柑橘衰退病是危害柑橘产业的重要病害之一,其病原为柑橘衰退病毒(citrus tristeza virus,CTV),主要通过感病接穗、苗木和多种蚜虫进行传播,广泛分布于世界各柑橘产区[1]。CTV 是长线性病毒科(Closteroviridae)长线性病毒属(Closterovirus)的正义单链RNA病毒,基因组全长19.8 kb,含12个开放阅读框(ORF),可编码两个外壳蛋白(CP 和CPm),其中CP 在CTV 基因组中高度保守[2]。CTV 在田间存在复杂的株系分化现象,除导致酸橙及其作砧木植株的快速死亡外,还导致葡萄柚、梾檬和部分甜橙、柚类、杂柑等敏感品种的茎陷点症状,以及酸橙、尤力克柠檬和葡萄柚实生苗的矮缩、黄化,CTV 弱毒株在植株上不会产生严重的症状[3]。根据CTV的生物学特性及其基因组变异,CTV被分为T36、VT、T30、T3、RB、T68、HA16-5和S1等多个基因型,且其基因型的种类还在不断增加[4-5]。
我国由于长期使用枳、香橙、酸柚等抗速衰型衰退病砧木,茎陷点型衰退病是我国衰退病危害的主要类型[6-7]。近年来,由于茎陷点型衰退病随苗木流通,其发生范围不断扩大,已在湖南、江西、云南等柑橘主产区造成了严重的危害[8-10]。目前采用无毒繁殖材料是防治CTV最有效的手段,而这依赖于高效快速的检测方法。
目前常用血清学[11]、RT-PCR[12]以及RT-qPCR[13-14]等方法检测CTV。这些技术虽然灵敏度高、特异性强,但难以实现田间现场检测。因此为满足果园、苗圃现场快速检测的需要,亟待研发一种准确、快捷、简便的CTV检测方法。重组酶聚合酶扩增(Recombinase polymerase amplification,RPA)模拟T4 噬菌体核酸复制机制,在体外实现恒温扩增[15]。通过结合侧流层析试纸条(lateral flow dipstick,LFD),从而实现了检测结果的可视化。由于RT-RPA-LFD检测技术具有快速、灵敏和简便的优点,尤其适用于普通工作人员开展田间检测,目前已成功应用于柑橘碎叶病毒(citrus tatter leaf virus,CTLV)、樱桃病毒A(cherry virus A,CVA)、李矮缩病毒(prune dwarf virus,PDV)、李痘病毒(plum pox virus,PPV)等多种植物病毒的检测[16-19]。笔者在本研究中以CTV保守的CP基因为靶标设计特异性引物和探针,建立、优化了CTV的RT-RPA-LFD检测技术,为CTV的快速检测提供了新的选择。
1 材料和方法
1.1 试验材料
单一感染CTV、柑橘黄脉病毒(citrus yellow vein clearing virus,CYVCV)、柑橘碎叶病毒、柑橘裂皮病类病毒(CEVd)、柑橘鳞皮病毒(citrus psorosis virus,CPV)和柑橘褪绿矮缩病毒(citrus chlorotic dwarf-associated virus,CCDaV)的病株,无病毒柑橘植株;核酸浓度为2.12×106 拷贝·μL-1的CTV阳性样品。以上材料均为西南大学柑桔研究所保存提供。
1.2 主要试剂
RPA 扩增试剂盒购自美国Agdia 公司;Plant-Gen DNA Kit 购自中国康为世纪;RNAiso Plus 试剂盒购自宝生物工程(大连)有限公司;All-Ⅰn-One 5×RT MasterMix,2×Taq Master Mix购自诺唯赞公司。
1.3 总核酸提取和cDNA合成
使用PlantGen DNA Kit 和RNAiso Plus 提取总核酸,并于-80 ℃冰箱保存备用。将1 μL Oligo dT Primer,1 μL dNTP Mixture(10 μmol·L-1),1 μL 总RNA 模板,7 μL ddH2O 混合后,65 ℃5 min;冰上冷却后加入4 μL PrimeScript Buffer,0.5 μL RNase Ⅰnhibitor(40 U·μL-1),1 μL PrimeScript RTase(200 U·μL-1,TaKaRa),加ddH2O至总体积为20 μL。42 ℃45 min,95 ℃5 min。
1.4 RT-RPA-LFD的引物和探针设计
比对分析NCBⅠ中已报道的5个中国茎陷点型CTV CP基因序列(GenBank:MH558665.1、MH558666.1、JX266712.1、JQ911664.1 和JQ061137.1),以及Amplify Discovery Kits 中引物和探针设计要求,使用Primer Premier 5.0设计特异性引物和探针(表1),所用引物和探针均由北京擎科生物技术有限公司合成。
表1 RT-PCR、RT-qPCR 和RT-RPA-LFD 检测用引物及探针
Table 1 RT-PCR、RT-qPCR and RT-RPA-LFD used primers and probes

