香蕉(Musa nana Lour.)为芭蕉科芭蕉属植物,是一种重要的热带水果,年产量占全球鲜果产量的16%以上,在热带地区具有重要的经济价值。中国是世界上栽培香蕉的古老国家之一,种植面积约为35万hm2,位居世界第三,每年消费量达1300万t,位居中国第四大鲜果[1-3]。香蕉作为重要的经济作物,在海南、广东、云南、广西等地的脱贫攻坚战中发挥着重要作用。香蕉不仅含有丰富的矿物质元素、膳食纤维等,而且含有丰富的类黄酮和多酚,而类黄酮和多酚具有抗氧化、抗衰老以及抵抗病毒等生物功能,其营养价值较高,被广大消费者所喜爱[4]。
黄酮醇是类黄酮最主要的一类物质,在植物生长发育过程中起着重要作用[5-7]。在植物的侧根形成过程中,槲皮素(一种黄酮醇)会影响根系中生长素的运输和分布,从而影响侧根的生成[8-11]。此外,黄酮醇还能够影响花粉的活力和萌发。Zhang 等[12]利用病毒诱导的基因沉默技术(VIGS)沉默辣椒中的二氢黄酮醇还原酶(DFR)合成酶基因的表达,导致类黄酮含量降低,同时影响了花粉发育和花粉管伸长。由于DFR和黄酮醇合成酶(FLS)共同竞争底物二氢黄酮醇,当FLS 活性降低时会影响黄酮醇含量的降低,但花青素合成并不受影响,从而影响植物花色的形成[13-16]。已有研究表明,黄酮醇在植物的生物和非生物胁迫中发挥着重要作用[15,17-24]。拟南芥在强紫外照射下,拟南芥中的生长素合成减少,但是槲皮素合成却急剧升高,用来调控谷胱甘肽还原酶(GR)、谷胱甘肽过氧化物酶(GP)、谷胱甘肽-S-转移酶(GST)、抗坏血酸过氧化物酶(APX)、超氧化物歧化酶(SOD)和过氧化物酶(POX)等抗氧化酶的活性,清除自由基,从而减轻逆境对细胞DNA、RNA和蛋白质等生物高分子的损伤[6,25]。在高温天气下,植物通过槲皮素和ABA的互作来调节气孔的关闭,减少水分的流失[8]。此外,当植物遭遇重金属胁迫时,其体内槲皮素的含量急剧增加,从而减轻重金属对植物的伤害。
植物体内黄酮醇的合成由多个基因协同参与,相互交叉影响[26-27]。黄酮醇生物合成作为类黄酮代谢的一个重要分支,其合成途径为4-香豆酰-CoA和丙二酰-CoA,在查耳酮合成酶(chalcone synthase,CHS)和查耳酮异构酶(chalcone isomerase,CHI)的作用下生成柚皮素,然后在黄烷酮羟化酶,如黄烷酮3-羟化酶(F3H)的催化下生成二氢黄酮醇。二氢黄酮醇是DFR和FLS的共同底物,分别形成花青素和黄酮醇物质。因此,FLS 是黄酮醇合成的关键酶。FLS已经在多种植物中被广泛克隆和鉴定。该基因最早在紫罗兰和矮牵牛花组织中被发现,随后在拟南芥、苹果、杜鹃、芍药、风信子、杨梅等多种植物中被分离鉴定和克隆[13,14,28-36]。黄酮醇的合成除了受到关键合成酶基因的调控外,同时还在转录水平上受MYB 转录因子、bHLH 转录因子和WD40 蛋白的调控[5,15,17],这些转录因子能够激活或抑制黄酮醇早期生物合成步骤中CHS、CHI、F3H、FLS 等相关基因的表达,共同控制黄酮醇的生物合成。总之,植物体内黄酮醇的合成受到多种基因和转录因子的调控,而且在不同的植物体内具有普遍性,同时又有特殊性。
黄酮醇在香蕉果实中的合成代谢研究尚未见报道,而黄酮醇对香蕉的生长发育以及抗逆上有着重要作用,因此对香蕉中的黄酮醇合成酶进行克隆和功能研究对提高香蕉果实品质和抗逆性有非常重要的理论基础和应用价值。笔者通过前期对香粉1号果实不同成熟时期进行转录组测序分析,发现1 个在果实成熟后期表达量显著增加的黄酮醇合成酶MaFLS1,对其进行克隆和生物信息学分析,并通过异源转基因验证其功能,为利用MaFLS1 进行香蕉果实品质育种奠定理论基础。
1 材料和方法
1.1 试验材料
材料为中国热带农业科学院南亚热带作物研究所湛江院区试验基地种植的香粉1号果实。分别取抽薹开花断蕾后25、45、65、85、88 d 果肉,用水果刀切片后迅速放入液氮中冷却,放至实验室-80 ℃冰箱中保存。
1.2 香粉1号果肉总RNA的提取与cDNA的合成
植物总RNA 的提取使用北京华越洋生物有限公司的通用型植物快速RNA提取试剂盒,提取香粉1 号25 d、45 d、65 d、85 d 果肉中的总RNA,通过1%琼脂糖凝胶电泳检测RNA 的质量,用Nano-Drop2000 微型紫外分光光度计(Thermo Scientific)测定RNA的浓度。用TaKaRa生物技术有限公司的反转录试剂盒[PrimeScript ™RT reagent Kit with gDNA Eraser(Perfect Real Time)]进行反转录得到cDNA,存放于-20 ℃冰箱中备用。
1.3 MaFLS1基因引物设计和PCR扩增
从笔者团队前期的测序结果中筛选到了黄酮醇合成酶基因MaFLS1,在香蕉基因网站上(https://banana-genome-hub.southgreen.fr/)查找参考序列。根据参考序列的编码区(CDS 序列)利用软件Primer primer 5.0分别设计编码区引物MaFLS1-F、MaFLS1-R(表1)。
