随着消费者购买力和消费观念的演变,果实大小、色泽、果面光洁度等外在品质已成为影响消费者购买决策的主要因素[1],其中果皮底色对果实外观有显著影响,桃果实色泽和香味直接影响其感官品质,也是影响消费者选择的重要因素[2]。
叶绿素是植物体的重要代谢产物[3],同样也是决定果皮底色的关键因素。叶绿素降解是植物叶片衰老和果实成熟的一个重要代谢过程,叶绿素由多步骤途径分解,其降解产生叶片变黄及果皮颜色变化,展现果实成熟的状态[4],所以全面、深入地了解叶绿素降解及其关键基因的调控机制,可以加深对桃果实成熟时色泽变化的理解。近年来,植物中叶绿素的主要降解途径已有较多的研究[5-6]。叶绿素降解途径中叶绿素b首先被NYC1和NOL两种同工酶还原生成7-羟甲基叶绿素a;随后,7-羟甲基叶绿素a 被HCAR 还原为叶绿素a[7];进而,叶绿素a被叶绿素酶(CLH)催化形成脱植基叶绿素[8],在金属螯合物作用下先除去镁离子生成脱镁叶绿素,生成的Pheina 则在脱镁叶绿素酶(PPH)作用下除去植基[9];之后PAO能将脱镁基叶绿酸a(Pheidea)降解为不稳定的红色叶绿素代谢产物(RCC)后,最终在RCCR的催化下形成无色的初生荧光叶绿素代谢产物(pFCC)[10]。
PAO 是控制叶绿素降解的重要基因[11-12],PAO催化的卟啉环氧化开环是叶绿素降解的关键步骤,因而这条降解途径被称为PAO 降解途径。枣中ZjPAO1 和ZjPAO2 基因在果实不同发育阶段均有表达,并会随着果实发育表达量升高[13]。水稻中发现一个单碱基突变导致PAO 编码蛋白变化,从而导致水稻中活性氧积累和叶绿素降解受阻[14]。SGR 和SGRL 基因是叶绿素降解调控途径中具有里程碑意义的基因。SGR 与SGRL 基因与叶绿素降解相关,可以通过招募叶绿素降解基因形成复合体,结合到光系统Ⅱ上,从而导致叶绿素降解。但相比于SGR,SGRL 在植物衰老前表达量较高,随后表达量开始下降[15]。枳砧红绵蜜柚嫁接黄化苗中净光合速率、叶绿素含量降低,SGR 表达量升高[16]。在小麦中发现在自然衰老7 d 时CLH 表达量升高,之后下降[17]。在大白菜中,MeJA 处理显著诱导叶绿素降解相关基因BrPAO1、BrNYC1、BrPPH1 和BrSGR1 的表达[18]。在柑橘中发现,宗橙中CsSGRaSTOP丧失了降解叶绿素的功能,导致其果皮呈棕褐色[19]。在香蕉中发现MaERF012 在果肉和果皮中差异表达,并与果实成熟密切相关,MaERF012 激活叶绿素降解基因MaSGR的表达从而导致香蕉果皮叶绿素降解[20]。拟南芥在叶片自然衰老过程中,CCGs 剧烈上调表达且CCEs 蛋白逐渐累积,导致衰老细胞中叶绿素的大量降解[21]。
桃果皮色泽是影响食用价值和经济价值的重要因素之一,叶绿素降解及相关基因调控在部分物种中有一定的研究,但在桃中叶绿素降解及相关基因的表达规律尚未见报道[22]。笔者在本研究中通过荧光定量技术对4 个桃品种/品系果皮叶绿素降解基因表达规律进行研究,并系统分析了叶绿素降解相关基因表达规律与叶绿素水平和色差之间的关系。
1 材料和方法
1.1 试验材料
以4个不同成熟期的桃品种/品系春美、2-50、中蟠102、中桃7 号为研究材料(图1),其中,春美和2-50 在6 月中旬成熟,中蟠102 和中桃7 号分别在7 月上旬和8 月上旬成熟。供试材料来自中国农业科学院郑州果树研究所桃品种圃,株行距为1.0 m×4.0 m,2017 年定植,常规管理。2022 年5—8 月,于果实成熟前30 d开始,采摘树体外围中上部大小均匀、无病虫害、成熟度一致的果实30 个,带回实验室使用尼康700d 相机于自然光下以黑色植绒布为背景拍照。以10 个果实为1 个样本,取样进行3 次重复,取样间隔7 d,在果实转色期每隔4 d 取样1 次,削取表皮进行液氮速冻,保存至-80 ℃冰箱备用。
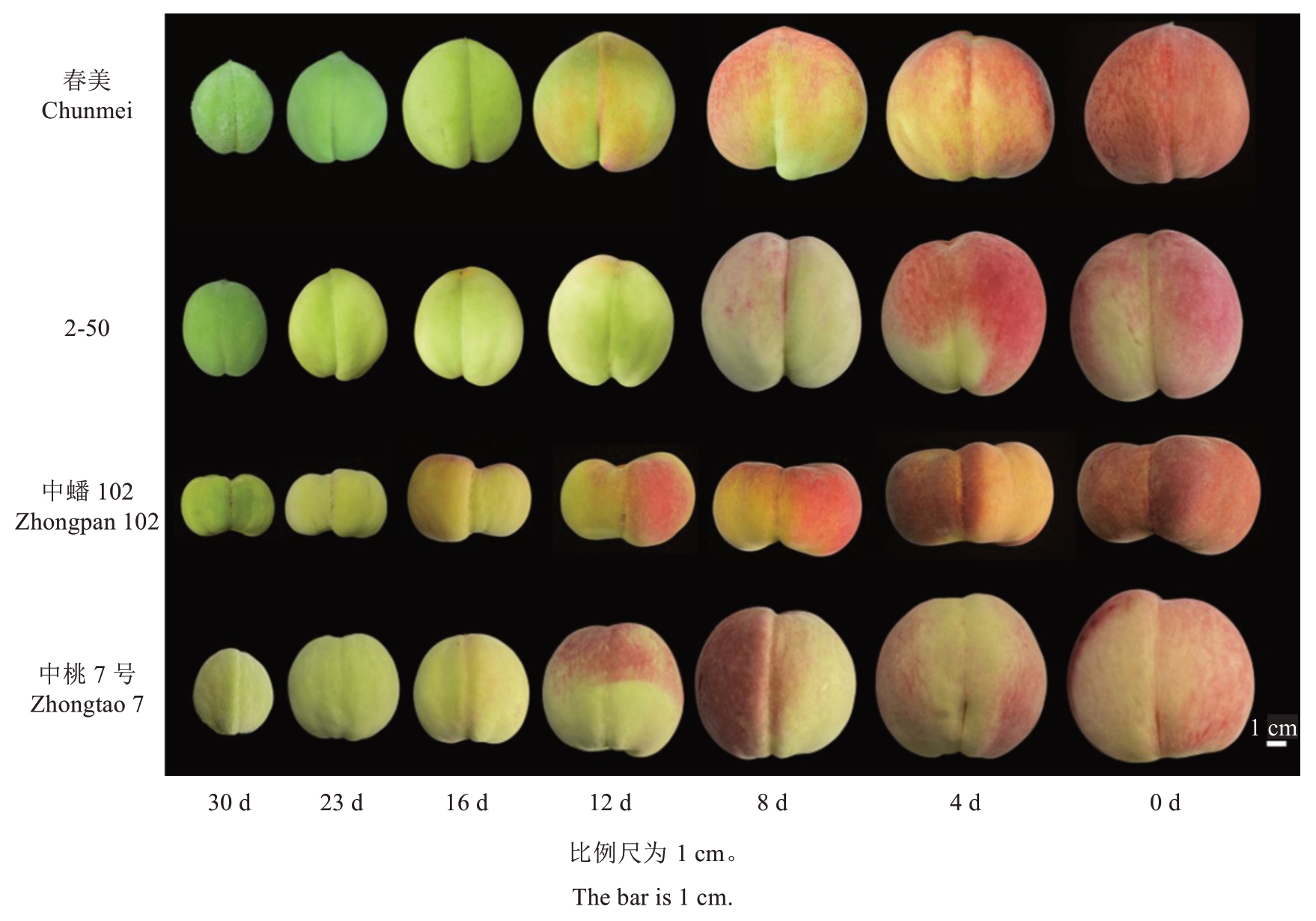
图1 成熟前30~0 d 的果实表型Fig.1 Phenotypes of fruits 30-0 days before maturation
