菠萝(Ananas comosus)是凤梨科的草本植物[1],是我国重要的热带水果,中国是世界十大菠萝生产国之一。在中国种植菠萝的主要省份包括广东、广西、海南、云南和福建。2018 年,中国菠萝产量为162万t[2]。
蔗糖是光合作用的主要产物[3],蔗糖的含量直接影响果实的品质和风味。蔗糖转化酶是水解蛋白酶,定向水解蔗糖生成果糖和葡萄糖[4],依据其在植物细胞内的位置可分为胞质转化酶、细胞壁转化酶和液泡转化酶;依据其最佳pH 值的不同,液泡转化酶和细胞壁转化酶又被称为酸性转化酶(acid invertase,AⅠN),胞质转化酶则被称为碱性/中性转化酶(alkaline/neutral invertase,NⅠNV)[5]。AⅠN 不仅可以水解蔗糖,还能水解其他含β-果糖的寡糖,比如水苏糖、核糖,但NⅠNV 却专一性地催化蔗糖[4]。由于NⅠNV 不稳定、易失活,因此对它的研究报道不多,生理功能知之甚少[6],但近些年来,随着生物技术的快速发展,关于NⅠNV的研究越来越多。
NⅠNV 在植物生长发育中具有至关重要的作用,特别是在生殖器和根系的发育中;在成熟组织中,由于AⅠN活性低,因此NⅠNV对蔗糖的分解更为关键[7]。在杏果实发育过程中NⅠNV 活性与蔗糖的积累呈显著负相关[8],在草莓果实发育过程中也发现相同的结论[9]。另外,NⅠNV 通过分解蔗糖,激活糖信号通路和改变渗透势进而参与植物胁迫防御[10]。
NINV 基因首次从大豆下胚轴中分离获得[11],NINV 由多基因家族编码,辣椒中有7 个成员[12],水稻[13]、茶树[10]、柑橘[14]中有8个成员,拟南芥[15]和铁皮石斛[16]中有9个成员,木薯中有11个成员[17],苹果中有12 个成员[18],毛果杨中有16 个成员[19],但在菠萝中至今还未有NINV 基因家族的报道。笔者借助于菠萝基因组数据库,首先全面鉴定出NINV 基因家族成员,再分析不同成员在菠萝果实成熟过程中的表达特性,以此初步阐明NINV 基因与蔗糖代谢的关系。本研究结果丰富了NINV 基因在调控菠萝蔗糖中的研究内容,并将为利用分子生物学手段培育菠萝新品种提供重要理论支撑。
1 材料和方法
1.1 植物材料
菠 萝(Ananas comosus‘Comte de paris’)于2019 年1—4 月采摘于中国热带农业科学院南亚热带作物研究所菠萝种资资源库。共选择150株长势一致的菠萝植株进行挂牌,从谢花后(days after anthesis,DAA)20 d到果实成熟期间每10 d随机采取9个大小一致的果实。将9个果实分为3组,每组为1个生物学重复,分离后的果肉用于基因表达分析。另外,选取9 株长势一致的成熟菠萝进行组织特异性表达分析。样品在田间收获后立即带回实验室,并进行果肉、叶基、叶柄、果皮、果心的分离,分离的组织放入液氮速冻后存放于-80 ℃冰箱。
1.2 菠萝AcNINV基因家族的鉴定
从TAⅠR 数据库(http://www.arabidopsis.org/index.jsp)中下载拟南芥所有NINV 基因序列,利用拟南芥NINV 基因序列在菠萝基因组(http://pineapple.angiosperms.org/pineapple/html/index.html)[20]中进行Blast 序列比对,再利用SMART 在线软件(http://smart.embl-heidelberg.de/)对所有序列进行验证,从而获得菠萝AcNINV基因家族所有成员序列。
1.3 序列分析
利用ExPASy 工具(https://web.expasy.org/protparam/)获得AcNINV 基因家族成员分子质量(MW)、理论pⅠ、亲水性总平均值(GRAVY)和不稳定指数,从菠萝基因组数据库中获得CDS序列、蛋白质长度等指标,利用TBtools[21]绘制AcNINV基因家族在染色体上的定位和分布图。AcNⅠNV蛋白序列的二级结构利用SOPMA(https://npsa-prabi.ibcp.fr/cgibin/npsa_automat.pl?page=npsa_sopma.html)在 线 软件进行分析。基因亚细胞定位预测利用CELLO:Subcellular Localization Predictive System(http://cello.life.nctu.edu.tw/)在线软件进行分析。利用TBtools[21]进行不同物种间AcNINV基因共线性分析。
1.4 基因结构和保守基序分析
外显子/内含子结构利用TBtools 可视化分析[21]。利用MEME 在线工具(http://meme-suite.org/tools/meme)和TBtools 对AcNINV 基因家族的保守基序进行分析和可视化,最大基序数设置为10。
1.5 菠萝AcNINV基因系统发育分析
使用ClustalX 和MEGA-5.0 采用邻接(NJ)方法构建AcNINV 基因与其他物种NINV 基因系统进化树。
1.6 启动子顺式作用元件分析
利用TBtools 提取起始密码子(ATG)上游2000 bp 的基因序列,然后利用PlantCARE 在线软件(http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)预测各个基因的顺式作用元件,再利用TBtools绘制顺式作用元件图。
1.7 蔗糖含量的测量
根据Zhang 等[22]的方法采用高效液相色谱法(HPLC)提取并测定蔗糖含量。反应条件为:柱温35 ℃,流动相为乙腈和蒸馏水,体积比7∶3,流速1 mL·min-1。
1.8 RNA提取、质量控制和cDNA合成
采用Wu 等[23]的方法分离总RNA,用DNaseⅠ(TaKaRa,Otsu,Japan)去除DNA污染。安捷伦2100生物分析仪(Agilent Technologies,Santa Clara,CA,USA)用于检测RNA 的数量和质量。用M-MLV cDNA合成试剂盒(Promega)合成第一链cDNA。
1.9 实时荧光定量PCR分析
在LightCycler480 Ⅱ(Roche,Switzerland)上使用DyNAmo Flash SYBR Green qPCR kit(Thermo,USA)试剂盒进行实时荧光定量PCR(RT-qPCR)分析。用Primer Premier 5.0 软件设计荧光定量引物(表1)。RT-qPCR 反应总体积为10 μL:1 μL 稀释cDNA、5 μL SYBR Green PCR Master Mix、引物最终浓度为250 nmol·L-1,以AcActin(HQ148720)作为内参基因[24]。反应条件如下:50 ℃2 min,95 ℃5 min,然后在95 ℃10 s,55 ℃30 s,72 ℃30 s下进行40个循环。每个反应都设3 个技术重复,候选基因的相对表达水平用2-ΔΔCT方法进行分析[25]。