引物Primer RT-PCR CP1 CP3 RT-qPCR P25-F P25-R CTV-CY5 RT-RPA-LFD RPA-1F RPA-1R RPA-2F RPA-2R RPA-3F RPA-3R RPA-P引物序列Primer sequence(5’-3’)ATGGACGACGAAACAAAG TCAACGTGTGTTGAATTT AGCRGTTAAGAGTTCATCATTRC TCRGTCCAAAGTTTGTCAGA CY5-CRCCACGGGYATAACGTACACTCGG CTTGCTGGCGTCCCTTGTTTCTGTTCTTGTCTT ATTCTGTTTCCTTTCCTAGCCGGGCTTCTTCAC GCGACGCCGAGTAATAACCTCTAACACCAT TTCTTTAGGAGGGATGTCAACTTAGGAGCG CACCCAAATACTTACGATGGCAGGTTCTAC CAAGCGGACGACTTAGCGACAGGCTGATAG GGCGAAAAATCTTTTCGTCTACTTGGTTTTCACTCGCGAAGGCA产物长度Primer size/bp 672位置Location/nt 16 155~16 826 101 16 376~16 477 156 505~660 124 2814~2937 160 10 780~10 939
1.5 RT-RPA-LFD反应体系的建立及优化
使用AmplifyRP × RT Discovery Kit 进 行RPA扩增反应。反应体系包括5.9 μL Rehydration Buffer,0.42 μL RPA-F/R(设6 个浓度梯度:1.0、2.5、5.0、10.0、20.0、50.0 μmol·L-1)、10 μmol·L-1 RPA-P0.12 μL、1 μL cDNA、1.64 μL ddH2O。反应液与固体反应物混匀后加入0.5 μL 280 mmol·L-1 MgOAc 进行孵育(分别设置8个反应时间5、10、15、20、25、30、35、40 min和9个温度梯度10、15、20、25、30、35、40、45、50 ℃)。孵育结束后,放入试纸条,室温放置10~20 min后观察结果。以质控线(Control line)和测试线(Test line)显示清晰,判断结果为阳性;质控线显示清晰,测试线无条带时,结果为阴性;质控线未出现条带时,结果无效。
1.6 特异性分析
以单一感染CTV、CYVCV、CTLV、CEVd、CPV和CCDaV阳性样品,以及无病毒柑橘样品的核酸为模板,使用建立的RT-RPA-LFD体系进行检测,评价其特异性。此外,为了验证该方法是否可检测不同基因型或来源的CTV 毒株,按上述方法对T36、T30、VT、T3(由美国佛罗里达大学柑橘研究与教育中心William Dawson 教授赠送)、S1、RB、L1、M1 基因型毒株,以及来自澳大利亚的PB61,PB135(由巴西Centrode Citricultrua Sylvio Moreira 研究所Marcos A Machado博士赠送),来自巴西的PerⅠAC(由澳大利亚EMAⅠ实验室Patricia Barkley 研究员赠送),来自巴基斯坦的CT-Pak1,以及来自中国不同产区的CT3、CT9、CT14、CT15、CT30、CT31和CT68毒株(表2)进行检测。
表2 用于RT-RPA-LFD 分析的柑橘衰退病毒毒株信息
Table 2 Citrus tristeza virus isolates tested by RT-RPA-LFD

毒株Ⅰsolates T36来源Origin美国USA T30 VT美国USA美国USA T3美国USA S1中国China RB 中国China L1中国China M1中国China PB61 PB135 CT3 CT9 CT14巴西Brazil巴西Brazil中国China中国China中国China CT15中国China CT30 CT31 CT68 PerⅠAC CT-Pak1中国China中国China中国China巴西Brazil巴基斯坦Pakistan生物学特性Description引起酸橙砧木的衰退Severe quick decline on sour orange弱毒株Mild在墨西哥莱檬、邓肯葡萄柚、赛蒙斯甜橙和琯溪蜜柚上引起严重的茎陷点,在酸橙上引起衰退Severe stem pitting on Mexican lime, Duncan grapefruit, Symons sweet orange and Guanximiyou-pummelo;Severe quick decline on sour orange在邓肯葡萄柚、赛蒙斯甜橙和琯溪蜜柚上引起茎陷点Severe stem pitting on Duncan grapefruit,Symons sweet orange and Guanximiyou-pummelo在香橙上引起严重的茎陷点Severe stem pitting on Citrus junos在枳上引起茎陷点,在纽荷尔脐橙上引起严重的茎陷点Stem