1.2 色差检测
用色差仪(美能达CR-400,柯尼卡美能达)评价果皮底色,颜色用CIE a*、b*标尺表示。随机选取果实赤道区域的四个不同点取平均值,记录a*和b*值[23-24]。
1.3 叶绿素含量测定
桃果皮叶绿素含量采用紫外分光光度法测定[25-26]。取去离子水清洗过的桃果皮0.4 g,用研磨机磨碎,放入10 mL 离心管中,加入6 mL 95%乙醇溶液,避光提取叶绿素至果皮完全变白。分别测定叶绿素溶液在665 nm 和649 nm 波长下的吸光度,通过公式1~3计算叶绿素含量(w,后同):

1.4 RNA提取
根据多糖多酚植物总RNA 提取试剂盒(DP441,天根生化科技有限公司,北京,中国)说明书提取桃总RNA。利用1%琼脂糖凝胶电泳检测RNA 的质量和纯度,取1 µL RNA 利用微量紫外分光光度计NanoDrop2000(Thermo Scientific,麻省)测定浓度。取1 ng RNA 参照FastKing cDNA 第一链合成试剂盒说明书(天根生化科技有限公司,北京,中国)进行反转录,放置在-20 ℃冰箱保存进行后续的实时荧光定量PCR。
1.5 实时荧光定量检测
从基因组数据库(https://phytozome-next.jgi.doe.gov/)下载相关基因的CDS(Coding sequence,编码序列);利用NCBI 网站(https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome)设计特异性荧光定量引物,引物长度在20 bp 左右,GC含量为40%~60%,引物Tm值为58 ℃~62 ℃,扩增片段大小为80~150 bp;PCR 体系为15.1 µL:cDNA 1µL、Mix 7.5µL、引物F 0.3µL、引物R 0.3µL、水6 µL。PCR 程序设置为:95 ℃预变性5 min;95 ℃变性30 s、60 ℃退火30 s、72 ℃延伸30 s,扩增循环数为40,设置3 次技术重复。引物序列见表1。实时荧光定量PCR 所用的仪器为罗氏480,所用试剂盒为SYBR Real-time PCR Premixtur(eBiotake)。选取Actin(ppa007228mg)为桃内参基因[27],最后按照2-ΔΔCT方法进行结果计算[28]。
表1 实时定量PCR 反应中各基因的引物序列
Table 1 Primers sequence for real-time quantitative PCR reaction

基因名称Gene name Actin登记号GenBank ppa007228m PpNYC1 ppa010004m PpNOL ppa005304m PpHCAR ppa004221m PpCLH1 ppa009825m PpCLH2 ppa009788m PpPPH ppa019738m PpPAO ppa009783m PpRCCR ppa004339m PpSGR ppa010416m PpSGRL引物序列Primer sequence(5′–3′)GATTCCGGTGCCCAGAAGT CCAGCAGCTTCCATTCCAA ATCGTGTGGTTGTCGCTTCT CAGGTGCTTAGAGGAGGCAC ATACGGGGCAACAAAGCGTA ACCATTCCTGGCGACAAGTT CAGTGGAAATCGCCAACCAT AACTTTGGGGCAGGTTCAGG CATGCCAAAACTGCCCTGTC AGGATATGGGGCCTGGTTCT TCTCACGGCTTCATTGTCGT TGAACATGGGGTGGAAGCAA AGACTCGGGGCTTAGTAGCA CGCTCCGTCTCTGACAAACT AGGCAACCCACGGATTACTG AGTCTTCCCTGGTGCCATTG ACATCCGCAGTGTTGTGTCT ATCCAGCCAAATTCCCAGCA GCTGTTGCTTCCCACCATTG TGTTTCTTGGGTTTGGCCCT TGACGTGGTTGCAGAATGGA GCCAGGTCCAGCATGAGATT ppa014909m