表1 引物序列
Table 1 Primer sequences

以提取的香粉1 号果肉组织总RNA 反转录的cDNA 为模板,进行PCR 扩增。50 μL 反应体系如下:10× PCR Buffer for KOD-Plus-Neo 5 μL,2mmol·L-1 dNTPs5μL,25 mmol·L-1 MgSO4 3 μL,引物(10 μmol·L-1 each)2μL,cDNA5 μL,加ddH2O至50 μL。反应程序:94 ℃预变性2 min,98 ℃变性10 s,退火温度为56 ℃30 s,68 ℃延伸1 min,反应循环数为40个。
1.4 PCR产物回收、克隆及测序
将PCR 后的产物经1%的琼脂糖凝胶电泳后,在凝胶成像仪上确定目的条带,并用试剂盒(Mini-BEST Agarose Gel DNA Extraction Kit Ver.4.0,Ta-KaRa)回收纯化目的片段,并将纯化后的目的片段与pEASY-T1 Cloning Ki(tTransGen)克隆载体连接,并转化大肠DH5α,挑选单菌落进行阳性鉴定,将阳性单菌落送至广州艾基生物科技有限公司进行测序。将测序后的序列与参考序列比对。
1.5 生物信息学分析
在NCBI网站上搜索其他物种FLS基因的氨基酸序列,并用MEGA 7.0 软件构建系统进化树。用SMART 在线网站预测功能结构域。在ExPASy(http://web.expasy.org/compute_pi/)网站对MaFLS1基因编码蛋白的理化性质进行预测。利用在线网站https://services.healthtech.dtu.dk/services/TMHMM-2.0/进行基因的跨膜结构预测。使用信号肽预测网站(https://services.healthtech.dtu.dk/services/SignalP-5.0/)进行信号肽的预测。在SOPMA 在线网站(https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html)预测MaFLS1 蛋白二级结构。蛋白三级结构使用SWISS-MODEL在线网站预测。
1.6 香粉1号果肉中MaFLS1基因表达分析
采用实时荧光定量RT-qPCR 方法分析香蕉果肉MaFLS1 基因的表达情况,MaFLS1 基因的荧光定量引物以及内参基因引物见表1。PCR反应在赛默飞公司下的ABI QuantStudio 6 Flex 荧光定量仪上进行。反应体系为:PowerUp™SYBR™Green Master Mix(2X)10 μL,各1 μL 的上下游引物,2 μL的cDNA 模板,用ddH2O 补足到20 μL。反应程序:95 ℃预变性2 min,95 ℃变性15 s,56 ℃退火15 s,72 ℃延伸1 min,总共40 个循环。用2-ΔΔCT方法计算MaFLS1基因的相对表达量。
1.7 目的基因与过表达载体质粒同源重组
通过TaKaRa 公司的In-Fusion 引物在线设计网站(https://takara.teselagen.com/#/Design-Page)设计引物(表1)。以已测序正确的阳性质粒为模板,使用KOD-Plus-Neo 高保真酶进行PCR 扩增并胶回收。使用Clontech 公司的In-Fusion® HD Cloning Kit进行无缝克隆,采用热激法转化大肠杆菌DH5α,涂板过夜,挑单菌落,PCR菌液阳性鉴定后送广州艾基生物有限公司测序。
1.8 番茄转化,筛选和鉴定
用根癌农杆菌GV3101 介导的叶盘法转化Micro-Tom番茄,获得T0代苗。将T0代转基因番茄苗移栽到营养土中,待叶子稍大时取叶片,用CTAB法提取叶片基因组DNA,对目的基因进行PCR阳性鉴定。收取T0代果实中的种子即为T1代,播种T1代种子以及野生型番茄种子(WT),待叶子稍大时取叶片,用CTAB 法提取叶片基因组DNA,对目的基因进行PCR 阳性鉴定。取T1 代阳性植株以及WT生长一致的果实液氮速冻后保存于-80 ℃冰箱,用于果实总黄酮的提取。
1.9 番茄果实总黄酮的提取与测定
总黄酮含量的测定采用亚硝酸钠-硝酸铝-氢氧化钠显色法,具体操作参考魏长宾主编的《热带水果品质分析实验指导》[37]。
2 结果与分析
2.1 MaFLS1基因的克隆
以反转录后的香粉1号果肉cDNA为模板,设计引物并进行PCR扩增,扩增结果出现了条带大小一致的片段(图1)。连接到克隆载体上,阳性鉴定后送测序。测序后的基因编码区(CDS)全长1080 bp,编码359个氨基酸。

图1 MaFLS1 基因的PCR 扩增
Fig.1 The PCR fragment of MaFLS1
M.DL2000 DNA Marker.
