuals of first self-crossing from‘Tunisiruanzi’pomegranate[C]//CAO Shangyin,LI Haoxian. The Research Progress of Pomegranate (Ⅲ). Beijing:China Forestry Publishing House,2018:225-236.
[21] 高磊,李慧,郑焕,陶建敏.果树中花色苷的生物合成及其调控机制研究进展[J].江苏农业学报,2022,38(1):258-267.GAO Lei,LI Hui,ZHENG Huan,TAO Jianmin. Advances in biosynthesis and regulation mechanism of anthocyanins in fruit trees[J]. Jiangsu Journal of Agricultural Sciences,2022,38(1):258-267.
[22] 招雪晴,杨静,沈雨,苑兆和.石榴PgUGT 基因表达特性与重组表达分析[J].西北植物学报,2022,42(3):390-397.ZHAO Xueqing,YANG Jing,SHEN Yu,YUAN Zhaohe. Expression profiles and recombinant expression of PgUGT in pomegranate[J]. Acta Botanica Boreali- Occidentalia Sinica,2022,42(3):390-397.
[23] 招雪晴,沈雨,苑兆和.石榴UFGT 基因克隆与表达特征[C]//李好先,曹尚银.中国石榴研究进展(四).北京:中国林业出版社,2022:227-236.HAO Xueqing,SHEN Yu,YUAN Zhaohe. Cloning and expression analysis of pomegranate UFGT gene[C]//LI Haoxian,CAO Shangyin.The Research Progress of Pomegranate (Ⅳ). Beijing:China Forestry Publishing House,2022:227-236.
[24] 招雪晴,苑兆和.石榴果色合成相关基因CHS 和CHI 的表达特性分析[J].基因组学与应用生物学,2019,38(5):2175-2182.ZHAO Xueqing,YUAN Zhaohe.Expression profiles of fruit color related genes CHS and CHI in Punica granatum L.[J]. Genomics and Applied Biology,2019,38(5):2175-2182.
[25] ATTANAYAKE R,EESWARAN R,RAJAPAKSHA R,WEERAKKODY P,BANDARANAYAKE P C G.Biochemical composition and expression of anthocyanin biosynthetic genes of a yellow peeled and pinkish ariled pomegranate (Punica granatum L.) cultivar are differentially regulated in response to agro-climatic conditions[J]. Journal of Agricultural and Food Chemistry,2018,66(33):8761-8771.
[26] YUAN Z H,FANG Y M,ZHANG T K,FEI Z J,HAN F M,LIU C Y,LIU M,XIAO W,ZHANG W J,WU S,ZHANG M W,JU Y H,XU H L,DAI H,LIU Y J,CHEN Y H,WANG L L,ZHOU J Q,GUAN D,YAN M,XIA Y H,HUANG X B,LIU D Y,WEI H M,ZHENG H K. The pomegranate (Punica granatum L.)genome provides insights into fruit quality and ovule developmental biology[J]. Plant Biotechnology Journal,2018,16(7):1363-1374.
[27] 景宗慧,尹梦娇,王前,鲍锞,周佩娜,刘潺潺,吴啟南.芡叶类黄酮-3’-羟化酶(F3’H)基因表达分析及功能验证[J].中国中药杂志,2021,46(18):4712-4720.JING Zonghui,YIN Mengjiao,WANG Qian,BAO Ke,ZHOU Peina,LIU Chanchan,WU Qinan. Expression profiling and functional verification of flavonoid 3’-hydroxylase gene from leaves of Euryale ferox[J]. China Journal of Chinese Materia Medica,2021,46(18):4712-4720.
[28] 侯杰,佟玲,崔国新,许志茹,李玉花.植物类黄酮3’-羟化酶(F3’H)基因的研究进展[J]. 植物生理学报,2011,47(7):641-647.HOU Jie,TONG Ling,CUI Guoxin,XU Zhiru,LI Yuhua. Research advances of plant flavonoid 3’-hydroxylase (F3’H)gene[J].Plant Physiology Journal,2011,47(7):641-647.
[29] NITARSKA D,BOEHM R,DEBENER T,LUCACIU R C,HALBWIRTH H. First genome edited poinsettias:Targeted mutagenesis of flavonoid 3’-hydroxylase using CRISPR/Cas9 results in a colour shift[J]. Plant Cell,Tissue and Organ Culture,2021,147(1):49-60.