1.6 数据分析
试验数据用SPSS 软件分析,用Excel 2003 软件制作图表。
2 结果与分析
2.1 桃果实发育后期果皮色差变化规律
2.1.1 a*值的变化 4 个品种/品系a*值(图2)均呈现为S形,在成熟前23~12 d色差a*上升较快。在果实成熟前12 d,桃果实绿色不断减弱,果实开始着红色,色差a*值不断上升。在成熟前12~8 d,4 个品种果实色差a*值均由负转正或接近于0,认为此时是果实重要的转色期。
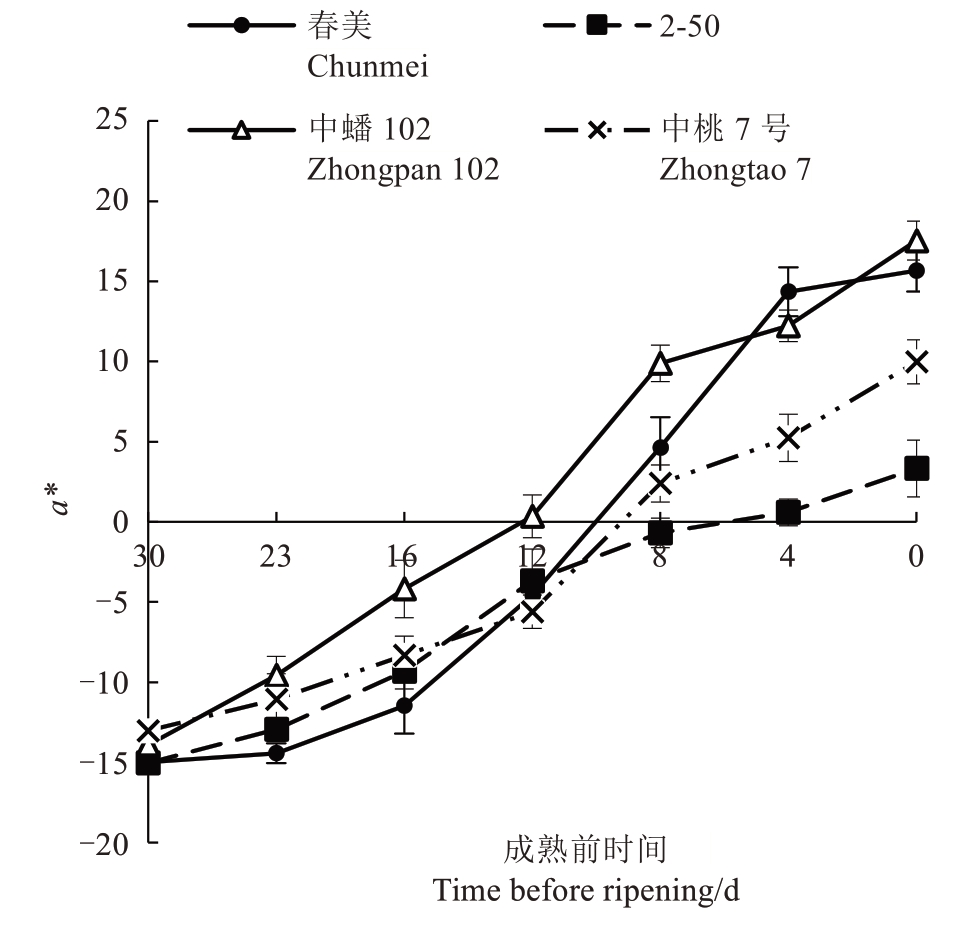
图2 4 个品种/品系果实成熟前果皮色差a*值的变化
Fig.2 Changes in peel color difference a*values of four peach varieties/strains before fruit ripening
2.1.2 b*值的变化 中蟠102 和中桃7 号果实b*值(图3)在成熟前30~16 d 升高,之后随着果实发育不断下降。春美和2-50 果实b*值在成熟前30~23 d 升高,随后快速下降。春美、2-50、中桃7 号在果实成熟时b*值为20~30。
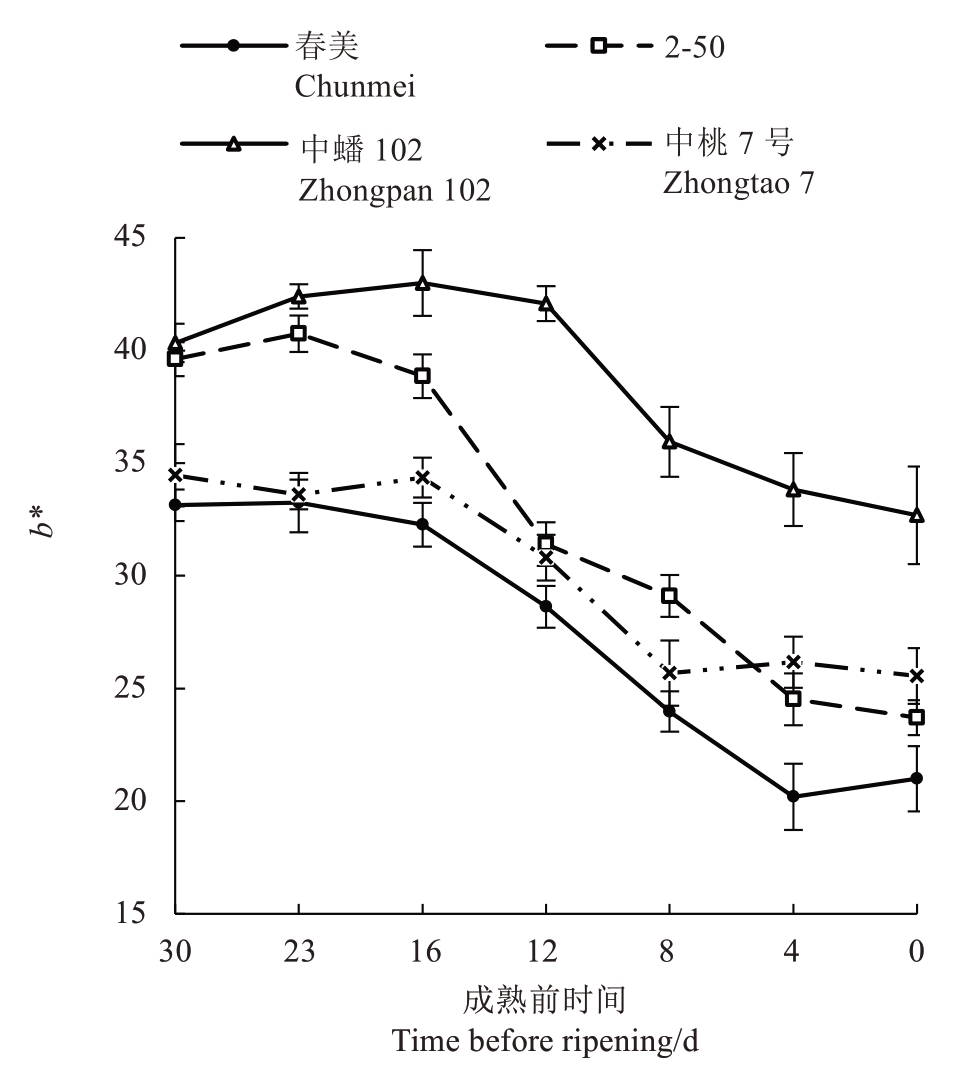
图3 4 个品种/品系果实成熟前果皮色差b*值的变化
Fig.3 Changes in peel color difference b*values of four peach varieties/strains before fruit ripening
2.2 果实发育后期叶绿素含量变化
4 个品种/品系的叶绿素含量(图4)随着果实的发育表现出明显的下降趋势,叶绿素含量在成熟前30~16 d 快速下降。在果实成熟前12~8 d,各品种的叶绿素含量(w)均在8 μg·g-1以下,从图1 可以看到这时各品种有明显着色,但果实着色还不完全。在果实成熟前8 d至果实成熟,4个桃品种/品系的叶绿素含量下降较为缓慢。中蟠102和2-50在成熟前8 d时叶绿素含量与成熟时的叶绿素含量差异不大。

图4 4 个桃品种/品系果实成熟前叶绿素含量的变化
Fig.4 Changes in chlorophyll content of four peach varieties/strains before fruit ripening
2.3 果实发育后期叶绿素降解相关基因的表达分析
通过实时荧光定量PCR 方法,测定了4 个桃品种/品系中与叶绿素降解相关的10个基因(PpNYC1、PpNOL、PpHCAR、PpCLH1、PpCLH2、PpPPH、PpPAO、PpRCCR、PpSGR、PpSGRL)的表达量,并绘制热图进行聚类分析。如图5 所示,PpCLH1、PpPPH、PpPAO、PpRCCR、PpSGR、PpSGRL 基因在春美材料中高表达,PpNYC1在中蟠102中的表达量较高,PpNOL 在中桃7 号中表达量最高,PpCLH2 则在中蟠102 和中桃7 号高表达,而PpHCAR 在4 个桃品种/品系中无明显表达差异。

图5 叶绿素降解基因相对表达量
Fig.5 Relative expression of chlorophyll degradation gene

图5 (续) Fig.5 (Continued)
不同时间段内,每个基因的表达量通常会随着果实发育不断变化。PpNYC1 在中蟠102 中成熟前4 d时有升高,PpNOL 在中桃7 号中明显先上调后下调,PpSGRL 在中桃7 号成熟前12 d 有短暂升高。PpHCAR、PpPPH、PpPAO、PpRCCR 随着果实发育在成熟前12~4 d表达量上升,在果实成熟时表达量下降,PpHCAR、PpRCCR 在3 个桃品种/品系中无明显差异,在2-50中表达模式有所不同。PpPPH 和PpPAO在春美中分别在成熟前8 d 和成熟前4 d 有明显升高。