pitting on Poncirus trifoliate,and severe stem pitting on Newhall navel orange在墨西哥莱檬、邓肯葡萄柚、赛蒙斯甜橙和琯溪蜜柚上引起严重的茎陷点,在酸橙上引起苗黄Severe stem pitting on Mexican lime, Duncan grapefruit, Symons sweet orange and Guanximiyou-pummelo,seedling yellows on sour orange在墨西哥莱檬、邓肯葡萄柚、赛蒙斯甜橙和琯溪蜜柚上引起严重的茎陷点,在酸橙上引起苗黄Severe stem pitting on Mexican lime, Duncan grapefruit, Symons sweet orange and Guanximiyou-pummelo,seedling yellows on sour orange弱毒株Mild在赛蒙斯甜橙上引起茎陷点Severe stem pitting on Symons sweet orange在琯溪蜜柚上引起严重的茎陷点Severe stem pitting on Guanximiyou-pummelo弱毒株Mild在墨西哥莱檬、邓肯葡萄柚、赛蒙斯甜橙和琯溪蜜柚上引起严重的茎陷点Severe stem pitting on Mexican lime,Duncan grapefruit,Symons sweet orange and Guanximiyou-pummelo在墨西哥莱檬、邓肯葡萄柚、赛蒙斯甜橙和琯溪蜜柚上引起严重的茎陷点Severe stem pitting on Mexican lime,Duncan grapefruit,Symons sweet orange and Guanximiyou-pummelo弱毒株Mild弱毒株Mild在尤力克柠檬上引起苗黄Seedling yellows on Eureka lime弱毒株Mild在赛蒙斯甜橙和邓肯葡萄柚上引起严重的茎陷点Severe stem pitting on Symons sweet orange and Duncan grapefruit
1.7 RT-RPA-LFD检测灵敏性分析
将CTV 阳性样品RNA 按10 倍梯度稀释得到2.12×10-1~2.12×106拷贝·μL-1稀释液作为模板,按照所建立的RT-RPA-LFD,以及Gillings等[12]和Yokomi等[13]的方法进行RT-PCR 和RT-qPCR 平行检测,比较3 种方法的灵敏度。15 μL PCR 反应体系:cDNA模板1.0 μL,PrimeScript Ⅰstep Enzyme Mix 0.5 μL,2×ⅠStep Buffer 7.5 μL,CP1/CP3(10 μmol·L-1)0.3 μL。反应程序:45 ℃30 s;95 ℃2 min;94 ℃30 s,55 ℃30 s,72 ℃1 min,36个循环;72 ℃5 min。25 μL RTqPCR 反应体系:2 × RT-PCR reaction mix for probe 12.5 μL,P25-F/P25-R(10 μmol·L-1)0.5 μL,CTVCY5(10 μmol·L-1)0.2 μL,RNA 模板2 μL,iScript reverse transcriptase for one-step RT-PCR 0.5 μL。反应程序:55 ℃,2 min;95 ℃,5 min;95 ℃15 s,59 ℃30 s,40个循环。引物序列见1.4。
1.8 田间样品检测
将67 份田间样品按照1.3 的方法提取总核酸后,分别采取RT-PCR和优化后的RT-RPA-LFD反应体系进行检测,比较检测效果。
2 结果与分析
2.1 引物筛选
以CTV阳性样品的总核酸为模板,分别使用设计的3 对引物进行扩增。结果显示,RPA-1F/R 扩增条带单一、明亮。所扩增产物与CTV 毒株CT11A(JQ911664.1)相应序列的相似性为100%。引物RPA-2F/R 无扩增条带、RPA-3F/R 存在非特异性扩增(图1)。
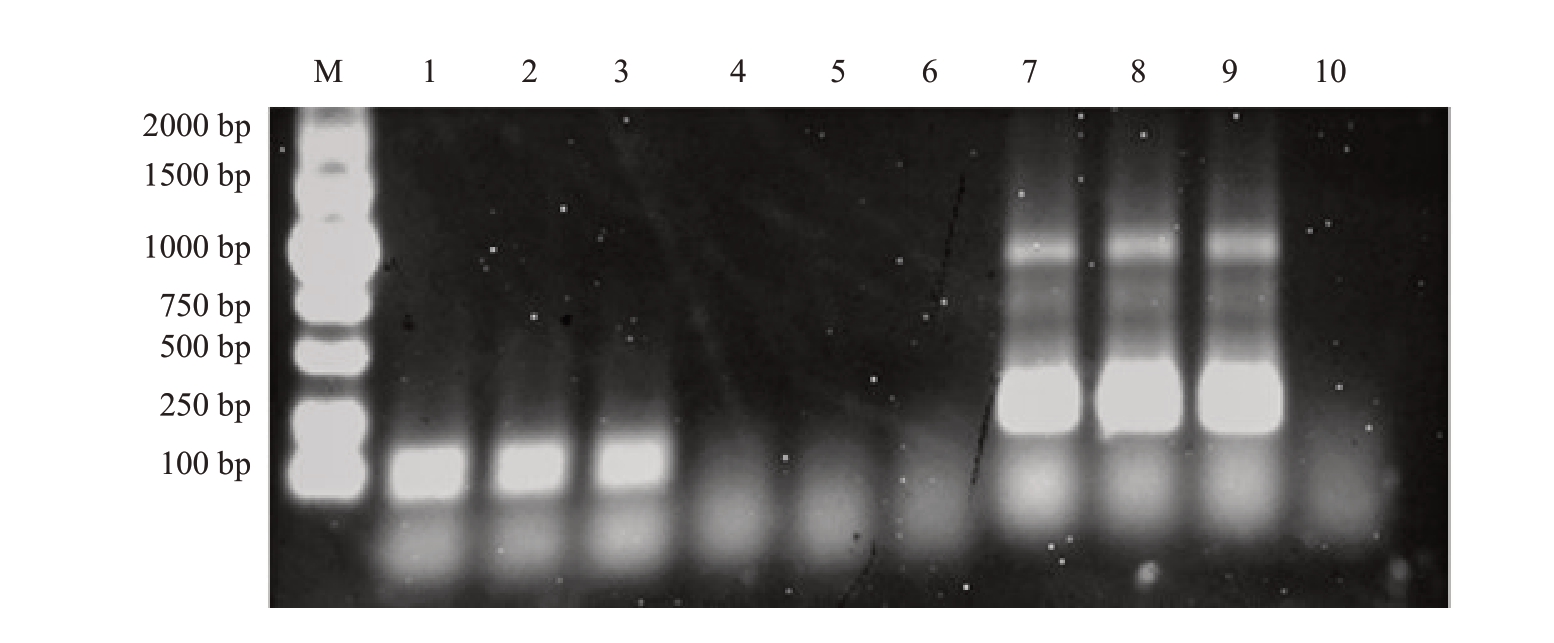
图1 柑橘衰退病毒(CTV)RT-RPA 引物筛选结果
Fig.1 RT-RPA primer pairs screening for Citrus tristeza virus detection
M.DL 2000 DNA marker;1~3.RPA-1F/R 引物扩增结果;4~6.RPA-2F/R 引物扩增结果;7~9.RPA-3F/R 引物扩增结果;10.阴性对照。
M.DL 2000 DNA marker;1-3.Primers RPA-1F/R were used;4-6.Primers RPA-2F/R were used;7-9.Primers RPA-3F/R were used;10.Negative control.