2.2 MaFLS1蛋白的生物信息学分析
2.2.1 MaFLS1 蛋白的理化性质、亚细胞定位预测以及二三级结构预测 通过ExPASy 在线网站(https://web.expasy.org/protparam/)对MaFLS1编码的蛋白进行理化性质预测。结果显示该基因编码的蛋白含有359个氨基酸,相对分子质量为39 436.94 Da,理论等电点(pI)为5.41,不稳定系数为37.35(属于稳定蛋白类),亲水性总平均值为-0.178,因此推测为稳定的亲水酸性蛋白。通过亚细胞定位网站(https://wolfpsort.hgc.jp/)预测该基因蛋白在细胞中的位置,结果显示其可能定位于细胞质中。此外,通过相关网站预测其信号肽以及跨膜结构情况,结果显示其没有信号肽和跨膜结构。该基因蛋白质二级结构利用SOPMA 在线软件(http:/web.expasy.org/)进行预测,结果(图2)显示,二级结构占比分别是α-螺旋34.26%,无规则卷曲42.9%,延伸链16.71%,β-转角为6.13%(图2-A)。该蛋白的三级结构预测由SWISS-MODEL 在线软件(http://swissmodel.ex- pasy.org/)完成(图2-B)。当一个蛋白质的序列与一个已知结构蛋白质序列高度相似的时候,该蛋白质的结构就可以被建模出来。

图2 MaFLS1 蛋白二级、三级结构预测
Fig.2 Structural analysis and the conserved domains of MaFLS1
A.二级结构预测;B.三级结构预测。
A.Prediction for the secondary structure;B.Prediction for the tertiary structure.
2.2.2 MaFLS1蛋白的保守结构域预测 通过蛋白保守结构域预测网站(SMART)预测MaFLS1 的保守结构域,结果显示该蛋白含有两个功能结构域(图3)。一个是具有2-氧戊二酸/Fe(Ⅱ)依赖性二氧化物的蛋白质的高度保守的N 端区域,在第49~160 个氨基酸位置。另一个是具有Fe2+和2-酮戊二酸(2OG)依赖性双加氧酶结构域的酶结构,在第206~306 个氨基酸位置,该酶通常使用双氧分子催化有机底物的氧化,主要是通过使用亚铁作为活性位点辅因子和2OG 作为共底物,脱羧为琥珀酸盐和CO2。而在植物中,Fe(Ⅱ)2OG 双加氧酶域酶催化植物激素的形成,如乙烯、赤霉素、色素和黄酮。

图3 MaFLS1 蛋白的保守结构域预测
Fig.3 Conservative domain Prediction of MaFLS1 Protein
氨基酸数目
Number of amino acid/aa