由图5 可知,在4 个桃品种/品系中,PpCLH1 基因在成熟前12~8 d 时间段内的表达量高于其他基因,在果实成熟时表达量迅速下降,因为此时果实处于转色的关键期,因此推断PpCLH1 基因可能是桃果实转色时的重要调控基因。随着果实发育,果皮叶绿素降解,果实完全成熟,PpSGR 基因的表达量显著升高。进一步推测PpSGR 基因有可能也是桃果实成熟前造成4 个桃品种/品系叶绿素降解的主要基因。
根据聚类分析结果(图6)在春美和2-50 中PpSGR 与PpCLH1、PpPPH 聚为一类,在中蟠102 和中桃7号中PpSGR与PpCLH1、PpRCCR聚为一类。在春美和中蟠102 中PpSGRL 和PpCLH2 聚为一类,在2-50 和中桃7 号中PpSGRL、PpNYC1 和PpCLH2 聚为一类。因此PpSGRL、PpNYC1 和PpCLH2 可能是果实早期叶绿素降解的关键基因,PpSGR 与PpCLH1、PpPPH、PpRCCR 可能是桃果实成熟前造成不同桃品种/品系叶绿素降解差异的主要基因。

图6 4 个桃品种/品系叶绿素降解基因的聚类分析
Fig.6 Cluster analysis of chlorophyll degradation genes in different peach varieties
对叶绿素含量与其降解基因进行相关性分析(表2),发现PpCLH2 基因表达与春美和2-50 中叶绿素含量呈显著正相关,PpSGR 与春美、中桃7 号的叶绿素含量呈显著负相关,PpSGRL 与春美、2-50、中蟠102 的叶绿素含量呈显著正相关。
表2 叶绿素降解基因相对表达量与叶绿素含量的相关系数
Table 2 The correlation coefficient between the relative expression level of chlorophyll degradation genes and chlorophyll content
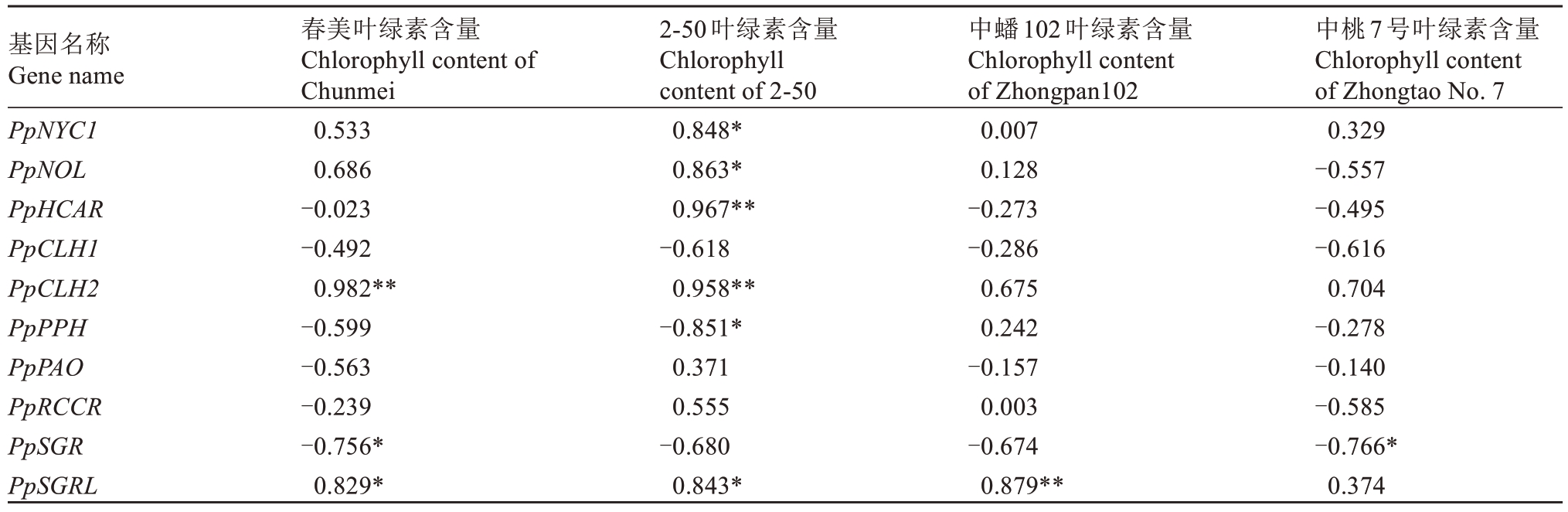
注:*和**分别表示相关性达到0.05 和0.01 显著水平。
Note:*and**respectively indicate that the correlation reaches a significant level of 0.05 and 0.01.
基因名称Gene name PpNYC1 PpNOL PpHCAR PpCLH1 PpCLH2 PpPPH PpPAO PpRCCR PpSGR PpSGRL中桃7号叶绿素含量Chlorophyll content of Zhongtao No.7 0.329-0.557-0.495-0.616 0.704-0.278-0.140-0.585-0.766*0.374春美叶绿素含量Chlorophyll content of Chunmei 0.533 0.686-0.023-0.492 0.982**-0.599-0.563-0.239-0.756*0.829*2-50叶绿素含量Chlorophyll content of 2-50 0.848*0.863*0.967**-0.618 0.958**-0.851*0.371 0.555-0.680 0.843*中蟠102叶绿素含量Chlorophyll content of Zhongpan102 0.007 0.128-0.273-0.286 0.675 0.242-0.157 0.003-0.674 0.879**
qRT-PCR 分析结果表明,成熟前12~8 d 时,PpCLH1、PpPAO 基因的表达量显著。而PpSGR 基因在果实成熟时表达量明显上调,聚类分析显示在春美和2-50 中PpSGR 与PpCLH1、PpPPH 聚为一类,在中蟠102 和中桃7 号中PpSGR 与PpCLH1、PpRCCR 聚为一类。相关性分析显示PpSGR 与春美、中桃7 号的叶绿素含量呈显著负相关。初步表明PpCLH1、PpSGR 基因是果实成熟前4 个桃品种/品系果皮叶绿素降解的关键基因。