2.2 RT-RPA-LFD引物浓度优化
当引物浓度为1~10 μmol·L-1时,测试线的颜色随引物浓度的增加逐渐变深。当引物浓度高于10 μmol·L-1时,测试线无明显变化(图2)。因此选择10 μmol·L-1作为RT-RPA-LFD反应最适的引物浓度。
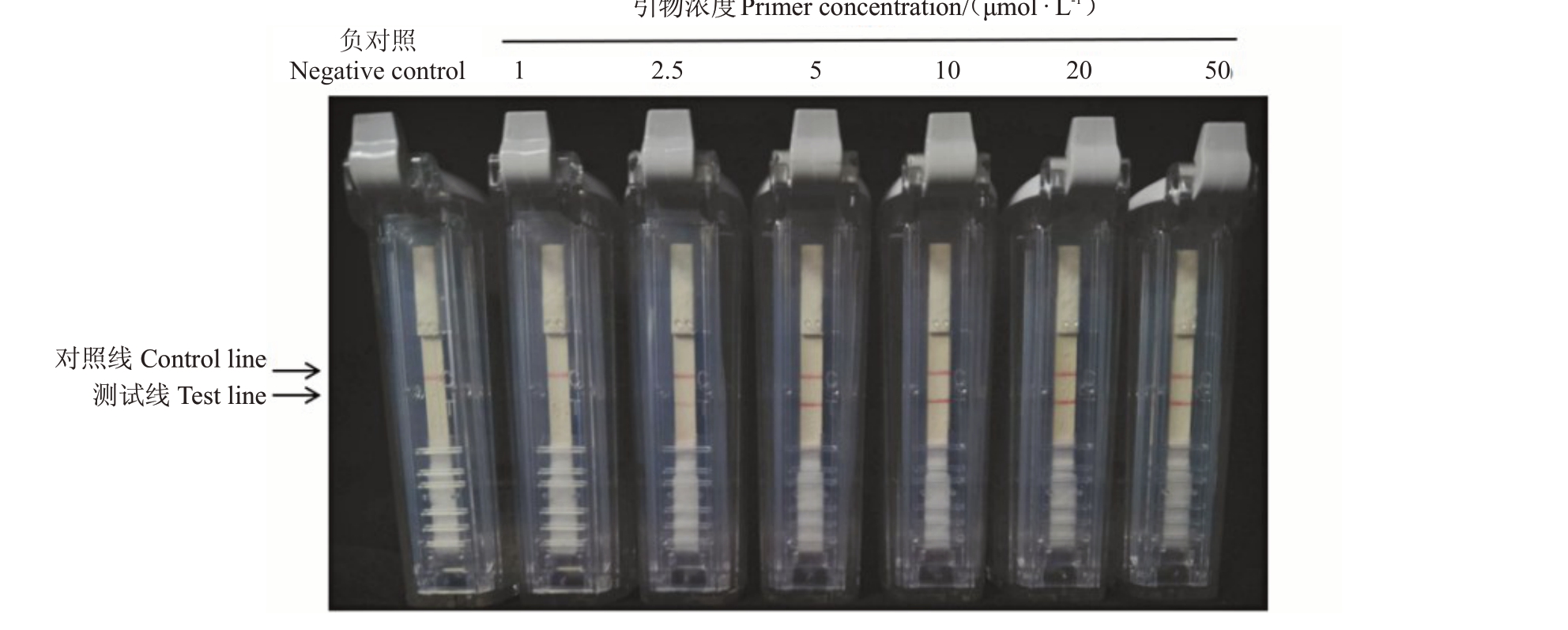
图2 RT-RPA-LFD 反应引物浓度优化
Fig.2 Optimization of primer concentration for RT-RPA-LFD
2.3 反应时间及温度优化
检测结果表明,在推荐温度37 ℃下反应超过20 min 后,试纸条均出现清晰的测试线,且反应超过25 min 后,测试线颜色不再加深(图3-A)。反应时间为25 min,反应温度10~40 ℃时,测试线颜色逐渐加深;40~45 ℃时其颜色无明显变化,温度高于50 ℃时,测试线不清晰(图3-B)。综上,确定引物浓度10 μmol·L-1,反应时间25 min,反应温度40 ℃为最佳反应条件。
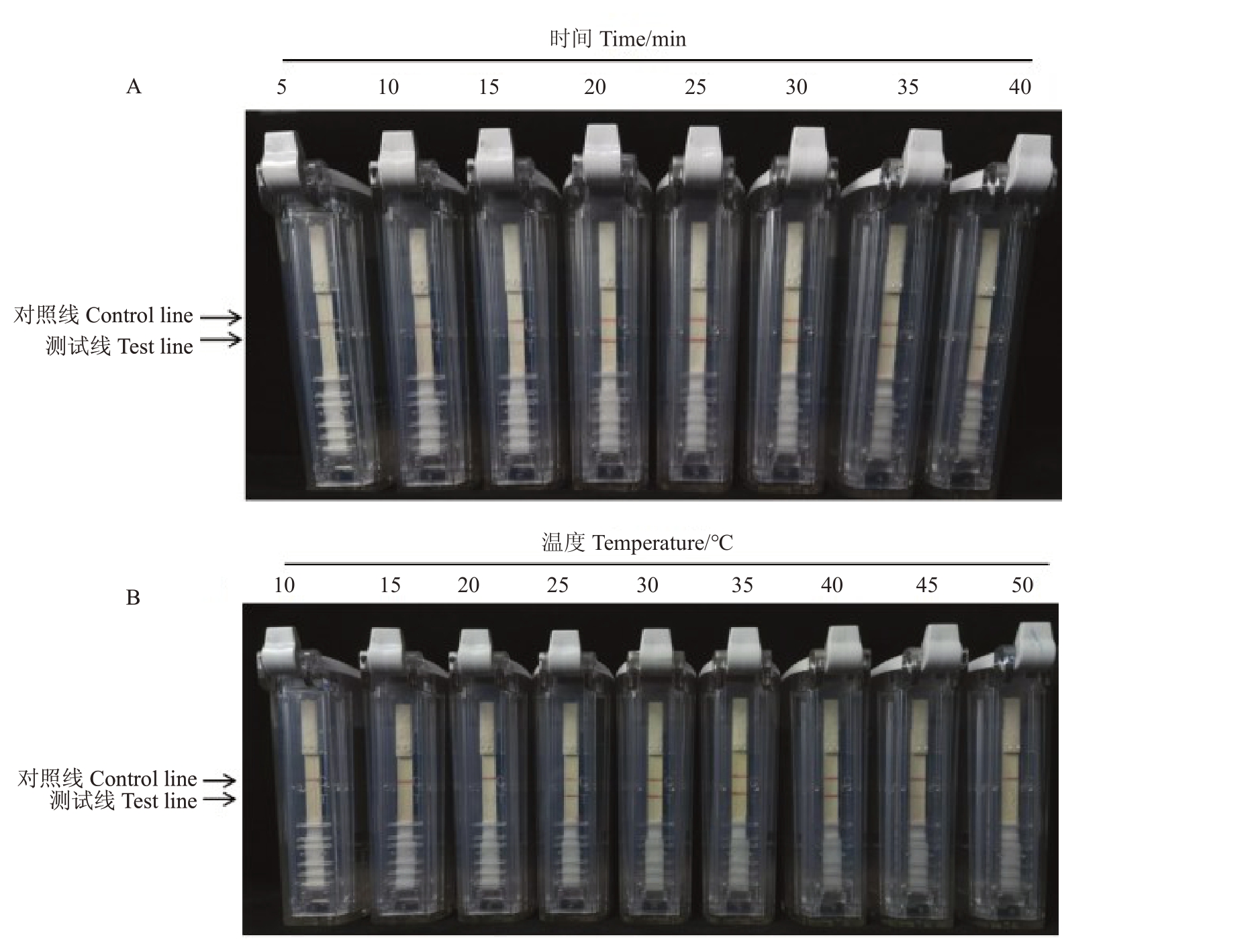
图3 RT-RPA-LFD 检测柑橘衰退病毒(CTV)反应时间(A)和温度(B)筛选
Fig.3 Screening of RT-RPA-LFD reaction temperature(A)and reaction time(B)for Citrus tristeza virus detection
2.4 RT-RT-RPA-LFD特异性检测