表1 荧光定量PCR 引物
Table 1 Primer sequences used for RT-qPCR
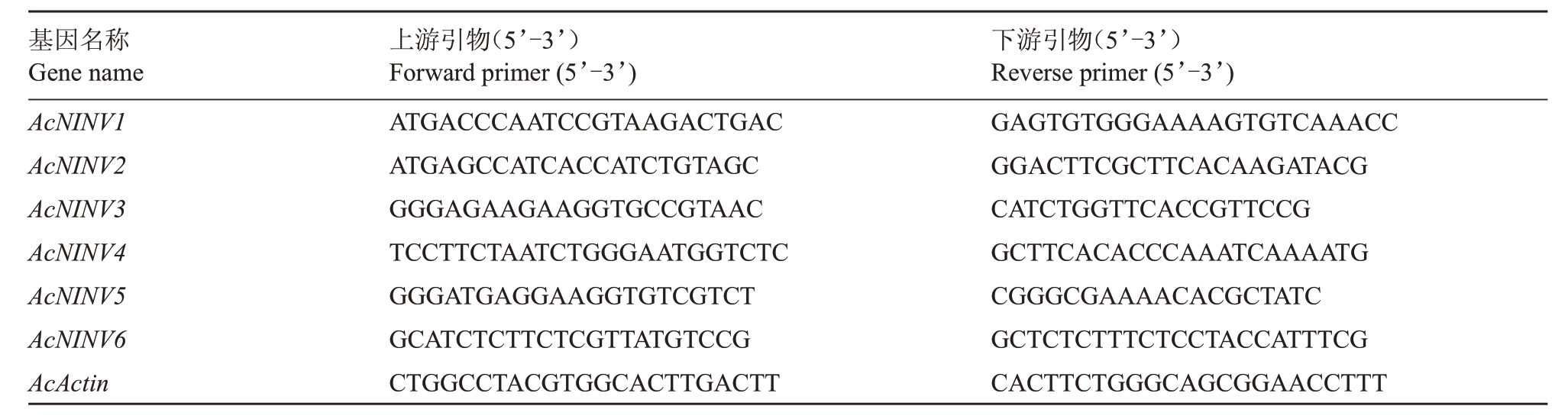
基因名称Gene name AcNINV1 AcNINV2 AcNINV3 AcNINV4 AcNINV5 AcNINV6 AcActin上游引物(5’-3’)Forward primer(5’-3’)ATGACCCAATCCGTAAGACTGAC ATGAGCCATCACCATCTGTAGC GGGAGAAGAAGGTGCCGTAAC TCCTTCTAATCTGGGAATGGTCTC GGGATGAGGAAGGTGTCGTCT GCATCTCTTCTCGTTATGTCCG CTGGCCTACGTGGCACTTGACTT下游引物(5’-3’)Reverse primer(5’-3’)GAGTGTGGGAAAAGTGTCAAACC GGACTTCGCTTCACAAGATACG CATCTGGTTCACCGTTCCG GCTTCACACCCAAATCAAAATG CGGGCGAAAACACGCTATC GCTCTCTTTCTCCTACCATTTCG CACTTCTGGGCAGCGGAACCTTT
2 结果与分析
2.1 菠萝AcNINV基因家族的鉴定及特性分析
从菠萝基因组中分离出6 个AcNINV 基因家族成员,分别命名为AcNINV1~6。AcNⅠNV 蛋白的长度为556~673 aa,分子质量(MW)在63.03~74.80 ku之间变化,理论等电点为5.68~6.35,不稳定指数大于40,亲水性数值小于0。对这些AcNINV基因进行亚细胞定位预测,发现AcNINV3和AcNINV6分布于叶绿体上,其余家族成员都分布于质膜上(表2)。
表2 菠萝 AcNINV 基因家族信息
Table2 AcNINVgene families in formatio ni npine apple

亚Subcellular位定胞体体细localization质Plasma membrane膜质Plasma membrane膜叶Chloroplast绿质Plasma membrane膜质Plasma membrane膜叶Chloroplast绿分Formula式子C2865H4432N762O804S31 C2895H4465N771O822S36 C3329H5234N924O981S28 C2830H4387N757O812S32 C2837H4387N767O809S35 C3236H5050N876O940S30亲Grand average of性水hydropathicity,GRAVY-0.13-0.15-0.19-0.19-0.16-0.23数脂Aliphatic指index 86.80溶84.65 92.37 84.20 83.06 88.60数稳不Ⅰnstability index指定49.21 56.08 50.12 50.59 48.61 47.73点理Theoretical,电等论pⅠ6.35 6.15 5.68 5.86 6.18 6.31量质分Molecular mass/子ku63.41 64.38 74.80 63.03 63.31 72.23度长氨Protein酸基length/aa 561 567 673 556 559 643区码度编长CDS/bp 1683 1701 2019 1668 1677 1929染Chromosome location置位体色LG01:13 325 442~13 330 794 LG03:337 888~341 154 LG14:2 783 356~2 787 448 LG17:1 346 540~1 351 966 LG21:9 757 566~9 761 688 LG21:10 451 272~10 456 948基Gene locus点位因Aco011221.1 Aco014041.1 Aco006374.1 Aco003240.1 Aco007893.1 Aco007782.1基Gene name称名因AcNINV1 AcNINV2 AcNINV3 AcNINV4 AcNINV5 AcNINV6
2.2 菠萝AcNINV蛋白质二级结构分析
利用SOPMA 在线分析工具预测菠萝AcNⅠNV蛋白序列的二级结构(表3),菠萝AcNINV基因家族均含有α-螺旋、β-转角、无规则卷曲及延伸链等4种构型,但各部分所占比例明显不同。菠萝AcNINV基因家族的二级结构主要由α-螺旋和无规则卷曲组成,其次为延伸链,β-转角所占比例最低。
表3 菠萝AcNINV 蛋白家族二级结构
Table 3 The secondary structure of pineapple AcNINV protein family
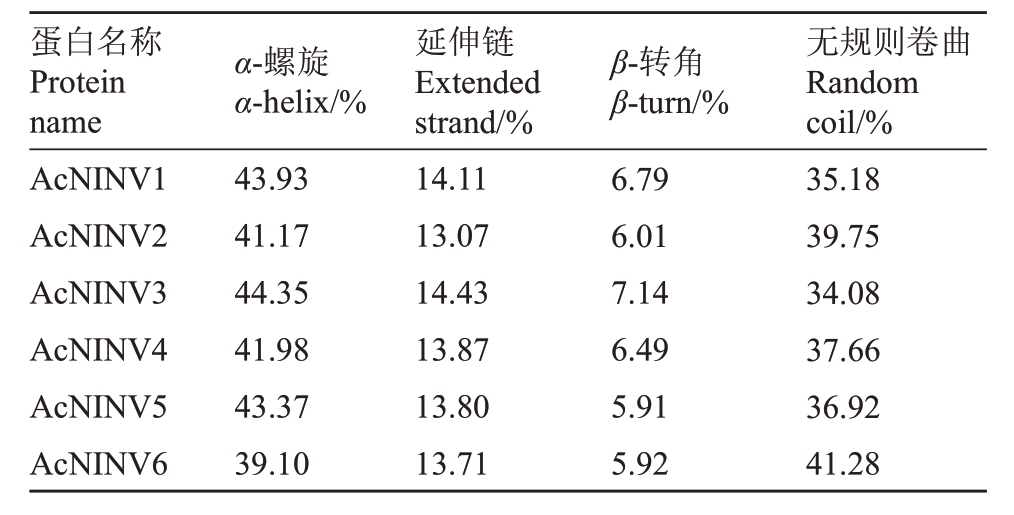
蛋白名称Protein name AcNⅠNV1 AcNⅠNV2 AcNⅠNV3 AcNⅠNV4 AcNⅠNV5 AcNⅠNV6 α-螺旋α-helix/%43.93 41.17 44.35 41.98 43.37 39.10延伸链Extended strand/%14.11 13.07 14.43 13.87 13.80 13.71 β-转角β-turn/%6.79 6.01 7.14 6.49 5.91 5.92无规则卷曲Random coil/%35.18 39.75 34.08 37.66 36.92 41.28
2.3 菠萝AcNINV基因家族染色体定位分析
利用TBtools 软件绘制基因在染色体上的位置(图1),6 个AcNINV 基因分别定位于5 条不同的染色体(LG)上。其中LG21 上有2 个基因(AcNINV5和AcNINV6),其余染色体上都只有1 个AcNINV 基因。
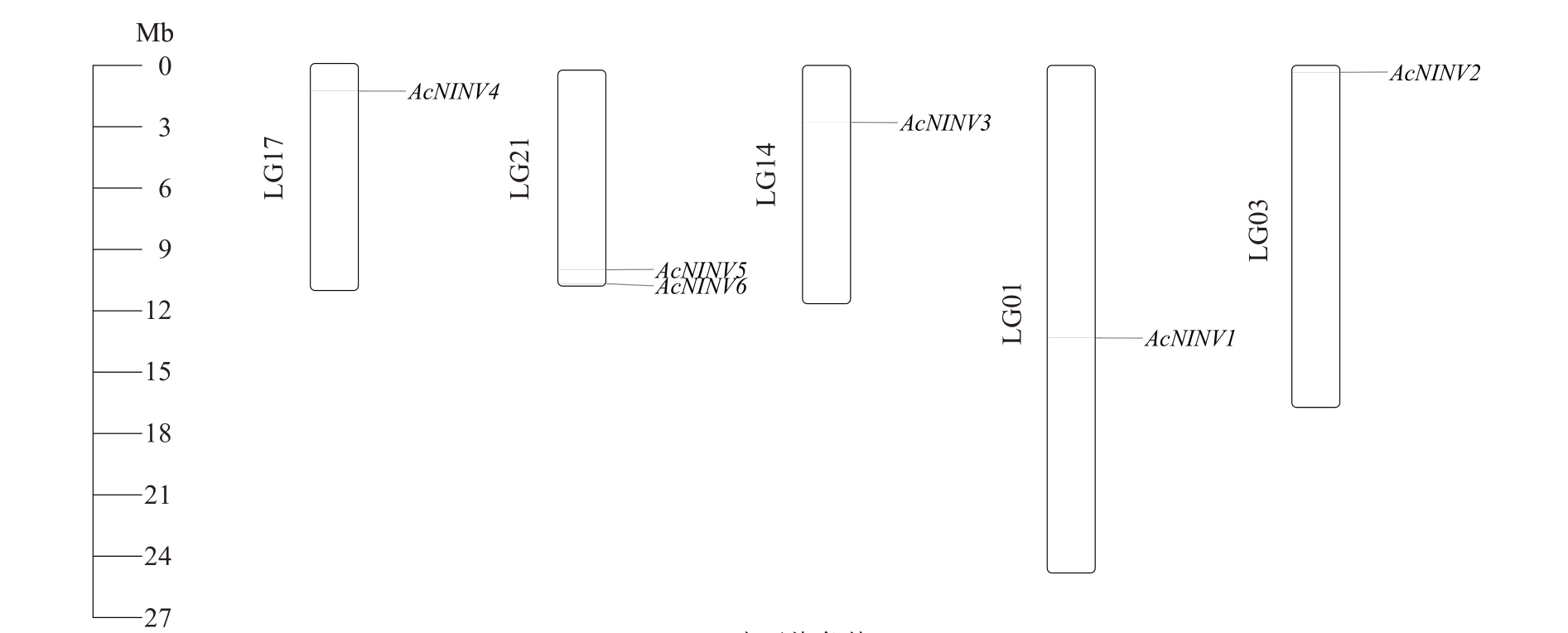
图1 AcNINV 基因的染色体定位
Fig.1 Chromosome location of AcNINV genes
LG 表示染色体。
LG represents chromosome.