3 讨 论
长期以来,CLH 被认为是参与叶绿素降解的第一种酶,与叶绿素的降解密切相关。有研究表明,乙烯处理的柑橘果皮中CLH 酶活力提高了2.5~4 倍,同时果皮明显褪绿[29]。樊艳燕等[30]发现BoCLH1(叶绿素酶1)在青花菜初始时检测到有较高表达,后期表达量很低,试验结果表明BoCLH1 主要在青花菜初始衰老中起降解叶绿素的作用。胡椒中发现叶绿素酶活性与叶绿素含量之间存在负相关关系,并在收获后的衰老过程中促进叶绿素的降解[31]。笔者在本试验中发现,桃PpCLH1 在果实成熟过程中表达量先升高后下降,在果实成熟前12~8 d 表达量较高,此时也是果实转色的关键时期,因此PpCLH1可能是桃果实转色时叶绿素降解的关键基因。在草地早熟禾叶片衰老时,CLH 基因的表达会导致叶绿素降解[32]。在荔枝中成熟过程中LcCLH 表达量先升高后下降,LcPAO、LcSGR 的表达量随着果实发育逐渐升高,相比于叶绿素降解更彻底的糯米滋,妃子笑中LcCLH、LcPAO、LcSGR的表达量明显更低[33]。
在高等植物中,脱镁叶绿酸a 氧化酶(PAO)基因表达量和酶活性的变化与叶绿素含量降解有关[12]。此外SGR 普遍参与了植物叶片衰老过程中叶绿素分解,也在植物其他器官的生长发育中有重要作用,如果实成熟和发育[34]。在大豆成熟前PPH、PAO、SGR 三个基因相对表达量显著提高(p<0.05),并且在褪绿材料科丰14中的表达量远高于滞绿材料北农108 中的表达量,因此推测叶绿素降解基因PPH、PAO、SGR 表达量低是北农108 滞绿的重要原因之一[35]。在本试验中PpPPH 和PpPAO 在春美中分别在8 DBM 和4 DBM 有明显升高,在其他4个品种/品系中也都有明显升高。雷波脐橙及其早熟单株果实也发现PAO 在果实各发育时期都有稳定表达,CLH、PPH、NYC1、SGR1 的表达先上调后下调[36]。青瓯柑和瓯柑的试验结果表明在花后210 d、240 d,瓯柑中NYC1、CLH、PPH、PAO、SGR 的表达量均高于青瓯柑,差异显著,这可能是青瓯柑叶绿素降解缓慢的原因[37]。笔者在本研究中通过聚类分析,在春美和2-50 中PpSGR 与PpCLH1、PpPPH 聚为一类,在中蟠102 和中桃7 号中PpSGR 与PpCLH1、PpRCCR 聚为一类。这说明在桃中叶绿素降解受多个叶绿素降解基因的影响。
大豆中GmSGR 丢失可能导致大豆种子从黄色变为绿色[38]。拟南芥在叶片衰老过程中,SGR1正向介导叶绿素降解,而过表达SGRL 的拟南芥植物表现出早期叶片黄变[15]。叶绿素降解导致桃果皮褪绿是桃果实成熟最明显的标志之一。有研究表明SlSGR1在调节番茄叶片和果实的叶绿素降解中起着关键作用[39]。与野生型(WT)果实相比,SlSGR1 敲除无效系明显显示出混浊的棕色,叶绿素水平明显更高[17],表明SlSGR1 影响叶绿素降解。笔者课题组发现PpSGR 基因在果实成熟时表达量较高,并且春美、中桃7号叶绿素含量与PpSGR表达量呈负相关,所以PpSGR 是桃果实成熟期叶绿素降解的关键调控基因。目前在其他物种中叶绿素降解基因的表达规律与功能的研究已经有很多,但在桃中还少见报道。因此,进一步解析桃中叶绿素降解基因调控果实成熟前叶绿素降解的机制具有重要意义。
4 结 论
桃果实成熟前12~8 d 是桃果实关键的转色期,此时PpCLH1基因相比绿色时有明显上调。在果实成熟前8~0 d,随着果皮叶绿素降解,PpSGR 表达量上升,表明PpSGR 是桃果实成熟前叶绿素降解的关键基因。本研究为探索桃果实发育过程中叶绿素降解的分子机制提供了参考。
[1] DELGADO C,CRISOSTO G M,HEYMANN H,CRISOSTO C H. Determining the primary drivers of liking to predict consumers’acceptance of fresh nectarines and peaches[J]. Journal of Food Science,2013,78(4):S605-S614.