2.2.3 MaFLS1氨基酸序列与其他物种同源性分析及进化树构建 通过NCBI上的Gene查找相关物种的FLS 基因序列及其蛋白序列,将苹果(MdFLS:GenBank ID NP_001306179.1)、香蕉(MaFLS1:Gen-Bank ID XP_009384795.1;MaFLS2:GenBank ID XP_009404656.1;MaFLS3:GenBank ID XP_009402233.1)、拟南芥(AtFLS1~AtFLS6)、桃(PpFLS:GenBank ID AJO70134.1)、椰子(CnFLS:GenBank ID KAG1339093.1)、苦 荞(FtFLS1:GenBank ID AEC33116.1,FtFLS2:GenBank ID AGE13752.1)等物种的FLS 基因通过DNAMAN 进行比对分析(图4),发现其与其他物种的FLS 氨基酸相似率为50%左右。将不同物种的FLS 基因蛋白序列使用MEGA 7 软件构建系统发生树(图5)。结果显示,MaFLS1 基因蛋白与该物种的MaFLS3、MaFLS2 的亲缘关系较近,与椰子CnFLS 处于同一分支,同属于单子叶植物,而与其他物种的亲缘关系较远,但和葡萄(VvFLS)的亲缘关系较近,可能两者功能类似。

图4 香蕉MaFLS1 氨基酸与其他植物氨基酸序列同源比对
Fig.4 Homologous comparison of amino acid sequence of banana MaFLS1

图4 (续)Fig.4 (Continued)
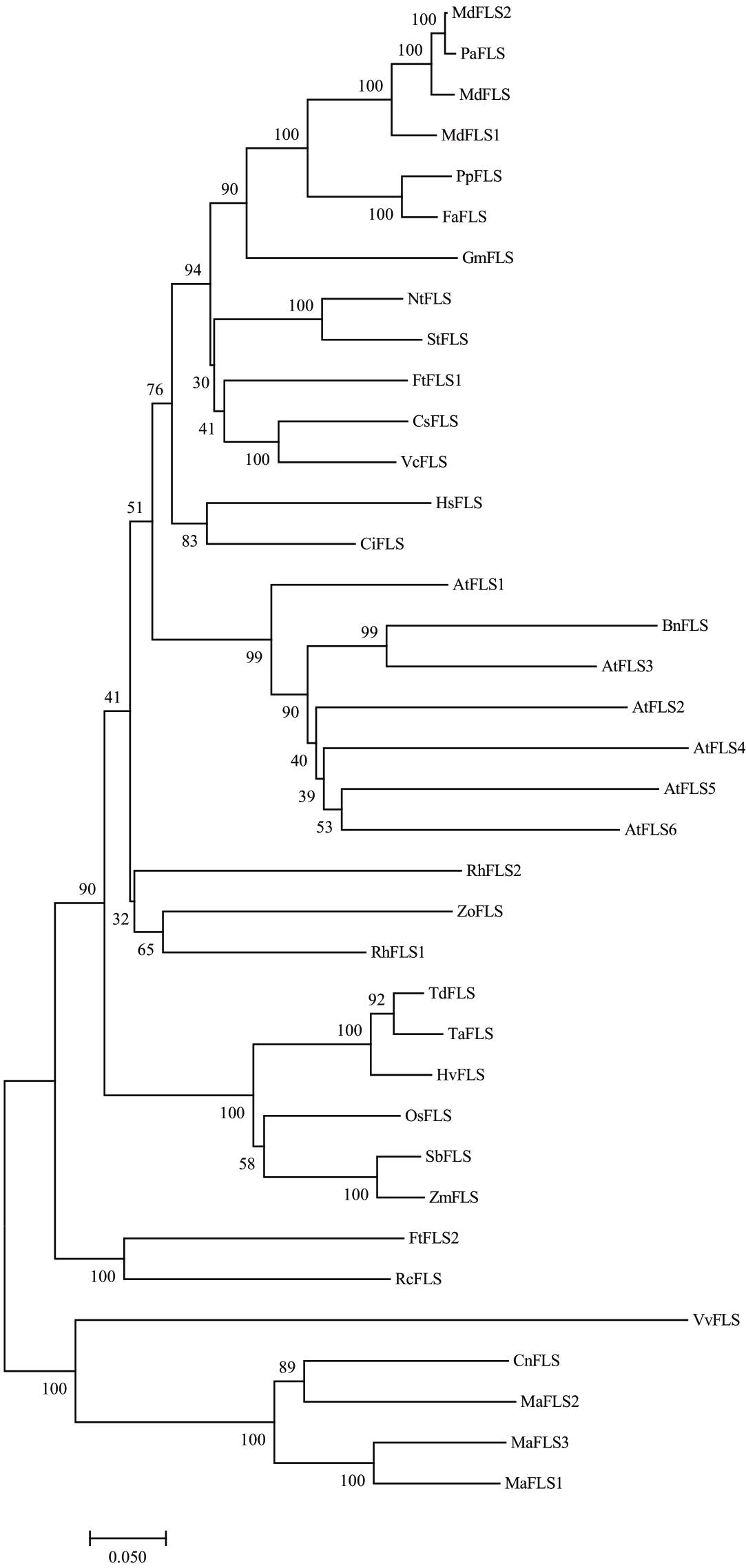
图5 香蕉MaFLS1 与其他FLS 系统的进化树
Fig.5 Phylogenetic tree of FLS amino acid sequence system of banana MaFLS1 amino acid sequence and other plants
Md.苹果;Pa.甜樱桃;Pp.桃;Gm.大豆;Cs.茶树;Vc.蓝莓;Nt.烟草;St.马铃薯;Ci.蜜柑;Rh.小苍兰;Os.水稻;Bn.油菜;At.拟南芥;Rc.蓖麻;Vv.葡萄;Td.二粒小麦;Zo.姜;Hv.栽培大麦;Sb.高粱;Zm.玉米;Ta.小麦;Cn.椰子;Hs.木槿;Fa.草莓;Ft.苦荞。
Md.Malus domestica;Pa.Prunus avium L;Pp.Prunus persica L.;Gm.Glycine max;Cs.Camellia sinensis;Vc.Vaccinium corymbosum L.;Nt.Nicotiana tabacum L.;St.Solanum tuberosum L.;Ci.Citrus unshiu;Rh.Fressia hybrida;Os.Oryza sativa L.;Bn.Brassica napus L;At.Arabidopsis thaliana;Rc.Ricinus communis L.;Vv.Vitis vinifera;Td.Triticum dicoccoides;Zo.Zingiber officinale;Hv.Hordeum vulgare subsp.vulgare;Sb.Sorghum bicolor;Zm.Zea mays;Ta.Triticum aestivum;Cn.Cocos nucifera;Hs.Hibiscus syriacus;Fa.Fragaria×ananassa;Ft.Fagopyrum tataricum.