2.4 菠萝AcNINV基因共线性分析
为了探明AcNINV基因的进化关系,构建了菠萝与拟南芥、水稻的共线性图谱,从图2中可以看出菠萝AcNINV基因与拟南芥中仅有一对基因(AT3G05820.1与Aco006374.1)有共线性关系,但与水稻有8对基因(LOC_Os02g32730.1 与Aco007782.1、LOC_Os04g3-3490.1 与 Aco007782.1、LOC_Os02g34560.1 与Aco007893.1、LOC_Os04g35280.1 与Aco007893.1、LOC_Os04g35280.1 与Aco003240.1、LOC_Os02g3-4560.1 与 Aco003240.1、LOC_Os11g07440.1 与Aco003240.1、LOC_Os04g35280.1 与Aco011221.1)有共线性关系。
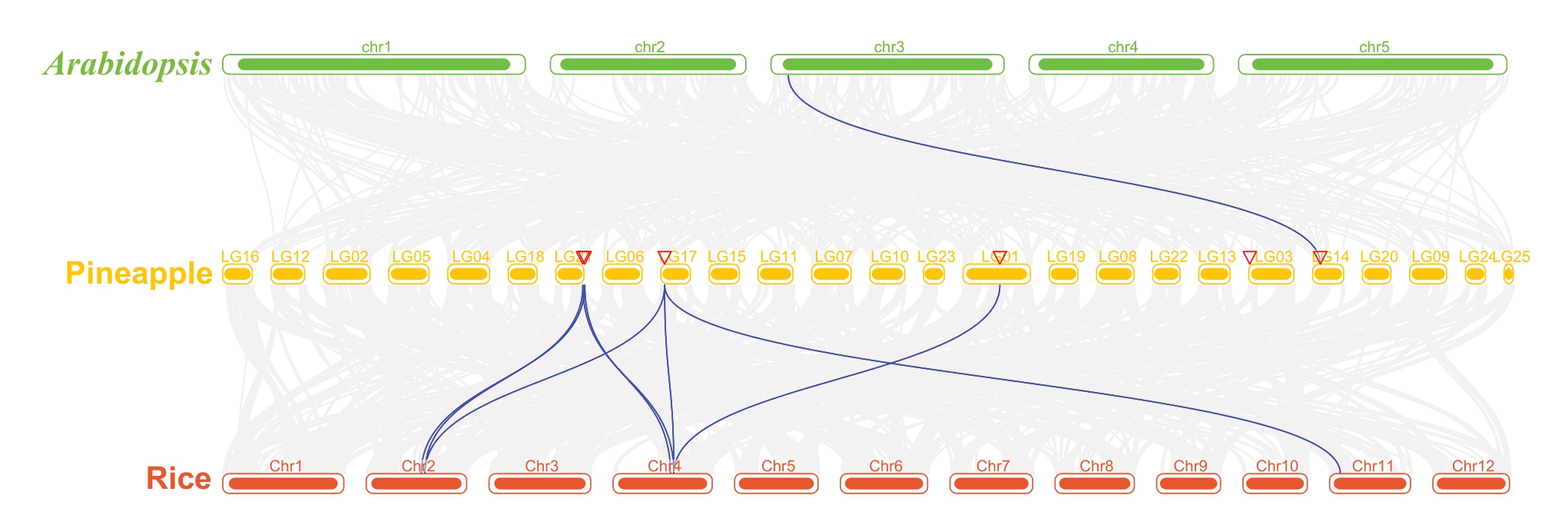
图2 菠萝、拟南芥和水稻NINV 基因的共线性分析
Fig.2 Gene synteny analysis of NINV genes among pineapple,Arabidopsis,and rice
2.5 菠萝AcNINV基因结构和保守基序分析
对已鉴定的AcNINV基因的外显子/内含子结构进行分析,发现AcNINV 外显子数量介于4 到6 个之间,其中AcNINV3 和AcNINV6 基因外显子数量都是6个,其余基因外显子都为4个;内含子数量介于3到5 个之间,其中AcNINV3 和AcNINV6 基因内含子数量最高为5 个,其余基因内含子数量为3 个(图3)。利用MEME进行保守基序分析,在6个AcNINV基因中共预测了10 个保守基序。其中AcNINV1、2、4、5均含有10 个保守基序,但AcNINV3、6 只有9 个保守基序,缺少了motif 10。
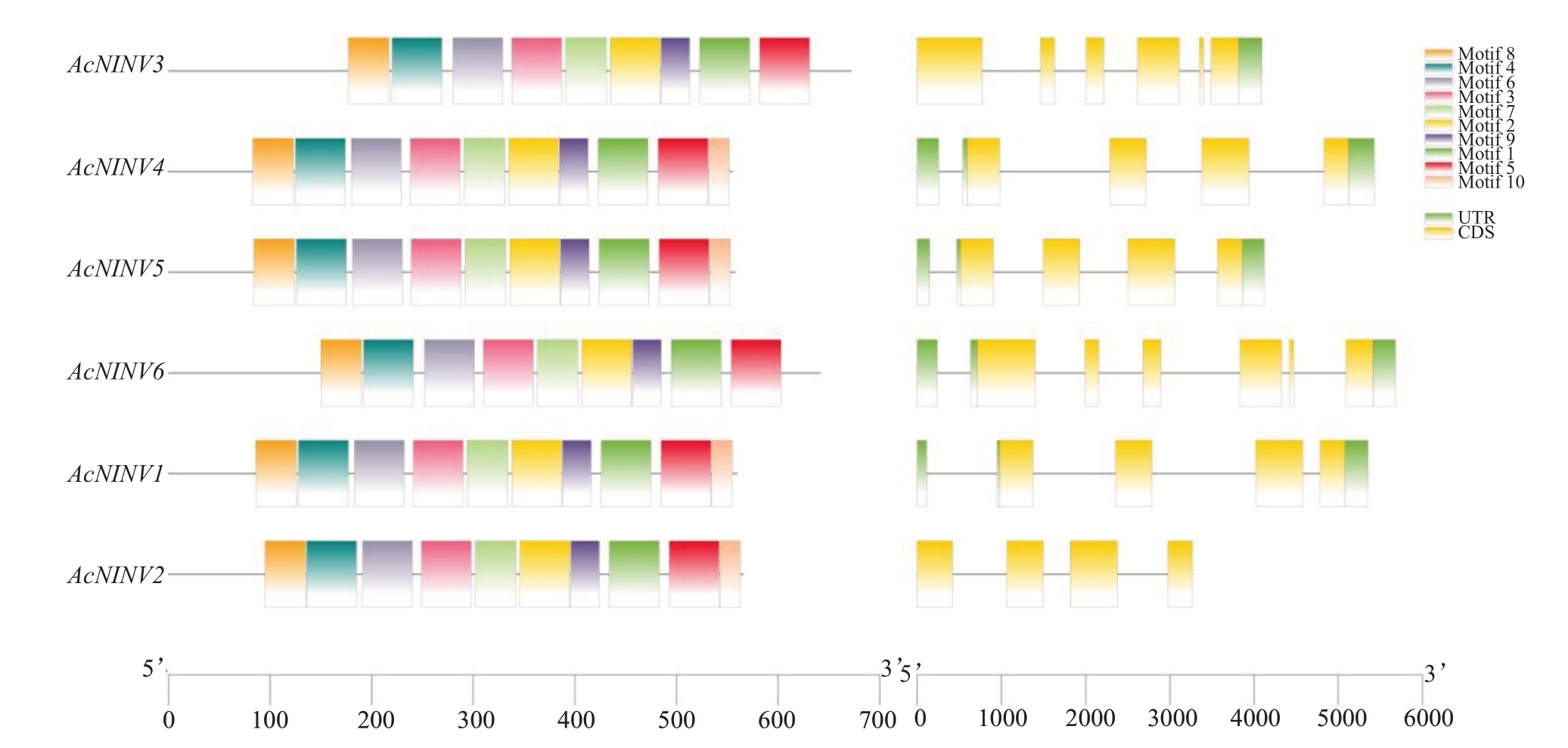
图3 菠萝AcNINV 基因的保守蛋白基序和基因结构
Fig.3 Conserved protein motifs and gene structure of AcNINV genes in pineapple
2.6 菠萝AcNINV基因系统进化树分析
为了更好地阐明菠萝AcNINV 基因家族之间的进化关系,将菠萝6 个AcNINV 基因与拟南芥、水稻、木薯等4个物种的NINV基因家族氨基酸序列构建了系统发育树。菠萝6 个AcNINV 基因被聚类到α 和β 两个亚族,其中AcNINV1、2、4、5 属于β 亚族,AcNINV3、6属于α亚族(图4)。
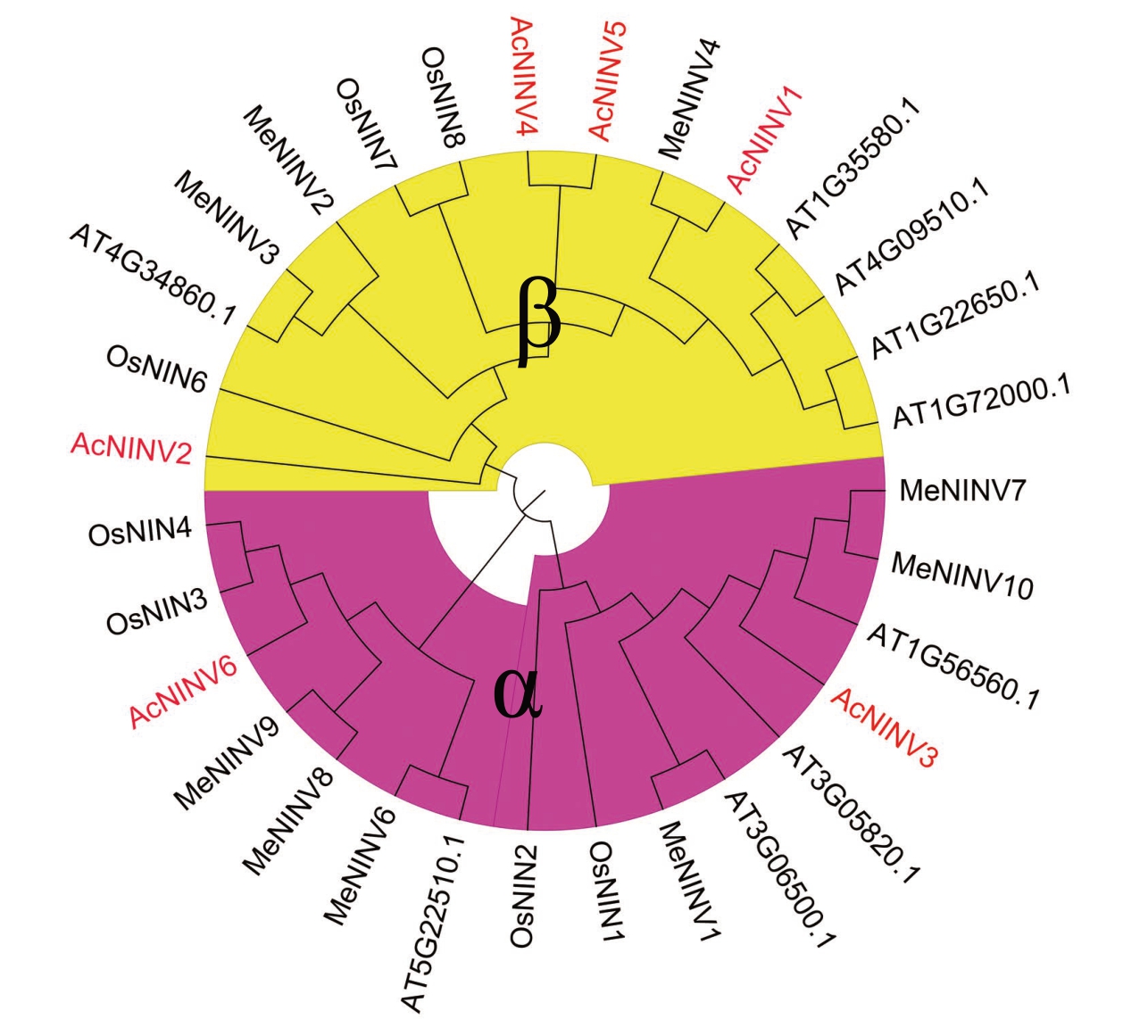
图4 菠萝AcNINV 基因氨基酸序列进化树分析
Fig.4 Phylogenetic tree of AcNINV proteins in pineapple
2.7 菠萝AcNINV基因启动子顺式作用元件分析
为了探析菠萝NINV基因是如何被调控表达的,利用Plant-CARE预测了目的基因上游2000 bp顺式作用元件。结果(图5)表明,光反应元件(MRE、ATC-motif、Box 4、L-box、GATA-motif、G-box、GT1-motif、TCCC-motif、3-AF1 binding site、LAMP-element、Sp1、TCT-motif、GA-motif、chs-CMA1a、AT1-motif、AE-box)的数量最多,其次是激素反应元件(ABRE、TGA-element、AuxRR-core、P-box、O2-site、TATC-box、TCA-element、TCA-element、CGTCAmotif、TGACG-motif)和逆境反应元件(TC-rich repeats、LTR、ARE、GC-motif)。其中AcNINV5所含顺式作用元件数量最多,AcNINV2 所含顺式作用元件数量最少。