[2] KELLEY K M,PRIMROSE R,CRASSWELLER R,HAYES J E,MARINI R. Consumer peach preferences and purchasing behavior:A mixed methods study[J]. Journal of the Science of Food and Agriculture,2016,96(7):2451-2461.
[3] TANAKA A,TANAKA R. Chlorophyll metabolism[J]. Current Opinion in Plant Biology,2006,9(3):248-255.
[4] ECKHARDT U,GRIMM B,HÖRTENSTEINER S. Recent advances in chlorophyll biosynthesis and breakdown in higher plants[J].Plant Molecular Biology,2004,56(1):1-14.
[5] HÖRTENSTEINER S,KRÄUTLER B. Chlorophyll breakdown in higher plants[J]. Biochimica et Biophysica Acta-Bioenergetics,2011,1807(8):977-988.
[6] MÜLLER T,MOSER S,ONGANIA K H,PRUZINSKA A,HÖRTENSTEINER S,KRÄUTLER B.A divergent path of chlorophyll breakdown in the model plant Arabidopsis thaliana[J].Chembiochem,2006,7(1):40-42.
[7] SATO Y,MORITA R,NISHIMURA M,YAMAGUCHI H,KUSABA M. Mendel’s green cotyledon gene encodes a positive regulator of the chlorophyll-degrading pathway[J]. Proceedings of the National Academy of Sciences of the United States of America,2007,104(35):14169-14174.
[8] KUNIEDA T,AMANO T,SHIOI Y.Search for chlorophyll degradation enzyme,Mg-dechelatase,from extracts of Chenopodium album with native and artificial substrates[J]. Plant Science,2005,169(1):177-183.
[9] SCHELBERT S,AUBRY S,BURLA B,AGNE B,KESSLER F,KRUPINSKA K,HÖRTENSTEINER S. Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis[J]. The Plant Cell,2009,21(3):767-785.
[10] FUKASAWA A,SUZUKI Y,TERAI H,YAMAUCHI N.Effects of postharvest ethanol vapor treatment on activities and gene expression of chlorophyll catabolic enzymes in broccoli florets[J].Postharvest Biology and Technology,2010,55(2):97-102.