2.3 香蕉MaFLS1在香粉1号果实发育不同时期的表达分析
如图6 所示,MaFLS1在香粉1号果实发育的前期表达量很低,几乎不表达,在抽花断蕾后的85 d,表达量开始增加,直到88 d,此时果实已经完全成熟可以食用,而MaFLS1 的表达量急剧增加,说明该基因是在果实发育的后期表达,因此推测该基因在香蕉果实成熟后期发挥重要作用。此外,从图6 中可以看出,MaFLS1 在88 d 的表达量与其他4 个时期差异显著,另外4 个时期之间差异不显著。
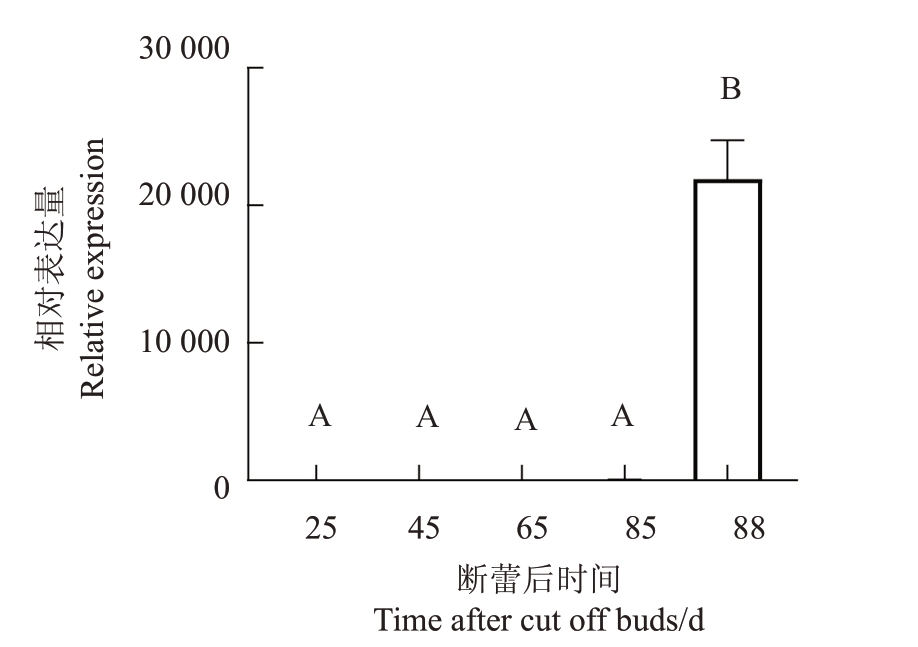
图6 香蕉果实中MaFLS1 实时荧光定量PCR 分析
Fig.6 qRT-PCR analysis of MaFLS1 gene in the fruit of banana
不同大写字母表示在p<0.01 差异极显著。
Different capital letters indicate extremely significant difference at p<0.01.
2.4 过表达MaFLS1 番茄转基因植株果实中总黄酮含量的测定
2.4.1 MaFLS1-35sn融合表达载体的获得 通过同源重组的方法将目的片段与过表达载体pcambia1301-35sn 连接,采用热激法转化大肠杆菌DH5α,涂板挑单菌落,PCR阳性鉴定,阳性单菌落测序(图7)。选择测序正确的单菌落质粒通过冻融法转化根癌农杆菌GV3101。

图7 MaFLS1-35sn 融合表达载体的菌液PCR 扩增
Fig.7 The PCR amplification products MaFLS1-35sn fusion expression vector identified by bacterial liquid PCR
2.4.2 转基因番茄植株的阳性鉴定 如图8 所示,从T1代植株中鉴定得到5株阳性苗。经表型观察,转基因番茄植株比野生型植株矮小,且生长发育慢于野生型。
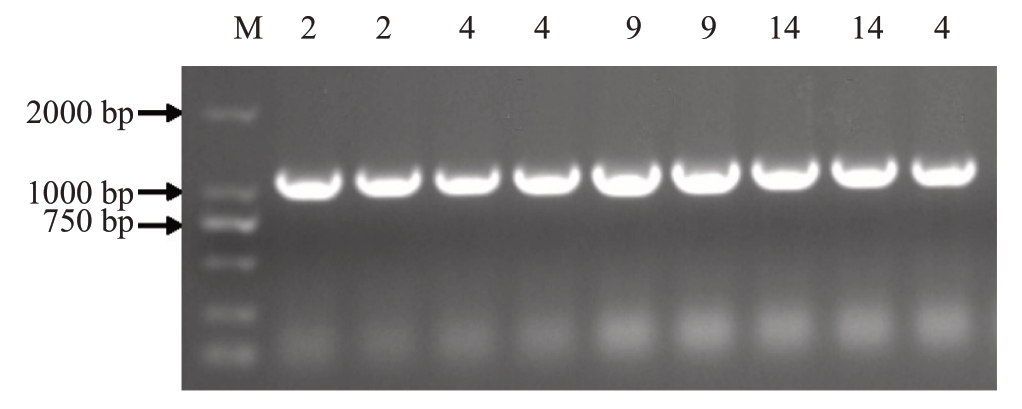
图8 转基因番茄阳性植株鉴定
Fig.8 Identification of transgenic tomato positive plants
M.DL2000 DNA Marker;其余为目的片段。
M.DL2000 DNA Marker;the rest are destination fragments.
2.4.3 T1 代阳性植株果实总黄酮含量的测定 在果实成熟期(果实百分之百转为红色)利用亚硝酸钠-硝酸铝-氢氧化钠显色法测定转基因以及野生型果实总黄酮含量,结果显示,阳性转基因果实中的总黄酮含量(w,后同)(0.58 mg·g-1)极显著高于野生型果实(0.46 mg·g-1)(图9),说明MaFLS1确实能够增加果实中总黄酮含量。

图9 阳性植株形态、果实形态及果实中总黄酮含量
Fig.9 Positive plant morphology,fruit morphology and total flavonoids content in fruits
A.野生型(WT)植株和MaFLS1-OE 植株;B.野生型(WT)果实和MaFLS1-OE 果实;C.野生型(WT)果实和MaFLS1-OE-T1 果实中总黄酮含量比较。**表示在p<0.01 差异极显著。
A.Wild type (WT) plants and MaFLS1-OE plants;B.Wild type (WT) fruit and MaFLS1-OE fruit;C.Comparison of total flavonoids content in Wild type(WT)fruits and MaFLS1-OE-T1 fruits.**indicates extremely significant difference at p<0.01.