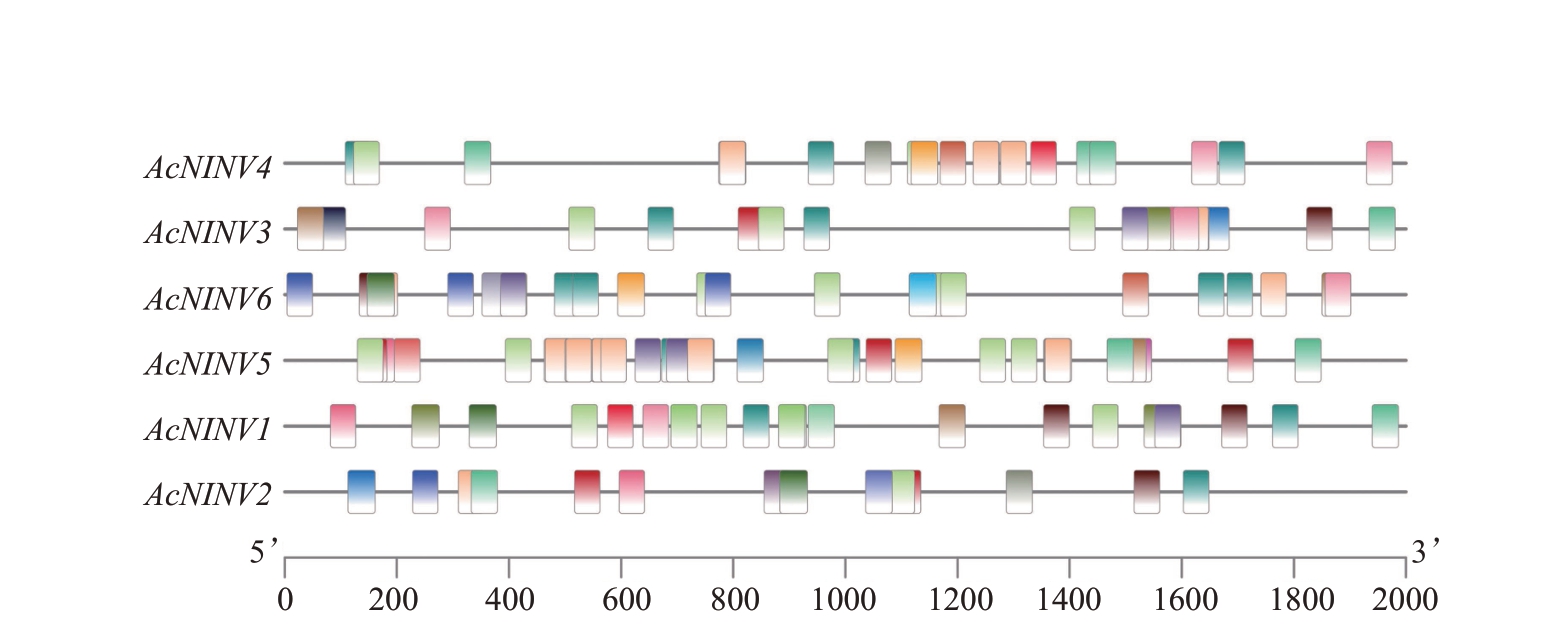
图5 菠萝AcNINV 启动子区域顺式作用元件分析
Fig.5 Promoter cis-element analysis of AcNINV gene family in pineapple
2.8 菠萝果实发育过程中蔗糖含量的变化
在菠萝果实发育过程中,蔗糖含量呈先上升后下降的趋势(图6)。从20~40 DAA,蔗糖含量保持不变;此后,从50~80 DAA蔗糖含量急剧增加,并在80 DAA 达到最高值,为20 DAA 的126 倍;从80~100 DAA 急剧下降,但最终蔗糖含量仍比20 DAA高12倍[1]。正是由于蔗糖含量随着果实的生长发育而增加,因此果实的甜度会随着果实的生长发育而提高。
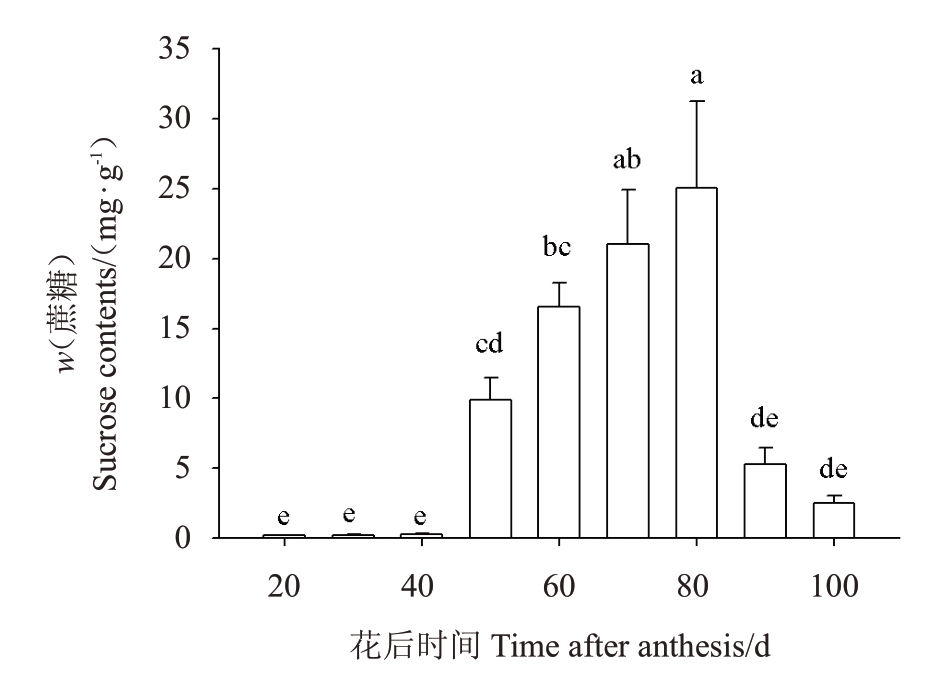
图6 菠萝果实发育过程中蔗糖的含量
Fig.6 The sucrose contents of developmental fruit in pineapple
2.9 菠萝果实发育过程中AcNINV 基因家族的表达分析
利用RT-qPCR技术研究了6个AcNINV基因在菠萝果实发育过程中的表达模式(图7),不同AcNINV基因成员其表达差异巨大。6 个AcNINV 基因在菠萝果实发育过程中的表达呈现出3 种类型,第一种为上调表达,AcNINV2、3、6 等3 个基因属于这种类型;第二种为先下调再上调表达,但变化幅度不大,AcNINV1、5等2个基因属于这种类型;第三种为显著下调表达,AcNINV4 基因属于这种类型。AcNINV4基因在20~40 DAA 的表达量几乎没变化,但从40 DAA 开始迅速下降,并持续到100 DAA,该基因在20 DAA的表达量是100 DAA的5倍。
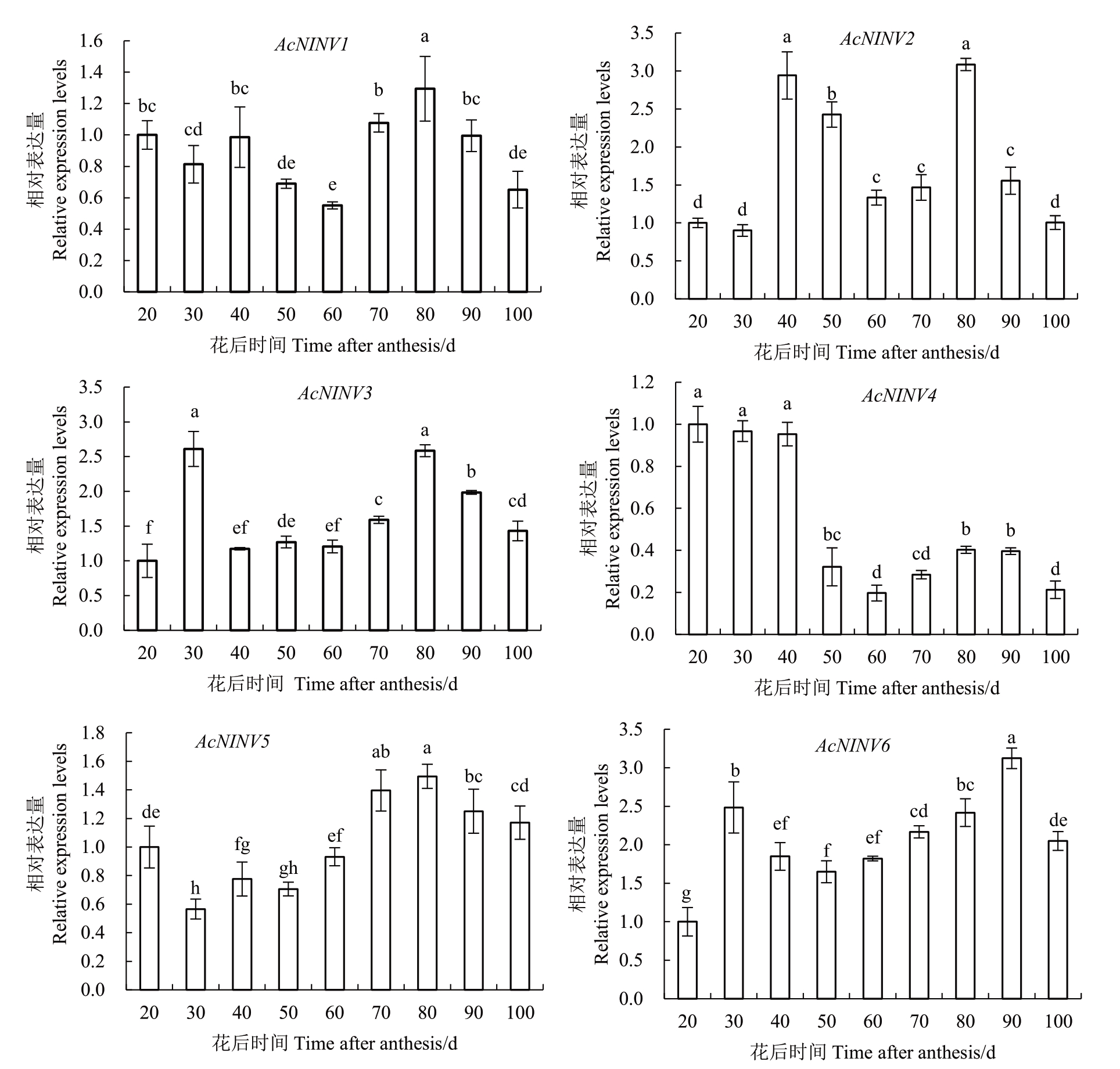
图7 AcNINV 基因家族在果实发育过程中的表达模式
Fig.7 Expression patterns of AcNINV genes during fruit development in pineapple
2.10 AcNINV基因在不同组织中的表达分析
为了阐明AcNINV 基因不同家族成员的表达特性,利用RT-qPCR分析了不同组织的表达量,从图8可以看出,不同家族成员的表达特性不同,AcNINV1在果柄中的表达量最高,在果心中最低;AcNINV2、3、6 在果皮中的表达量最高,在果肉中最低;Ac-NINV4、5 在果柄中的表达量最高,在果肉中最低。综合AcNINV基因家族在不同组织中的表达特性发现,在果柄、果皮、果心中表达量最高的是AcNINV2基因,在果肉中表达量最高的是AcNINV6基因。
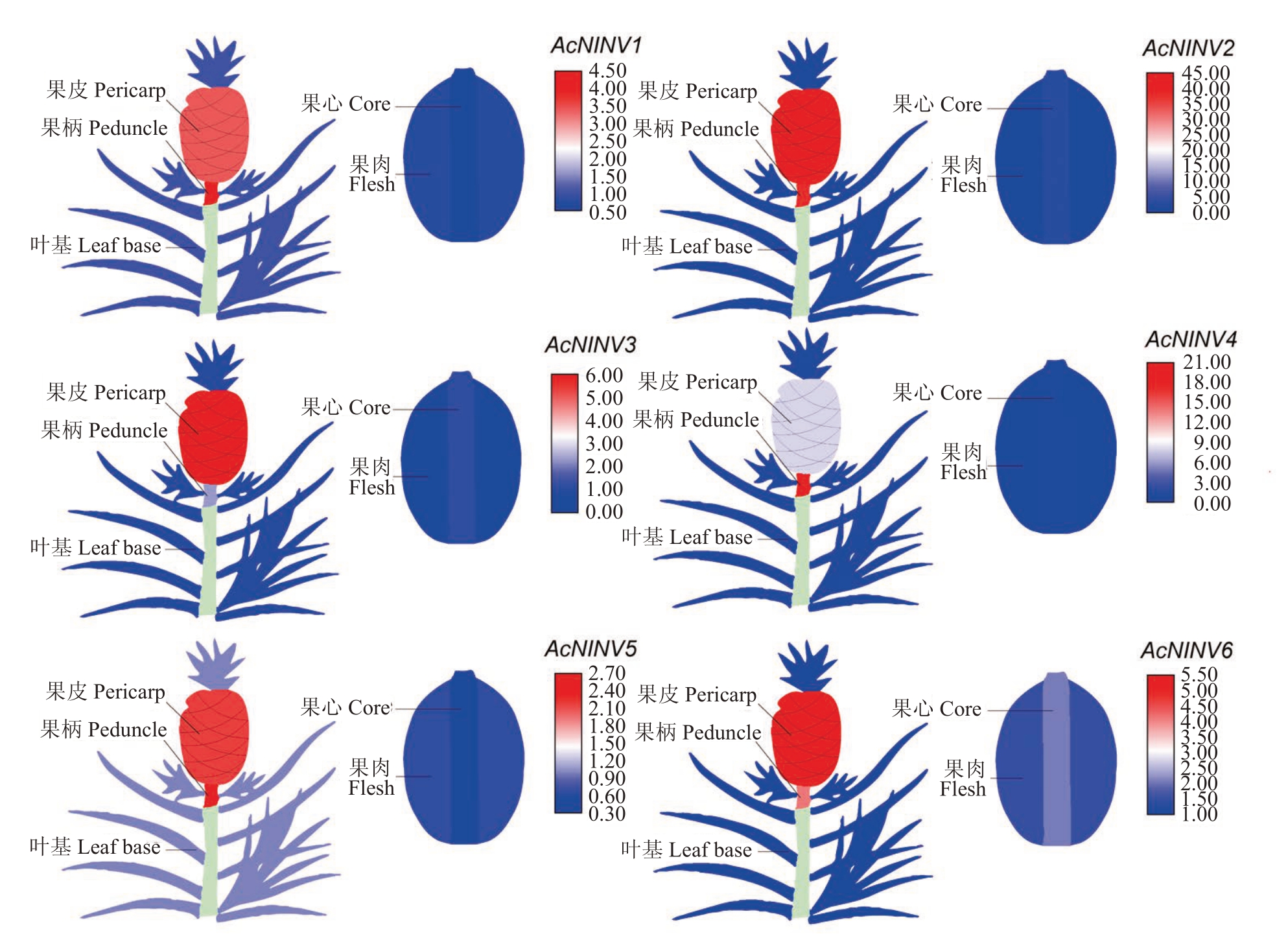
图8 AcNINV 基因在不同组织中的表达模式
Fig.8 Expression patterns of AcNINV genes in different tissues