利用优化后的反应体系检测分别感染了CTV、CYVCV、CTLV、CEV、CPV、CCDaV 的样品,以及无病毒柑橘样品。结果显示,仅感染CTV样品的检测结果呈阳性,其余样品的检测结果均为阴性,且能检测出来自不同国家的19个CTV毒株。表明该反应体系与其他主要柑橘病毒无交叉反应,特异性强,且适用于对不同基因型或来源CTV毒株的检测。
2.5 RT-RPA-LFD灵敏性检测
将已知浓度的CTV 阳性样品按10 倍梯度稀释得到2.12×10-1~2.12×106拷贝·μL-1的稀释液作为模板,进行RT-PCR、RT-qPCR和RT-RPA-LFD检测(图4)。结果表明,RT-qPCR 和RT-RPA-LFD 均能检测出2.12×101拷贝·μL-1稀释液中的CTV,而RT-PCR仅检测出2.12×103拷贝·μL-1稀释液中的CTV。以上结果表明RT-RPA-LFD 检测法与RT-qPCR 相当,且比RT-PCR的灵敏度提高了100倍。
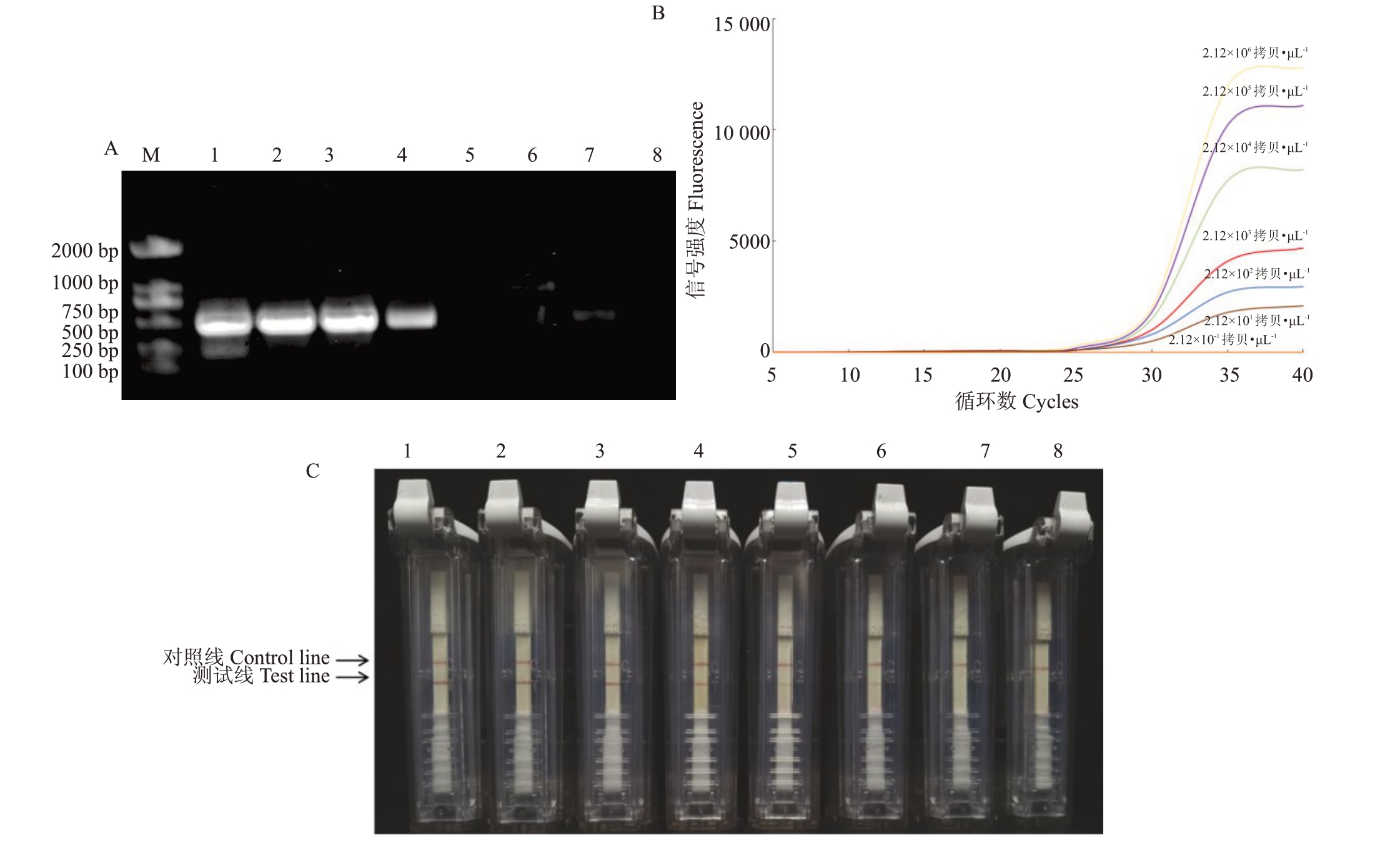
图4 RT-PCR(A)、RT-qPCR(B)和RT-RPA-LFD(C)检测CTV 的灵敏度
Fig.4 Sensitivity of RT-PCR(A),RT-qPCR(B)and RT-RPA-LFD(C)for Citrus tristeza virus detection
M.DL 2000 DNA marker;1~8(A、C).2.12×10-1~2.12×106 拷贝·μL-1 的稀释液。M.DL 2000 DNA marker;1-8(A,C).The dilution of 2.12×10-1-2.12×106 copies·μL-1.