[11] HÖRTENSTEINER S.Update on the biochemistry of chlorophyll breakdown[J].Plant Molecular Biology,2013,82(6):505-517.
[12] KUAI B K,CHEN J Y,HÖRTENSTEINER S. The biochemistry and molecular biology of chlorophyll breakdown[J]. Journal of Experimental Botany,2018,69(4):751-767.
[13] 李丽莉,樊丁宇,杨磊,靳娟,郝庆.枣PAO 基因的鉴定及在果实发育和贮藏过程中的表达分析[J].山东农业科学,2023,55(12):9-16.LI Lili,FAN Dingyu,YANG Lei,JIN Juan,HAO Qing.Identification and expression analysis of PAO gene during fruit development and storage of Ziziphus jujuba[J]. Shandong Agricultural Sciences,2023,55(12):9-16.
[14] SHEN J,GUO M J,WANG Y G,YUAN X Y,WEN Y Y,SONG X,DONG S Q,GUO P Y.Humic acid improves the physiological and photosynthetic characteristics of millet seedlings under drought stress[J]. Plant Signaling & Behavior,2020,15(8):1774212.
[15] SAKURABA Y,PARK S Y,KIM Y S,WANG S H,YOO S C,HÖRTENSTEINER S,PAEK N C.Arabidopsis STAY-GREEN2 is a negative regulator of chlorophyll degradation during leaf senescence[J].Molecular Plant,2014,7(8):1288-1302.
[16] 何文,王燕,陈清,徐世荣,汤浩茹,潘东明,王小蓉.枳砧红绵蜜柚嫁接黄化苗的光合特性及其CgSGR 基因克隆与表达分析[J].西北植物学报,2018,38(5):808-815.HE Wen,WANG Yan,CHEN Qing,XU Shirong,TANG Haoru,PAN Dongming,WANG Xiaorong. Photosynthetic characteristics of etiolated seedling of Citrus grandis (L.) Osbeck. cv.‘Hongmian Miyou’grafted to Poncirus trifoliata and expression of CgSGR[J]. Acta Botanica Boreali-Occidentalia Sinica,2018,38(5):808-815.
[17] 晋秀娟,孙丽丽,赵锴,ASHRAFUL I M,卢娟,王曙光,孙黛珍.小麦叶绿素酶基因家族的鉴定及其叶绿素降解过程中的功能预测[J].激光生物学报,2022,31(1):50-60.JIN Xiujuan,SUN Lili,ZHAO Kai,ASHRAFUL I M,LU Juan,WANG Shuguang,SUN Daizhen. Identification of CLH gene family and functional prediction of chlorophyll degradation in wheat[J].Acta Laser Biology Sinica,2022,31(1):50-60.
[18] LIU W,LI Q W,WANG Y,WU T,YANG Y F,ZHANG X Z,HAN Z H,XU X F.Ethylene response factor AtERF72 negatively regulates Arabidopsis thaliana response to iron deficiency[J].Biochemical and Biophysical Research Communications,2017,491(3):862-868.
[19] ZHU K J,ZHENG X J,YE J L,HUANG Y,CHEN H Y,MEI X H,XIE Z Z,CAO L X,ZENG Y L,LARKIN R M,XU Q,PEREZ-ROMAN E,TALÓN M,ZUMAJO-CARDONA C,WURTZEL E T,DENG X X. Regulation of carotenoid and chlorophyll pools in hesperidia,anatomically unique fruits found only in Citrus[J].Plant Physiology,2021,187(2):829-845.
[20] CHEN H C,LAI X H,WANG L H,LI X P,CHEN W X,ZHU X Y,SONG Z Y. Ethylene response factor MaERF012 modulates fruit ripening by regulating chlorophyll degradation and softening in banana[J].Foods,2022,11(23):3882.
[21] 窦锦慧,陈俊毅,鲁凡,吴声栋,蒯本科,张鼎宇.拟南芥叶片衰老前后叶绿素降解代谢基因表达和酶积累的模式及其影响因素分析[J].复旦学报(自然科学版),2023,62(5):565-574.DOU Jinhui,CHEN Junyi,LU Fan,WU Shengdong,KUAI Benke,ZHANG Dingyu.A preliminary study in the expression/accumulation pattern and regulation of chlorophyll catabolic genes/enzymes during leaf development in Arabidopsis thaliana[J].Journal of Fudan University(Natural Science),2023,62(5):565-574.
[22] 李靖雯,邹东方,黄莹莹,叶霞,冯建灿,李志谦.ERF 调控果蔬成熟过程颜色变化的研究进展[J].分子植物育种,2022,20(18):6236-6243.LI Jingwen,ZOU Dongfang,HUANG Yingying,YE Xia,FENG Jiancan,LI Zhiqian. Research progress of ERF in regulation of color variation in fruits and vegetables during maturation[J].Molecular Plant Breeding,2022,20(18):6236-6243.
[23] 李桂祥,马瑞娟,张斌斌,俞明亮,倪林箭.套袋对霞晖6 号桃果实发育过程中果皮色素含量和色差的影响[J].江苏农业学报,2012,28(6):1418-1423.LI Guixiang,MA Ruijuan,ZHANG Binbin,YU Mingliang,NI Linjian. Effect of bagging on peel pigment content and fruit chromatism of peach cultivar Xiahui 6[J].Jiangsu Journal of Agricultural Sciences,2012,28(6):1418-1423.
[24] 郭东花,白红,石佩,杨艳青,李高潮,范崇辉.不同时期套袋对“瑞光19 号”油桃果实挥发性成分及着色的影响[J].食品科学,2016,37(8):242-247.GUO Donghua,BAI Hong,SHI Pei,YANG Yanqing,LI Gaochao,FAN Chonghui. Effects of bagging at different stages on volatiles and color of“Ruiguang No. 19”nectarine fruits[J].Food Science,2016,37(8):242-247.