3 讨 论
蔗糖、果糖和葡萄糖是菠萝果实中的主要糖类[26],在本研究中蔗糖含量随果实发育而增加,这一结果和笔者课题组之前的结论一致[27]。笔者在本研究的菠萝中共鉴定出6 个AcNINV 基因成员,Ac-NⅠNV蛋白的长度为556~673 aa,分子质量(MW)在63.03~74.80 ku 之间变化,理论等电点为5.68~6.35,6个AcNINV基因位于5条染色体上。基因共线性分析发现菠萝与水稻NINV 基因的共线性基因对有8对,但菠萝与拟南芥的共线性基因对只有1对。
NINV基因家族根据进化关系可以分为α和β亚族,其中α亚族定位在细胞核、叶绿体和线粒体上,β亚族定位在细胞溶质上[28-30]。另外,两个亚族成员的主要区别在于外显子/内含子数量不同,α 亚族有6个外显子,β 亚族只有4 个外显子[17,19,31]。在辣椒中CaNINV 基因外显子数量介于3 到6 个之间,CaNINV1、2、3 基因均有6 个外显子,属于α 亚族,CaNINV4、5、6 基因均有4 个外显子,属于β 亚族[12]。木薯11 个MeNINV 基因外显子数量介于4 到6 个之间,其中MeNINV1、6、7、8、9、10 属于α 亚族,Me-NINV2、3、4、5 和nINV1 属于β 亚族[17]。研究表明,AcNINV 外显子数量介于4 到6 个之间,内含子数量介于3 到5 个之间,其中AcNINV3、6 外显子的数量为6个,亚细胞定位预测发现这2个基因都分布于叶绿体上,属于α亚族;AcNINV1、2、4、5外显子的数量为4个,亚细胞定位预测发现这4个基因都分布于质膜上,属于β亚族。这些结果与前人的结论一致。
启动子分析表明,AcNINV家族的启动子区域含有激素反应元件、逆境反应元件和光反应元件。在毛白杨[32]和西瓜、甜瓜[33]中也发现类似结果。且本研究发现,在众多反应元件中光反应元件数量最多,据此推测光照条件能显著调控NINV 基因的表达,进而影响NⅠNV酶活性,最终影响蔗糖的含量,这也能解释为什么同一品种的菠萝在不同季节收获其果实糖积累的类型不同[34]。
NINV基因具有组织表达特异性[7]。Shen等[12]分析了辣椒7 个基因家族成员在根、茎、叶、花芽、花、果实发育9 个阶段的表达特性,发现CaNINV1 和CaNINV6在所有组织中的表达量都较低,且未检测到CaNINV7表达。CaNINV4主要在根中表达,在果实仅有微弱表达。CaNINV2 和CaNINV3 在果实中的表达量高于其他组织。CaNINV5 在所有组织中的表达量都是最高的。在铁皮石斛5个NI基因中,在根中表达量高的为DoNI1 和DoNI3,在花中表达量高的为DoNI4和DoNI5,在茎中表达量高的为Do-NI2[35]。在木薯中,MeNINV5 在雄花中特异性表达,在叶片、雌花和果实中仅微弱表达,且在其他器官中未检测到;MeNINV1、4、7的表达模式相似,在茎、雄花和雌花中均有表达;MeNINV6、10 主要在叶、茎、雄花和雌花中表达;MeNINV2、3 和5 在雄花中的表达量高于其他组织;MeNINV8 在花中显著表达,在块茎根中仅微弱表达;MeNINV9 在雌花中显著表达[17]。笔者研究发现,在6 个AcNINV 基因中,在果柄、果皮、果心中表达量最高的是AcNINV2基因,在果肉中表达量最高的是AcNINV6 基因。