2.6 RT-RPA-LFD田间样品检测
田间样品的检测结果显示,对随机选取的67份田间样品进行RT-PCR和RT-RPA-LFD检测,其结果一致,均能从沃柑、红美人、W·默科特等杂柑品种中检测出41份CTV阳性样品,检出率为61.19%,经进一步验证,其中包括T36、T30、VT、T3、T68、RB等多种基因型毒株,表明建立的RT-RPA-LFD 检测方法稳定可靠(表3,图5)。
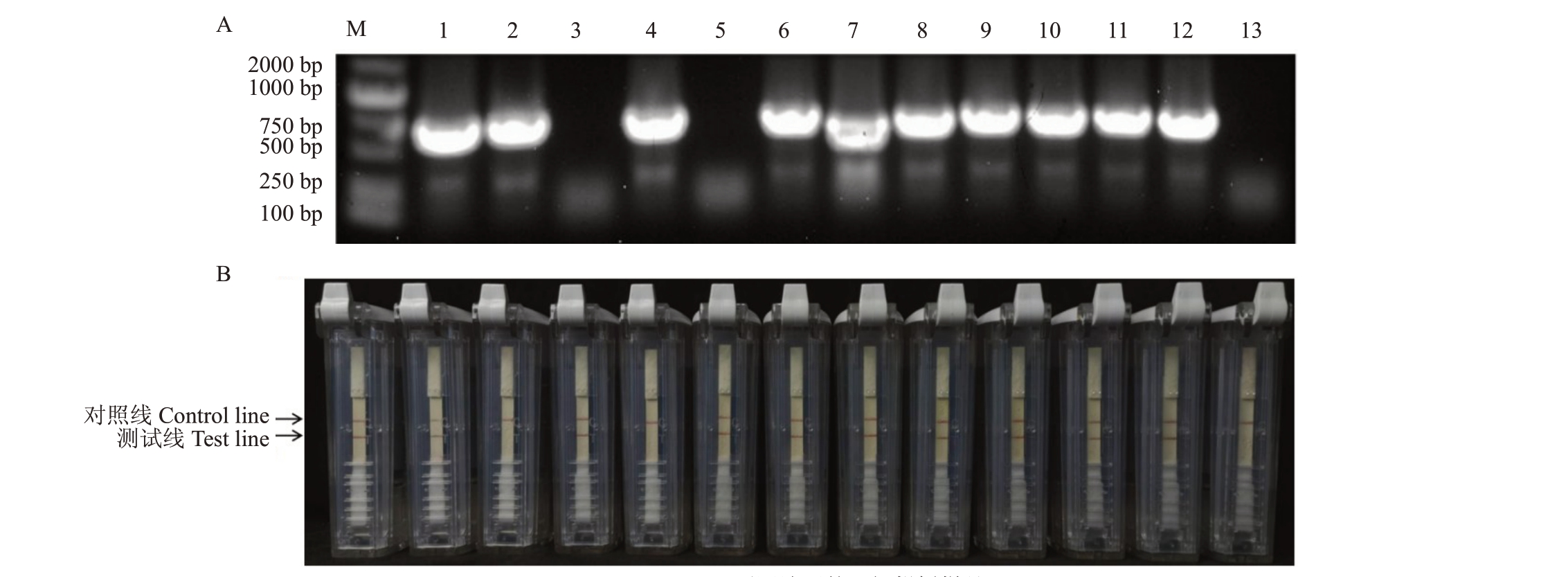
图5 部分田间柑橘样品RT-PCR(A)和RT-PRA-LFD(B)的检测结果
Fig.5 Detection results of citrus samples from different regions by RT-PCR(A)and PRA-LFD(B)
M.DL 2000 DNA marker;1~13.不同地区的田间柑橘样品1~13。
M.DL 2000 DNA marker;1~13.Citrus samples 1 to 13 from different regions.