[25] 项倩,吴磊,徐若涵,杨再强.不同温度下染病番茄叶片SPAD和叶绿素含量的相关性[J].北方园艺,2022(18):8-15.XIANG Qian,WU Lei,XU Ruohan,YANG Zaiqiang. Correlation between SPAD and chlorophyll content in infected tomato leaves at different temperatures[J]. Northern Horticulture,2022(18):8-15.
[26] 刘建新,丁华侨,田丹青,王炜勇,刘慧春.擎天凤梨苞片叶绿素代谢关键基因的分离及褪绿的分子机理[J].中国农业科学,2016,49(13):2593-2602.LIU Jianxin,DING Huaqiao,TIAN Danqing,WANG Weiyong,LIU Huichun. Isolation of chlorophyll metabolism key genes and molecular mechanism of green fade in Guzmania bracts discoloration process[J]. Scientia Agricultura Sinica,2016,49(13):2593-2602.
[27] BRANDI F,BAR E,MOURGUES F,HORVÁTH G,TURCSI E,GIULIANO G,LIVERANI A,TARTARINI S,LEWINSOHN E,ROSATI C.Study of‘Redhaven’peach and its white-fleshed mutant suggests a key role of CCD4 carotenoid dioxygenase in carotenoid and norisoprenoid volatile metabolism[J]. BMC Plant Biology,2011,11:24.
[28] SCHMITTGEN T D,LIVAK K J.Analyzing real-time PCR data by the comparative CT method[J]. Nature Protocols,2008,3:1101-1108.
[29] YAMAUCHI N,AKIYAMA Y,KAKO S,HASHINAGA F.Chlorophyll degradation in Wase satsuma mandarin (Citrus unshiu Marc.) fruit with on-tree maturation and ethylene treatment[J].Scientia Horticulturae,1997,71(1/2):35-42.
[30] 樊艳燕,刘玉梅,李占省,方智远,杨丽梅,庄木,张扬勇,孙培田. 青花菜衰老过程中叶绿素降解相关基因的表达分析[J].园艺学报,2015,42(7):1338-1346.FAN Yanyan,LIU Yumei,LI Zhansheng,FANG Zhiyuan,YANG Limei,ZHUANG Mu,ZHANG Yangyong,SUN Peitian.Analysis of the expression of chlorophyll degrading genes during senescence of broccoli[J]. Acta Horticulturae Sinica,2015,42(7):1338-1346.
[31] GUPTA S,GUPTA S M,SANE A P,KUMAR N. Chlorophyllase in Piper betle L. has a role in chlorophyll homeostasis and senescence dependent chlorophyll breakdown[J]. Molecular Biology Reports,2012,39(6):7133-7142.
[32] 张兰,滕珂,尹淑霞.草地早熟禾叶绿素酶1 基因PpCHL1 的克隆和表达分析[J].中国草地学报,2016,38(4):1-7.ZHANG Lan,TENG Ke,YIN Shuxia. Cloning and expression analysis of chlorophyllase 1 gene PpCHL1 from Poa pratensis L.[J].Chinese Journal of Grassland,2016,38(4):1-7.
[33] ZOU S C,ZHUO M G,ABBAS F,HU G B,WANG H C,HUANG X M.Transcription factor LcNAC002 coregulates chlorophyll degradation and anthocyanin biosynthesis in litchi[J].Plant Physiology,2023,192(3):1913-1927.
[34] 张巧丽,陈笛,宋艳萍,朱鸿亮,罗云波,曲桂芹.番茄果实叶绿素代谢转录调控网络研究进展[J]. 园艺学报,2023,50(9):2031-2047.ZHANG Qiaoli,CHEN Di,SONG Yanping,ZHU Hongliang,LUO Yunbo,QU Guiqin. Review on transcriptional regulation of chlorophyll metabolism network in tomato fruits[J].Acta Horticulturae Sinica,2023,50(9):2031-2047.
[35] 张鑫,莫翱伟,许鹏昊,张卿,王程,谢皓.持绿大豆叶片中叶绿素降解途径关键酶基因的表达分析[J].分子植物育种,2020,18(15):4871-4877.ZHANG Xin,MO Aowei,XU Penghao,ZHANG Qing,WANG Cheng,XIE Hao.Expression analysis of key enzyme gene in chlorophyll degradation pathway in stay green soybean leaves[J].Molecular Plant Breeding,2020,18(15):4871-4877.
[36] 王小佳.雷波脐橙及其早熟单株果实脱绿期的叶绿素降解代谢关键基因表达分析[D].雅安:四川农业大学,2020.WANG Xiaojia.Differential expression analysis of key genes for chlorophyll degradation and metabolism of Leibo navel orange and its early-ripening fruit mutation[D]. Yaan:Sichuan Agricultural University,2020.
[37] 郑丽.青瓯柑PAO 基因的克隆及分析[D].武汉:华中农业大学,2013.ZHENG Li. Cloning and analysis PAO gene in the stay-green mutant of ougan mandarine (Citrus suavissima)[D]. Wuhan:Huazhong Agricultural University,2013.
[38] WANG C,GAO L,LI R Z,WANG Y,LIU Y Y,ZHANG X,XIE H.High-throughput sequencing reveals the molecular mechanisms determining the stay-green characteristic in soybeans[J].Journal of Biosciences,2020,45(1):103.
[39] LUO Z D,ZHANG J H,LI J H,YANG C X,WANG T T,OUYANG B,LI H X,GIOVANNONI J,YE Z B. A STAYGREEN protein SlSGR1 regulates lycopene and β-carotene accumulation by interacting directly with SlPSY1 during ripening processes in tomato[J].The New Phytologist,2013,198(2):442-452.