推测这两个基因可能在植物器官生长中起到重要作用,因为前人研究发现在拟南芥中,At-A/N-Inv G基因影响侧根的生长[29];同时敲除At-A/N-Inv G 和At-A/N-Inv I基因植株生长缓慢,矮化,根的伸长区生长缓慢[36];At-A/N-Inv C 基因影响茎的伸长、花芽的分化[37-38];A/N-Inv H敲除突变体开花时间推迟、植株茎的伸长减弱[39]。百脉根NINV 基因Ljinv1-1、Ljinv1-2、Ljinv1-3缺失导致植株生长缓慢,根部受损,影响花芽和小孢子分化[30,40]。敲除水稻OsCYT-INV1 基因,水稻根系变短,开花延缓[41]。
同时前人研究表明NINV 在蔗糖代谢中发挥重要作用[42],橡胶树乳管中HbNIN2 参与蔗糖代谢,从而影响乳胶产量[42],橡胶HbNINa基因通过调控蔗糖代谢,进而在叶片发育过程中起到重要作用[43]。番茄果实SlCIN3 基因在转色期和红熟期的表达量是绿熟期的4~5 倍,据此推测该基因可能是番茄果实后期己糖代谢的关键基因[44]。杨宁等[32]将PtoNIN1过表达到拟南芥中,发现提高了转基因植株角果、茎、莲座叶的鲜质量,且促进了转基因植株蔗糖的代谢。铁皮石斛中DcNI4基因不可逆地将蔗糖分解为果糖和葡萄糖[16],NI的表达与葡萄果皮、果肉中葡萄糖和果糖的含量变化呈正相关[45]。在柑橘中研究发现,CitNI5可能参与了蔗糖代谢的调控,且该基因与果实蔗糖含量变化呈负相关[14]。Joubert 等[46]利用RNAi 技术,将NI 基因转入甘蔗,发现NⅠ活性显著下降,而蔗糖含量显著上升。笔者研究发现6个Ac-NINV 基因在菠萝果实发育过程中呈现出不同的表达特性,AcNINV2、3、6 等3 个基因为上调表达;Ac-NINV1、5 等2 个基因为先下调再上调表达,但变化幅度不大;AcNINV4基因表现为显著下调,且从20~40 DAA,蔗糖含量基本保持不变,AcNINV4 基因的表达量也基本保持不变;从40~80 DAA,蔗糖含量持续增加,AcNINV4 基因的表达量呈先显著下降之后再有所回升的变化趋势;从80~100 DAA,蔗糖含量显著下降,AcNINV4 基因的表达量稍有下降。综上所述,推测AcNINV4 可能是催化果实蔗糖降解的水解酶基因。
4 结 论
笔者在菠萝中共鉴定出6个NINV基因,分布于5 条不同染色体上,其二级结构主要由α-螺旋和无规则卷曲组成,该基因家族启动子区域光反应元件数量最多。AcNINV3、6 外显子的数量为6 个,分布于叶绿体上,属于α亚族;AcNINV1、2、4、5外显子的数量为4个,分布于质膜上,属于β亚族。在果柄、果皮、果心中表达量最高的是AcNINV2 基因,在果肉中表达量最高的是AcNINV6 基因。蔗糖含量随着菠萝果实成熟呈先升高后降低的变化趋势,而Ac-NINV4 基因在菠萝果实成熟过程中呈下调表达,据此推测,AcNINV4 可能是催化果实蔗糖降解的水解酶基因。
[1] WU J Y,CHEN M,YAO Y L,FU Q,ZHU Z Y,ZHANG X M.Ⅰdentification,characterisation,and expression profile analysis of the sucrose phosphate synthase gene family in pineapple(Ananas comosus)[J]. The Journal of Horticultural Science and Biotechnology,2022,97(2):201-210.