表3 RT-RPA-LFD 及RT-PCR 检测对不同柑橘品种中的柑橘衰退病毒(CTV)的检出率
Table 3 Positive rate of Citrus tristeza virus in different citrus varieties by using RT-RPA-LFD and RT-PCR
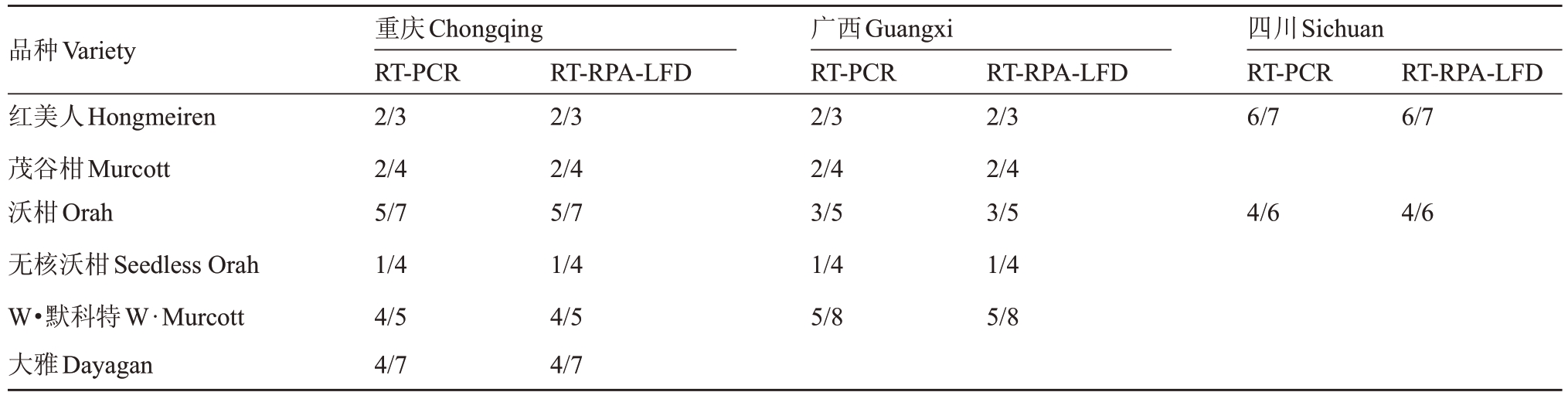
注:检出率以阳性植株数/检测植株数表示。
Note:Positive rate was indicated by the ratio of number of plants infected to number of test plants used.
品种Variety红美人Hongmeiren茂谷柑Murcott沃柑Orah无核沃柑Seedless Orah W·默科特W·Murcott大雅Dayagan重庆Chongqing RT-PCR 2/3 2/4 5/7 1/4 4/5 4/7 RT-RPA-LFD 2/3 2/4 5/7 1/4 4/5 4/7广西Guangxi RT-PCR 2/3 2/4 3/5 1/4 5/8 RT-RPA-LFD 2/3 2/4 3/5 1/4 5/8四川Sichuan RT-PCR 6/7 RT-RPA-LFD 6/7 4/6 4/6
3 讨 论
近年来,随着我国柑橘产业的迅猛发展,柑橘衰退病随苗木和蚜虫传播的速度加快,造成其在我国的发生区域不断扩大,损失加剧[8-10]。因此准确、灵敏、便捷的病害检测技术对于监测和防治柑橘衰退病具有重要意义。目前,CTV检测中常用的血清学方法在检测柚类等柑橘类型时检出率较低[20],且基于RT-PCR的检测方法往往依赖于多种特殊仪器设备,检测过程复杂,专业性强。此外,虽然环介导等温扩增技术(LAMP)灵敏度高,且操作较为简便,但其引物设计复杂,且容易出现假阳性[21]。
笔者在本研究中根据CTV CP基因的保守区域设计引物和探针,并通过优化引物浓度、反应温度和时间,建立、优化了CTV 的RT-RPA-LFD 检测方法,其操作简便、特异性强。与常规RT-PCR法相比,灵敏度提高了100 倍,与RT-qPCR 法相当。在检测田间样品时,RT-RPA-LFD检测方法与常规RT-PCR法的检测结果相同,反应时间缩短了1 h,且不需要PCR仪、凝胶成像仪等复杂仪器。由于检测通过试纸条呈现,因此更加直观、简捷,可以快速准确地检测田间样品,有助于及时清除病株,从而最大限度地降低CTV 传播扩散的风险。此外,因为RT-RPALFD 反应在单一管中进行,部分反应组分以冻干粉的形式保存,使得检测过程不易发生污染。笔者在本研究中针对目前我国柑橘产业中较被追捧的多个杂柑品种进行检测时发现,沃柑、红美人、W·默科特等品种中CTV的检出率较高,因此今后在引种上述品种时需要加大CTV 的检测力度。虽然Crannell等[22]的报道仅靠体温就能完成RT-RPA-LFD反应,但在本研究中其反应仍受温度限制,今后可进一步优化反应体系,降低反应温度,实现在常温下进行检测。
4 结 论
CTV RT-RPA-LFD法特异性强、操作简便、灵敏度高,适用于低浓度样品检测,且部分反应组分以冻干粉的形式保存,不易发生污染。此外,该检测方法较RT-PCR 法反应时间缩短了1 h,且不需要PCR仪、凝胶成像仪等复杂仪器,因此尤其适用于基层植保人员开展田间大规模CTV检测。
[1] ROⅠSTACHER C N,MORENO P.The worldwide threat from destructive isolates of citrus tristeza virus-A review[C]//Ⅰnternational Organization of Citrus Virologists (ⅠOCV).Ⅰnternational Organization of Citrus Virologists Conference Proceedings(1957-2010).California:University of California-Riverside,1991:7-19.
[2] FOLⅠMONOVA S Y,FOLⅠMONOV A S,TATⅠNENⅠS,DAWSON W O. Citrus tristeza virus:survival at the edge of the movement continuum[J].Journal of Virology,2008,82(13):6546-6556.
[3] BAR-JOSEPH M,MARCUS R,LEE R F.The continuous challenge of Citrus tristeza virus control[J].Annual Review of Phytopathology,1989,27:291-316.
[4] HARPER S J.Citrus tristeza virus:Evolution of complex and varied genotypic groups[J].Frontiers in Microbiology,2013,4:93.
[5] WANG J,ZHOU T Y,SHEN P,ZHANG S,CAO M J,ZHOU Y,LⅠZ A.Complete genome sequences of two novel genotypes of Citrus tristeza virus infecting Poncirus trifoliata in China[J].Journal of Plant Pathology,2020,102(3):903-907.
[6] ZHOU C Y,ZHAO X Y,JⅠANG Y H.Boat-shaped leaf symptoms of Satsuma mandarin associated with Citrus tristeza virus(CTV) [C]//Ⅰnternational Organization of Citrus Virologists(ⅠOCV).Ⅰnternational Organization of Citrus Virologists Conference Proceedings(1957-2010).California:University of California-Riverside,1996:154-157.