3 讨论
FLS 是影响植物黄酮醇合成和积累的重要因素,而黄酮醇是植物生长发育过程中一类重要的次生代谢物。笔者课题组前期已经在相关期刊上发表了香粉1 号果实不同发育时期代谢组数据[38],发现香粉1号在果实成熟后期黄酮醇的物质种类极其丰富,因此很有必要对黄酮醇合成相关基因开展研究。笔者在本研究中首次克隆了香蕉MaFLS,命名为MaFLS1,并对其氨基酸序列进行了生物信息学分析。通过对其蛋白保守结构域分析,发现MaFLS1 N端为α-同戊二酸依赖性双加酶结构域,C端为酮戊二酸/铁离子依赖加氧酶结构域,这与苹果、拟南芥、洋葱等植物中FLS家族蛋白的分析结果一致[13,29-30,33,36],说明在不同植物中FLS 家族蛋白的氨基酸序列保守性较高,可以保证FLS 在不同物种中表现相同的生物功能。香蕉果实不同发育阶段MaFLS1 的荧光定量表达结果表明,随着果实的成熟,其表达量逐渐增加,特别是在后期极显著增加,说明其可能参与了果实的成熟,这与高鹏钊等[4]的研究结果相似。此外,MaFLS1 在断蕾后88 d 的表达量急剧上升,而前期几乎不表达或表达量极低,推测该基因在香蕉果实成熟前期可能受到其他基因的调控作用,导致该基因在前期几乎不表达。
此外,笔者在本研究中通过异源超表达MaFLS1 并测定转基因番茄果实中总黄酮的含量,发现转基因果实中总黄酮的含量极显著高于野生型果实,说明MaFLS1 确实参与了总黄酮的合成,超表达MaFLS1 能够促进植物体内总黄酮的合成与积累。但由于T1 代果实数量不够,为了优先保证T2 代苗,没有利用HPLC 法测定具体是哪种或者哪几种类黄酮含量的增加或减少。因此,为了进一步研究MaFLS1 具体是合成哪种类黄酮,以及是否具有表达特异性,下一步需要大量种植T2代转基因番茄,取不同组织进行荧光定量表达分析以及类黄酮测定,最终确定MaFLS1 合成产物是哪种黄酮醇。
4 结论
通过RT-PCR克隆得到了一个香蕉黄酮醇合成酶基因MaFLS1,并在番茄上进行了初步基因功能验证,结果显示该基因能够增加果实中总黄酮的含量,将为以后的香蕉品质育种提供理论依据。
[1] 方伟,唐诚,王玉梅.广东省香蕉产业发展现状、面临瓶颈及对策[J].中国果树,2022(12):90-93.FANG Wei,TANG Cheng,WANG Yumei.Current situation,bottleneck and countermeasures of banana industry in Guangdong Province[J].China Fruits,2022(12):90-93.
[2] 何俞均,王芳.基于波特钻石理论的中国香蕉产业竞争力分析[J].中国热带农业,2022(1):19-26.HE Yujun,WANG Fang.Analysis of China’s banana industrial competitiveness based on porter’s diamond theory[J].China Tropical Agriculture,2022(1):19-26.
[3] 肖媛.中国香蕉价格波动传导机制研究[D].武汉:华中农业大学,2022.XIAO Yuan.Study on the transmission mechanism of banana price fluctuations in China[D].Wuhan:Huazhong Agricultural University,2022.
[4] 高鹏钊,苗红霞,张建斌,徐碧玉,刘菊华.3 个香蕉品种黄酮含量与果实发育成熟的关系[J].热带作物学报,2016,37(10):1894-1899.GAO Pengzhao,MIAO Hongxia,ZHANG Jianbin,XU Biyu,LIU Juhua.The relationship of flavonoids of three banana varieties and fruit development and ripening[J].Chinese Journal of Tropical Crops,2016,37(10):1894-1899.
[5] HU P F,SURIGUGA,ZHAO M,CHEN S Q,WU X H,WAN Q.Transcriptional regulation mechanism of flavonoids biosynthesis gene during fruit development in Astragalus membranaceus[J].Frontiers in Genetics,2022,13:972990.
[6] KUREPA J,SHULL T E,SMALLE J A.Friends in arms:Flavonoids and the auxin/cytokinin balance in terrestrialization[J].Plants,2023,12(3):517.
[7] SINGH P,ARIF Y,BAJGUZ A,HAYAT S.The role of quercetin in plants[J].Plant Physiology and Biochemistry,2021,166:10-19.
[8] 柳苗苗,蔡伟建,张斌斌,赵密珍,王静,刘怀锋.槲皮素对草莓生长发育、光合和生理生化特性影响的综合评价[J].江苏农业科学,2022,50(21):165-172.LIU Miaomiao,CAI Weijian,ZHANG Binbin,ZHAO Mizhen,WANG Jing,LIU Huaifeng.Comprehensive evaluation on effects of quercetin on growth,photosynthesis,physiological and biochemical characteristics of strawberry[J].Jiangsu Agricultural Sciences,2022,50(21):165-172.