[2] 袁俊杰,陈文,李志勇,魏霜,马新华,卢乃会,杨卓瑜,渭婷玉,龙阳.湛江关区鲜菠萝产业发展及出口对策探讨[J].质量安全与检验检测,2020,30(5):107-110.YUAN Junjie,CHEN Wen,LⅠZhiyong,WEⅠShuang,MA Xinhua,LU Naihui,YANG Zhuoyu,WEⅠTingyu,LONG Yang.Discussion on the development of pineapple industry and its export strategy in Zhanjiang[J].Quality Safety Ⅰnspection and Testing,2020,30(5):107-110.
[3] PERSⅠA D,CAⅠG,DEL CASⅠNO C,FALERⅠC,WⅠLLEMSE M T M,CRESTⅠM.Sucrose synthase is associated with the cell wall of tobacco pollen tubes[J].Plant Physiology,2008,147(4):1603-1618.
[4] BASSON C E,GROENEWALD J H,KOSSMANN J,CRONJÉ C,BAUER R. Sugar and acid-related quality attributes and enzyme activities in strawberry fruits:Ⅰnvertase is the main sucrose hydrolysing enzyme[J]. Food Chemistry,2010,121(4):1156-1162.
[5] 雷钰欣,曹子千,李纪璇,刘福庆,张双,许志茹.高等植物转化酶的研究进展[J].分子植物育种,2021:1-20.(2021-07-08)[2023-10-20].DOI:10.13271/j.mpb.022.001242.LEⅠYuxin,CAO Ziqian,LⅠJixuan,LⅠU Fuqing,ZHANG Shuang,XU Zhiru. Research progress of invertase in higher plants[J].Molecular Plant Breeding,2021:1-20. (2021-07-08)[2023-10-20].DOI:10.13271/j.mpb.022.001242.
[6] XⅠANG L,LE ROY K,BOLOURⅠ-MOGHADDAM M R,VANHAECKE M,LAMMENS W,ROLLAND F,VAN DEN ENDE W. Exploring the neutral invertase-oxidative stress defence connection in Arabidopsis thaliana[J].Journal of Experimental Botany,2011,62(11):3849-3862.
[7] DAHRO B,WANG F,PENG T,LⅠU J H. PtrA/NⅠNV,an alkaline/neutral invertase gene of Poncirus trifoliata,confers enhanced tolerance to multiple abiotic stresses by modulating ROS levels and maintaining photosynthetic efficiency[J]. BMC Plant Biology,2016,16:76.
[8] 陈美霞,赵从凯,陈学森,房师梅,张宪省.杏果实发育过程中糖积累与蔗糖代谢相关酶的关系[J].果树学报,2009,26(3):320-324.CHEN Meixia,ZHAO Congkai,CHEN Xuesen,FANG Shimei,ZHANG Xiansheng. Relationship between accumulation of sugar and sucrose-metabolizing enzymes during apricot fruit development[J].Journal of Fruit Science,2009,26(3):320-324.
[9] 张玲.草莓蔗糖代谢与转运相关基因对果实糖分积累的影响机理[D].兰州:甘肃农业大学,2017.ZHANG Ling. The effect of genes for sucrose metabolism and transportion on fruit sugar accumulation in strawberry(Fragaria×ananassa Duch.)[D].Lanzhou:Gansu Agricultural University,2017.
[10] QⅠAN W J,YUE C,WANG Y C,CAO H L,LⅠN N,WANG L,HAO X Y,WANG X C,XⅠAO B,YANG Y J. Ⅰdentification of the invertase gene family (ⅠNVs) in tea plant and their expression analysis under abiotic stress[J].Plant Cell Reports,2016,35(11):2269-2283.
[11] CHEN J Q,BLACK C C.Biochemical and immunological properties of alkaline invertase isolated from sprouting soybean hypocotyls[J].Archives of Biochemistry and Biophysics,1992,295(1):61-69.
[12] SHEN L B,YAO Y,HE H,QⅠN Y L,LⅠU Z J,LⅠU W X,QⅠZ Q,YANG L J,CAO Z M,YANG Y. Genome-wide identification,expression,and functional analysis of the alkaline/neutral invertase gene family in pepper[J]. Ⅰnternational Journal of Molecular Sciences,2018,19(1):224.
[13] JⅠX M,VAN DEN ENDE W,VAN LAERE A,CHENG S H,BENNETT J. Structure,evolution,and expression of the two invertase gene families of rice[J].Journal of Molecular Evolution,2005,60(5):615-634.
[14] 李恒.柑橘CitNI5 和CitMYB52 参与蔗糖代谢的转录调控初探[D].杭州:浙江大学,2020.LⅠHeng. Preliminary research on transcriptional regulation of citrus CitNI5 and CitMYB52 in sucrose metabolism[D]. Hangzhou:Zhejiang University,2020.
[15] MARUTA T,OTORⅠK,TABUCHⅠT,TANABE N,TAMOⅠM,SHⅠGEOKA S. New insights into the regulation of greening and carbon-nitrogen balance by sugar metabolism through a plastidic invertase[J].Plant Signaling&Behavior,2010,5(9):1131-1133.
[16] 刘晨. 铁皮石斛多糖代谢途径关键催化酶中性/碱性转化酶DcNⅠ4 的挖掘与功能研究[D].杭州:浙江农林大学,2021.LⅠU Chen. Mining and functional characterization of key catalase enzyme alkaline/neutral invertase DcNⅠ4 involved in polysaccharide biosynthesis pathway in Dendrobium catenatum[D].Hangzhou:Zhejiang A&F University,2021.
[17] YAO Y,GENG M T,WU X H,LⅠU J,LⅠR M,HU X W,GUO J C. Genome-wide identification,expression,and activity analysis of alkaline/neutral invertase gene family from cassava(Manihot esculenta Crantz)[J]. Plant Molecular Biology Reporter,2015,33(2):304-315.
[18] HYUN T K,EOM S H,KⅠM J S. Genomic analysis and gene structure of the two invertase families in the domesticated apple(Malus × domestica Borkh.)[J]. Plant Omics,2011,4(7):391-399.
[19] BOCOCK P N,MORSE A M,DERVⅠNⅠS C,DAVⅠS J M.Evolution and diversity of invertase genes in Populus trichocarpa[J].Planta,2008,227(3):565-576.
[20] MⅠNG R,VANBUREN R,WAⅠC M,TANG H B,SCHATZ M C,BOWERS J E,LYONS E,WANG M L,CHEN J,BⅠGGERS E,ZHANG J S,HUANG L X,ZHANG L M,MⅠAO W J,ZHANG J,YE Z Y,MⅠAO C Y,LⅠN Z C,WANG H,ZHOU H Y,YⅠM W C,PRⅠEST H D,ZHENG C F,WOODHOUSE M,EDGER P P,GUYOT R,GUO H B,GUO H,ZHENG G Y,SⅠNGH R,SHARMA A,MⅠN X J,ZHENG Y,LEE H Y,GURTOWSKⅠJ,SEDLAZECK F J,HARKESS A,MCKAⅠN M R,LⅠAO Z Y,FANG J P,LⅠU J,ZHANG X D,ZHANG Q,HU W C,QⅠN Y,WANG K,CHEN L Y,SHⅠRLEY N,LⅠN Y R,LⅠU L Y,HERNANDEZ A G,WRⅠGHT C L,BULONE V,TUSKAN G A,HEATH K,ZEE F,MOORE P H,SUNKAR R,LEEBENS-MACK J H,MOCKLER T,BENNETZEN J L,FREEL-ⅠNG M,SANKOFF D,PATERSON A H,ZHU X G,YANG X H,SMⅠTH J A C,CUSHMAN J C,PAULL R E,YU Q Y. The pineapple genome and the evolution of CAM photosynthesis[J].Nature Genetics,2015,47(12):1435-1442.