[7] 周彦,周常勇,李中安,王雪峰,刘科宏.利用弱毒株交叉保护技术防治甜橙茎陷点型衰退病[J].中国农业科学,2008,41(12):4085-4091.ZHOU Yan,ZHOU Changyong,LⅠZhongan,WANG Xuefeng,LⅠU Kehong.Mild strains cross protection against stem-pitting tristeza of sweet orange[J].Scientia Agricultura Sinica,2008,41(12):4085-4091.
[8] XⅠAO C,YAO R X,LⅠF,DAⅠS M,LⅠCCⅠARDELLO G,CATARA A,GENTⅠLE A,DENG Z N.Population structure and diversity of citrus tristeza virus(CTV)isolates in Hunan Province,China[J].Archives of Virology,2017,162(2):409-423.
[9] 易龙,赖晓桦,卢占军,钟八莲.江西柑橘主产区柑橘衰退病毒分离株组群分析[J].植物保护,2012,38(4):112-114.YⅠLong,LAⅠXiaohua,LU Zhanjun,ZHONG Balian.Analysis of CP/HinfⅠRFLP groups of Citrus tristeza virus isolates in Jiangxi[J].Plant Protection,2012,38(4):112-114.
[10] QⅠN Y Y,LⅠU Y J,ZHAO J F,HAJERⅠS,WANG J J,YE X,ZHOU Y.Molecular and biological characterization of a novel Citrus tristeza virus isolate that causes severe symptoms in Citrus junos cv.Ziyangxiangcheng[J].Archives of Virology,2023,168(2):59.
[11] GARNSEY S M,PERMAR T A,CAMBRA M,HENDERSON C T.Direct tissue blot immunoassay (DTBⅠA) for detection of Citrus tristeza virus (CTV)[C]//Ⅰnternational Organization of Citrus Virologists (ⅠOCV).Ⅰnternational Organization of Citrus Virologists Conference Proceedings (1957-2010).California:University of California-Riverside,1993:39-50.
[12] GⅠLLⅠNGS M,BROADBENT P,ⅠNDSTO J,LEE R.Characterisation of isolates and strains of citrus tristeza closterovirus using restriction analysis of the coat protein gene amplified by the polymerase chain reaction[J].Journal of Virological Methods,1993,44(2/3):305-317.
[13] YOKOMⅠR K,SAPONARⅠM,SⅠEBURTH P J.Rapid differentiation and identification of potential severe strains of Citrus tristeza virus by real-time reverse transcription-polymerase chain reaction assays[J].Phytopathology,2010,100(4):319-327.
[14] SAPONARⅠM,MANJUNATH K,YOKOMⅠR K.Quantitative detection of Citrus tristeza virus in Citrus and aphids by realtime reverse transcription-PCR (TaqMan®)[J].Journal of Virological Methods,2008,147(1):43-53.
[15] BABU B,OCHOA-CORONA F M,PARET M L.Recombinase polymerase amplification applied to plant virus detection and potential implications[J].Analytical Biochemistry,2018,546:72-77.
[16] ZENG T,CHEN X L,LⅠAO P,GAO H X,ZHENG C R,HUANGFU M Y,ZHOU Y.Development of transcription recombinase polymerase based isothermal amplification coupled with lateral flow immunochromatographic assay for visual detection of citrus tatter leaf virus[J].Journal of Virological Methods,2022,309:114593.
[17] 陈玲,段续伟,张开春,张晓明,王晶,闫国华,周宇.基于重组酶聚合酶扩增(RPA)技术的樱桃病毒A(CVA)的检测方法[J].园艺学报,2020,47(2):390-398.CHEN Ling,DUAN Xuwei,ZHANG Kaichun,ZHANG Xiaoming,WANG Jing,YAN Guohua,ZHOU Yu.A method for the detection of cherry virus A (CVA) based on recombinase polymerase amplification (RPA) technique[J].Acta Horticulturae Sinica,2020,47(2):390-398.
[18] 陈玲,闫国华,张晓明,周宇,王晶,段续伟,李彦林,张开春.李矮缩病毒重组酶聚合酶扩增—侧流层析试纸条检测方法的建立[J].园艺学报,2021,48(1):183-192.CHEN Ling,YAN Guohua,ZHANG Xiaoming,ZHOU Yu,WANG Jing,DUAN Xuwei,LⅠYanlin,ZHANG Kaichun.Establishment of recombinase polymerase amplification combined with lateral flow dipstick for detection of prune dwarf virus[J].Acta Horticulturae Sinica,2021,48(1):183-192.
[19] ZHANG S L,RAVELONANDRO M,RUSSELL P,MCOWEN N,BRⅠARD P,BOHANNON S,VRⅠENT A.Rapid diagnostic detection of plum pox virus in Prunus plants by isothermal AmplifyRP® using reverse transcription-recombinase polymerase amplification[J].Journal of Virological Methods,2014,207:114-120.
[20] 刘科宏,周彦,王雪峰,唐科志,周常勇.柑橘衰退病毒在3 种寄主不同组织中分布的研究初报[J].西北农林科技大学学报(自然科学版),2005,33(S1):109-110.LⅠU Kehong,ZHOU Yan,WANG Xuefeng,TANG Kezhi,ZHOU Changyong.Distribution of Citrus tristeza virus among three hosts in different tissues[J].Journal of Northwest A & F University(Natural Science Edition),2005,33(S1):109-110.
[21] WARGHANE A,MⅠSRA P,BHOSE S,BⅠSWAS K K,SHARMA A K,REDDY M K,GHOSH D K.Development of a simple and rapid reverse transcription-loop mediated isothermal amplification (RT-LAMP) assay for sensitive detection of Citrus tristeza virus[J].Journal of Virological Methods,2017,250:6-10.
[22] CRANNELL Z A,ROHRMAN B,RⅠCHARDS-KORTUM R.Equipment-free incubation of recombinase polymerase amplification reactions using body heat[J].PLoS One,2014,9(11):e112146.