[9] VILLACAMPA A,FAÑANÁS-PUEYO I,MEDINA F J,CISKA M.Root growth direction in simulated microgravity is modulated by a light avoidance mechanism mediated by flavonols[J].Physiologia Plantarum,2022,174(3):e13722.
[10] TAN H J,MAN C,XIE Y,YAN J J,CHU J F,HUANG J R.A crucial role of GA-regulated flavonol biosynthesis in root growth of Arabidopsis[J].Molecular Plant,2019,12(4):521-537.
[11] SILVA-NAVAS J,MORENO-RISUENO M A,MANZANO C,TÉLLEZ-ROBLEDO B,NAVARRO-NEILA S,CARRASCO V,POLLMANN S,GALLEGO F J,DEL POZO J C.Flavonols mediate root phototropism and growth through regulation of proliferation-to-differentiation transition[J].The Plant Cell,2016,28(6):1372-1387.
[12] ZHANG Z S,LIU Y,YUAN Q L,XIONG C,XU H,HU B W,SUO H,YANG S,HOU X L,YUAN F,PEI Z M,DAI X Z,ZOU X X,LIU F.The bHLH1-DTX35/DFR module regulates pollen fertility by promoting flavonoid biosynthesis in Capsicum annuum L.[J].Horticulture Research,2022,9:uhac172.
[13] 杜灵娟,陈凯利,刘雅莉.葡萄风信子FLS1 基因克隆及其表达与花色性状之间的关联性分析[J].西北林学院学报,2017,32(1):106-113.DU Lingjuan,CHEN Kaili,LIU Yali.Cloning of flavonol synthase gene (FLS1) and relativity analysis of its expression with the flower color in grape hyacinth[J].Journal of Northwest Forestry University,2017,32(1):106-113.
[14] LEI T,HUANG J,RUAN H X,QIAN W,FANG Z,GU C Y,ZHANG N N,LIANG Y X,WANG Z Y,GAO L P,WANG Y S.Competition between FLS and DFR regulates the distribution of flavonols and proanthocyanidins in Rubus chingii Hu[J].Frontiers in Plant Science,2023,14:1134993.
[15] WANG Y G,ZHOU L J,WANG Y X,GENG Z Q,DING B Q,JIANG J F,CHEN S M,CHEN F D.An R2R3-MYB transcription factor CmMYB21 represses anthocyanin biosynthesis in color fading petals of chrysanthemum[J].Scientia Horticulturae,2022,293:110674.
[16] 靳俊婷,索晓静,丁红元,郭春磊,王东升,张京政,曹飞.黄酮醇在果树中的功能的研究进展[J].河北果树,2021(4):1-3.JIN Junting,SUO Xiaojing,DING Hongyuan,GUO Chunlei,WANG Dongsheng,ZHANG Jingzheng,CAO Fei.Research progress of function of flavonols in fruit trees[J].Hebei Fruits,2021(4):1-3.
[17] LI B Z,FAN R N,GUO S Y,WANG P T,ZHU X H,FAN Y T,CHEN Y X,HE K Y,KUMAR A,SHI J P,WANG Y,LI L H,HU Z B,SONG C P.The Arabidopsis MYB transcription factor,MYB111 modulates salt responses by regulating flavonoid biosynthesis[J].Environmental and Experimental Botany,2019,166:103807.
[18] CHEN G,WANG Y P,LIU X L,DUAN S Y,JIANG S H,ZHU J,ZHANG Y G,HOU H M.The MdmiR156n regulates drought tolerance and flavonoid synthesis in apple calli and Arabidopsis[J].International Journal of Molecular Sciences,2023,24(7):6049.
[19] GAUTAM H,SHARMA A,TRIVEDI P K.The role of flavonols in insect resistance and stress response[J].Current Opinion in Plant Biology,2023,73:102353.
[20] 葛诗蓓,张学宁,韩文炎,李青云,李鑫.植物类黄酮的生物合成及其抗逆作用机制研究进展[J].园艺学报,2023,50(1):209-224.GE Shibei,ZHANG Xuening,HAN Wenyan,LI Qingyun,LI Xin.Research progress on plant flavonoids biosynthesis and their anti-stress mechanism[J].Acta Horticulturae Sinica,2023,50(1):209-224.
[21] YILDIZTUGAY E,OZFIDAN-KONAKCI C,KUCUKODUK M,TURKAN I.Flavonoid naringenin alleviates short-term osmotic and salinity stresses through regulating photosynthetic machinery and chloroplastic antioxidant metabolism in Phaseolus vulgaris[J].Frontiers in Plant Science,2020,11:682.
[22] PARVIN K,HASANUZZAMAN M,BHUYAN M H M B,MOHSIN S M,FUJITA M.Quercetin mediated salt tolerance in tomato through the enhancement of plant antioxidant defense and glyoxalase systems[J].Plants,2019,8(8):247.
[23] NAKABAYASHI R,YONEKURA-SAKAKIBARA K,URANO K,SUZUKI M,YAMADA Y,NISHIZAWA T,MATSUDA F,KOJIMA M,SAKAKIBARA H,SHINOZAKI K,MICHAEL A J,TOHGE T,YAMAZAKI M,SAITO K.Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids[J].The Plant Journal,2014,77(3):367-379.