[30] JEONG S T,GOTO-YAMAMOTO N,HASHIZUME K,ESAKA M. Expression of the flavonoid 3’-hydroxylase and flavonoid 3’,5’-hydroxylase genes and flavonoid composition in grape(Vitis vinifera)[J].Plant Science,2006,170(1):61-69.
[31] WEISSENSTEINER J,MOLITOR C,MARINOVIC S,FÜHRER L,HASSAN S W,HUTABARAT O S,SPORNBERGER A,STICH K,HAUSJELL J,SPADIUT O,HASELMAIR-GOSCH C,HALBWIRTH H. Molecular and enzymatic characterization of flavonoid 3’-hydroxylase of Malus× domestica[J].Plants,2021,10(9):1956.
[32] JIN Y,LIAO M Y,LI N,MA X Q,ZHANG H M,HAN J,LI D Z,YANG J F,LU X P,LONG G Y,DENG Z N,SHENG L.Weighted gene coexpression correlation network analysis reveals the potential molecular regulatory mechanism of citrate and anthocyanin accumulation between postharvest‘Bingtangcheng’and‘Tarocco’blood orange fruit[J].BMC Plant Biology,2023,23(1):296.
[33] 胡悦.石榴花青苷合成相关基因的克隆和PgF3’H 基因功能分析[D].郑州:河南农业大学,2019.HU Yue. Cloning of the genes related to the synthesis of pomegranate anthocyanin and functional analysis of PgF3’H gene[D].Zhengzhou:Henan Agricultural University,2019.
[34] PARK S,CHOI M J,LEE J Y,KIM J K,HA S H,LIM S H.Molecular and biochemical analysis of two rice flavonoid 3'-hydroxylase to evaluate their roles in flavonoid biosynthesis in rice grain[J]. International Journal of Molecular Sciences,2016,17(9):1549.
[35] JUNG Y J,LEE H J,KIM J H,KIM D H,KIM H K,CHO Y G,BAE S,KANG K K.CRISPR/Cas9-targeted mutagenesis of F3’H,DFR and LDOX,genes related to anthocyanin biosynthesis in black rice (Oryza sativa L.)[J]. Plant Biotechnology Reports,2019,13(5):521-531.
[36] TANAKA Y,BRUGLIERA F. Flower colour and cytochromes P450[J].Philosophical Transactions of the Royal Society of London.Series B,Biological Sciences,2013,368(1612):20120432.
[37] JI X H,WANG Y T,ZHANG R,WU S J,AN M M,LI M,WANG C Z,CHEN X L,ZHANG Y M,CHEN X S. Effect of auxin,cytokinin and nitrogen on anthocyanin biosynthesis in callus cultures of red-fleshed apple(Malus sieversii f.niedzwetzkyana)[J]. Plant Cell,Tissue and Organ Culture,2015,120(1):325-337.
[38] HIRATSUKA S,ONODERA H,KAWAI Y,KUBO T,ITOH H,WADA R. ABA and sugar effects on anthocyanin formation in grape berry cultured in vitro[J]. Scientia Horticulturae,2001,90(1/2):121-130.
[39] LI B J,GRIERSON D,SHI Y N,CHEN K S. Roles of abscisic acid in regulating ripening and quality of strawberry,a model non- climacteric fruit[J]. Horticulture Research,2022,9:uhac089.
[40] WANG Z H,YANG X M,WANG C Z,FENG L J,YIN Y L,LI J L. PgMYB1 positively regulates anthocyanin accumulation by activating PgGSTF6 in pomegranate[J]. International Journal of Molecular Sciences,2023,24(7):6366.
[41] YI D B,ZHANG H N,LAI B,LIU L Q,PAN X L,MA Z L,WANG Y C,XIE J H,SHI S Y,WEI Y Z. Integrative analysis of the coloring mechanism of red Longan pericarp through metabolome and transcriptome analyses[J]. Journal of Agricultural and Food Chemistry,2021,69(6):1806-1815.
[42] PENG Y Y,WANG K L,COONEY J M,WANG T C,ESPLEY R V,ALLAN A C. Differential regulation of the anthocyanin profile in purple kiwifruit (Actinidia species) [J]. Horticulture Research,2019,6:3.
[43] TODA K,AKASAKA M,DUBOUZET E G,KAWASAKI S,TAKAHASHI R. Structure of flavonoid 3’-hydroxylase gene for pubescence color in soybean[J]. Crop Science,2005,45(6):2212-2217.