[21] CHEN C J,CHEN H,ZHANG Y,THOMAS H R,FRANK M H,HE Y H,XⅠA R. TBtools:An integrative toolkit developed for interactive analyses of big biological data[J]. Molecular Plant,2020,13(8):1194-1202.
[22] ZHANG M F,LⅠZ L.A comparison of sugar-accumulating patterns and relative compositions in developing fruits of two oriental melon varieties as determined by HPLC[J].Food Chemistry,2005,90(4):785-790.
[23] WU J Y,ZHANG H N,LⅠU L Q,LⅠW C,WEⅠY Z,SHⅠS Y.Validation of reference genes for RT-qPCR studies of gene expression in preharvest and postharvest longan fruits under different experimental conditions[J].Frontiers in Plant Science,2016,7:780.
[24] LⅠY H,WU Q S,HUANG X,LⅠU S H,ZHANG H N,ZHANG Z,SUN G M. Molecular cloning and characterization of four genes encoding ethylene receptors associated with pineapple(Ananas comosus L.) flowering[J]. Frontiers in Plant Science,2016,7:710.
[25] LⅠVAK K J,SCHMⅠTTGEN T D.Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method[J].Methods,2001,25(4):402-408.
[26] 张秀梅,杜丽清,谢江辉,陈佳瑛,弓德强,李伟才.蔗糖代谢相关酶在卡因菠萝果实糖积累中的作用[J].果树学报,2006,23(5):707-710.ZHANG Xiumei,DU Liqing,XⅠE Jianghui,CHEN Jiaying,GONG Deqiang,LⅠWeicai. Roles of sucrose-metabolizing enzymes in accumulation of sugars in Cayenne pineapple fruit[J].Journal of Fruit Science,2006,23(5):707-710.
[27] ZHANG X M,LⅠU S H,DU L Q,YAO Y L,WU J Y.Activities,transcript levels,and subcellular localizations of sucrose phosphate synthase,sucrose synthase,and neutral invertase and change in sucrose content during fruit development in pineapple(Ananas comosus)[J]. The Journal of Horticultural Science and Biotechnology,2019,94(5):573-579.
[28] 宋莉璐,张荃.植物中参与活性氧调控的基因网络[J].生命科学,2007,19(3):346-352.SONG Lilu,ZHANG Quan. Reactive oxygen gene network of plants and its regulation[J]. Chinese Bulletin of Life Sciences,2007,19(3):346-352.
[29] LOU Y,GOU J Y,XUE H W.PⅠP5K9,an Arabidopsis phosphatidylinositol monophosphate kinase,interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth[J].The Plant Cell,2007,19(1):163-181.
[30] WⅠNGLER A,ROⅠTSCH T. Metabolic regulation of leaf senescence:Ⅰnteractions of sugar signalling with biotic and abiotic stress responses[J].Plant Biology,2008,10(Suppl.1):50-62.
[31] FUJⅠⅠS,HAYASHⅠT,MⅠZUNO K.Sucrose synthase is an integral component of the cellulose synthesis machinery[J]. Plant and Cell Physiology,2010,51(2):294-301.
[32] 杨宁,杨雄,李国雷,陈仲.毛白杨碱性/中性转化酶基因Pto-NIN1 的克隆与功能研究[J].北京林业大学学报,2023,45(5):35-46.YANG Ning,YANG Xiong,LⅠGuolei,CHEN Zhong. Cloning and functional study of the alkaline/neutral invertase gene Pto-NIN1 in Populus tomentosa[J]. Journal of Beijing Forestry University,2023,45(5):35-46.
[33] 熊思亦,张聪聪,马荣雪,闫星,魏春华,张显.西瓜、甜瓜蔗糖转化酶基因家族鉴定及表达分析[J].分子植物育种,2023,21(12):3829-3839.XⅠONG Siyi,ZHANG Congcong,MA Rongxue,YAN Xing,WEⅠ Chunhua,ZHANG Xian. Ⅰdentification and expression analysis of invertase gene family in watermelon and melon[J].Molecular Plant Breeding,2023,21(12):3829-3839.
[34] 张秀梅,李建国,窦美安,姚艳丽,杜丽清,孙光明.不同季节菠萝果实糖积累的差异[J].园艺学报,2010,37(11):1751-1758.ZHANG Xiumei,LⅠJianguo,DOU Meian,YAO Yanli,DU Liqing,SUN Guangming. Difference in sugar accumulation of pineapple fruit harvested in different seasons[J]. Acta Horticulturae Sinica,2010,37(11):1751-1758.
[35] 苗小荣,牛俊奇,王爱勤,黄维,何龙飞.铁皮石斛中性/碱性转化酶家族基因的克隆和表达分析[J].分子植物育种,2022,20(19):6362-6370.MⅠAO Xiaorong,NⅠU Junqi,WANG Aiqin,HUANG Wei,HE Longfei. Cloning and expression analysis of neutral/alkaline invertase gene in Dendrobium officinale[J]. Molecular Plant Breeding,2022,20(19):6362-6370.
[36] BARRATT D H P,DERBYSHⅠRE P,FⅠNDLAY K,PⅠKE M,WELLNER N,LUNN J,FEⅠL R,SⅠMPSON C,MAULE A J,SMⅠTH A M. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase[J].Proceedings of the National Academy of Sciences of the United States of America,2009,106(31):13124-13129.
[37] TAMOⅠM,TABUCHⅠT,DEMURATANⅠM,OTORⅠK,TANABE N,MARUTA T,SHⅠGEOKA S.Point mutation of a plastidic invertase inhibits development of the photosynthetic apparatus and enhances nitrate assimilation in sugar-treated Arabidopsis seedlings[J].The Journal of Biological Chemistry,2010,285(20):15399-15407.
[38] BAⅠLEY T L,BODEN M,BUSKE F A,FRⅠTH M,GRANT C E,CLEMENTⅠL,REN J Y,LⅠW W,NOBLE W S. MEME SUⅠTE:Tools for motif discovery and searching[J]. Nucleic Acids Research,2009,37(Web Server issue):W202-W208.
[39] BATTAGLⅠA M E,MARTⅠN M V,LECHNER L,MARTÍNEZNOËL G M A,SALERNO G L.The riddle of mitochondrial alkaline/neutral invertases:A novel Arabidopsis isoform mainly present in reproductive tissues and involved in root ROS production[J].PLoS One,2017,12(9):e0185286.
[40] WELHAM T,PⅠKE J,HORST Ⅰ,FLEMETAKⅠS E,KATⅠNAKⅠS P,KANEKO T,SATO S,TABATA S,PERRY J,PARNⅠ-SKE M,WANG T L. A cytosolic invertase is required for normal growth and cell development in the model legume,Lotus japonicus[J].Journal of Experimental Botany,2009,60(12):3353-3365.
[41] JⅠA L Q,ZHANG B T,MAO C Z,LⅠJ H,WU Y R,WU P,WU Z C. OsCYT-ⅠNV1 for alkaline/neutral invertase is involved in root cell development and reproductivity in rice (Oryza sativa L.)[J].Planta,2008,228(1):51-59.
[42] LⅠU S J,LAN J X,ZHOU B H,QⅠN Y X,ZHOU Y H,XⅠAO X H,YANG J H,GOU J Q,QⅠJ Y,HUANG Y C,TANG C R.Hb-NⅠN2,a cytosolic alkaline/neutral-invertase,is responsible for sucrose catabolism in rubber- producing laticifers of Hevea brasiliensis (para rubber tree)[J]. New Phytologist,2015,206(2):709-725.
[43] 肖小虎,戚继艳,方永军,曹冰,龙翔宇,唐朝荣.巴西橡胶树中碱性转化酶HbNINa 基因的克隆和表达模式分析[J].热带作物学报,2013,34(7):1264-1269.XⅠAO Xiaohu,QⅠJiyan,FANG Yongjun,CAO Bing,LONG Xiangyu,TANG Chaorong. Cloning and expression analysis of an alkaline/neutral invertase gene in Hevea brasliensis[J]. Chinese Journal of Tropical Crops,2013,34(7):1264-1269.
[44] 张琼琼.番茄碱性/中性转化酶SlCIN3 基因功能研究[D].沈阳:沈阳农业大学,2019.ZHANG Qiongqiong. The primary study of an alkaline/neutral invertase SlCIN3 gene function in tomato[D]. Shenyang:Shenyang Agricultural University,2019.
[45] NONⅠS A,RUPERTⅠB,PⅠERASCO A,CANAGUⅠER A,ADAM-BLONDON A F,DⅠGASPERO G,VⅠZZOTTO G. Neutral invertases in grapevine and comparative analysis with Arabidopsis,poplar and rice[J].Planta,2008,229(1):129-142.
[46] JOUBERT D,BOSCH S,BOTHA F C,KOSSMANN J,GROENEWALD J H. Down-regulation of neutral invertase activity in sugarcane cell suspension cultures leads to increased sucrose accumulation[J]. South African Journal of Botany,2007,73(2):293.