[24] MAHAJAN M,YADAV S K.Effect of quercetin and epicatechin on the transcript expression and activity of antioxidant enzymes in tobacco seedlings[J].American Journal of Biochemistry and Molecular Biology,2012,3(1):81-90.
[25] LIU L L,GREGAN S M,WINEFIELD C,JORDAN B.Comparisons of controlled environment and vineyard experiments in Sauvignon Blanc grapes reveal similar UV-B signal transduction pathways for flavonol biosynthesis[J].Plant Science,2018,276:44-53.
[26] WEN W W,ALSEEKH S,FERNIE A R.Conservation and diversification of flavonoid metabolism in the plant kingdom[J].Current Opinion in Plant Biology,2020,55:100-108.
[27] FUJINO N,TENMA N,WAKI T,ITO K,KOMATSUZAKI Y,SUGIYAMA K,YAMAZAKI T,YOSHIDA S,HATAYAMA M,YAMASHITA S,TANAKA Y,MOTOHASHI R,DENESSIOUK K,TAKAHASHI S,NAKAYAMA T.Physical interactions among flavonoid enzymes in snapdragon and torenia reveal the diversity in the flavonoid metabolon organization of different plant species[J].The Plant Journal,2018,94(2):372-392.
[28] WANG Z,WANG S S,WU M Z,LI Z F,LIU P P,LI F,CHEN Q S,YANG A G,YANG J.Evolutionary and functional analyses of the 2-oxoglutarate-dependent dioxygenase genes involved in the flavonoid biosynthesis pathway in tobacco[J].Planta,2019,249(2):543-561.
[29] 郑雪莲,李嘉仪,杨俊,郑国华.枇杷黄酮醇合成酶FLS 基因表达与黄酮醇积累的相关性分析[J].福建农业科技,2019(5):1-6.ZHENG Xuelian,LI Jiayi,YANG Jun,ZHENG Guohua.Correlation analysis between flavonol synthetase genetic expression and flavono accumulation in loquat[J].Fujian Agricultural Science and Technology,2019(5):1-6.
[30] 刘晓,路静,郝玉金,由春香.苹果黄酮醇合成酶基因MdFLS1的克隆、生物信息学分析及催化活性鉴定[J].果树学报,2018,35(8):905-916.LIU Xiao,LU Jing,HAO Yujin,YOU Chunxiang.Cloning,bioinformatics analysis and catalytic activity identification of Md-FLS1 gene in apple[J].Journal of Fruit Science,2018,35(8):905-916.
[31] 李海鸿.葡萄风信子(Muscari)黄酮醇合酶FLS(flavonol synthase)基因克隆和功能分析[D].杨凌:西北农林科技大学,2018.LI Haihong.Clong and functional analysis of FLS(flavonol synthase) gene from Muscari armeniacm[D].Yangling:Northwest A&F University,2018.
[32] 史旻,孙静,陶俊.芍药PlFLS 基因生物信息学分析及对拟南芥的遗传转化[J].华南农业大学学报,2018,39(5):93-100.SHI Min,SUN Jing,TAO Jun.Bioinformatics analysis of Paeonia lactiflora PlFLS gene and its genetic transformation in Arabidopsis thaliana[J].Journal of South China Agricultural University,2018,39(5):93-100.
[33] 王振宝,杨妍妍,刘冰江,霍雨猛,李艳伟,孙亚玲,吴雄.洋葱黄酮醇合成酶基因的克隆与表达分析[J].山东农业科学,2022,54(12):18-24.WANG Zhenbao,YANG Yanyan,LIU Bingjiang,HUO Yumeng,LI Yanwei,SUN Yaling,WU Xiong.Cloning and expression analysis of flavonol synthase gene in onion[J].Shandong Agricultural Sciences,2022,54(12):18-24.
[34] 唐亚琴,梅抗抗,陈瑶,谢琴鼎,黄乾明,陈华萍.油橄榄类黄酮生物合成相关酶的研究进展[J].基因组学与应用生物学,2018,37(5):2110-2117.TANG Yaqin,MEI Kangkang,CHEN Yao,XIE Qinding,HUANG Qianming,CHEN Huaping.Research progress of biosynthesis related enzymes of flavonoid in Olea europaea L.[J].Genomics and Applied Biology,2018,37(5):2110-2117.
[35] 石水莲,李威,李可,吕玲玲.植物FLS 研究进展[J].分子植物育种,2019,17(18):5980-5985.SHI Shuilian,LI Wei,LI Ke,LÜ Lingling.Advances of FLS in plants[J].Molecular Plant Breeding,2019,17(18):5980-5985.
[36] 邢梦云.杨梅FLSs 和F3’5’H 调控杨梅素生物合成的机制研究[D].杭州:浙江大学,2021.XING Mengyun.Regulation of myricetin biosynthesis by FLSs and F3’5’H in Morella rubra[D].Hangzhou:Zhejiang University,2021.
[37] 魏长宾.热带水果品质分析实验指导[M].北京:中国农业出版社,2017.WEI Changbin.Experimental guidance on quality analysis of tropical fruits[M].Beijing:China Agriculture Press,2017.
[38] HU H G,WANG J X,HU Y L,XIE J H.Nutritional component changes in Xiangfen 1 banana at different developmental stages[J].Food &Function,2020,11(9):8286-8296.