小G 蛋白(small GTPase)存在于多种真核生物中,是多个信号转导途径的中枢调节因子[1-3]。在结构上,小G 蛋白被分为5 个亚家族,包括Ras、Rho、Rab、Ran 和Arf。其中Rab、Arf 主要调控胞内囊泡运输,Ran 调控核质运输,而Ras 和Rho 起信号分子的作用[1,4]。动物和真菌中Rho 蛋白可分为Cdc42、Rac 等亚家族,而植物中仅存在一类特有的Rho 家族成员,被命名为ROP(Rho-related GTPase from plants);由于ROP与动物的Rac蛋白序列相似,所以也被为RAC[5]。拟南芥基因组编码11个ROP基因,分为4 组:第Ⅰ组(ROP8)、第Ⅱ组(ROP9~ROP11)、第Ⅲ组(ROP7)、第Ⅳ组(ROP1~ROP6)[6-7]。ROP1调控拟南芥花粉管顶端生长[5];ROP2调控根毛顶端生长,促进铺板细胞凸起区域形成;ROP4 和ROP6 引起根毛去极性生长[8]。ROP10 和ROP11 负调控ABA 反应,包括气孔的关闭、种子的萌发和根的伸长等[9-10]。这些研究结果都表明不同的ROP 成员具有不同的生理功能,并且参与调控众多细胞学过程。
ROP是细胞中重要的分子开关,可以在GDP结合的非活性形式与GTP 结合的活性形式之间不断转换[6,11]。ROP蛋白活性的上游调控因子,包括鸟苷酸交换因子(guanine nucleotide exchange factor,GEF)、GTP 酶激活蛋白(GTPase-activating protein,GAP)和鸟苷酸解离抑制因子(guanine nucleotide dissociation inhibitor,GDI)等。其中GEF 蛋白促进ROP蛋白由非活性态转变为活性态,从而激活ROP蛋白及其下游的信号转导途径。GAP 蛋白促进GTP的水解,使ROP蛋白变成与GDP结合的非活性形式。GDI 蛋白抑制非活性ROP 蛋白转变为活性形式,并将非活性的ROP 蛋白隔离于胞质中[12-13]。RIC(ROP-interactive CRIB motif containing protein)是ROP 蛋白重要的下游效应子,包含一个保守的CRIB(Cdc/Rac-interactive binding)结构。拟南芥基因组编码11个RIC基因,根据其序列相似性可分为5 组:第Ⅰ组(RIC9~RIC11)、第Ⅱ组(RIC6~RIC8)、第Ⅲ组(RIC1、RIC3)、第Ⅳ组(RIC5)以及第Ⅴ组(RIC2、RIC4)[14]。目前关于RIC 的研究主要集中在花粉管、铺板细胞和根细胞。在花粉管中,ROP1通过两个相互拮抗的下游途径相互制约对花粉管的生长进行调控:激活RIC4来促进花粉管顶端微丝的聚合,同时又激活RIC3提高顶端Ca2+浓度进而促进顶端微丝的解聚[15-16]。在铺板细胞中,RIC的功能与花粉管类似,但相对复杂一点的两个相互拮抗的途径调节了铺板细胞的嵌合生长:ROP2/4 激活RIC4 促进微丝的聚合,进而促进凸起区(lobe)的形成和发育;同时ROP2/4 使RIC1 失活,将RIC1 的活性限制在凹陷区(neck)。凹陷区的RIC1 被ROP6 激活,造成微管排列成横向有序列阵来抑制该区的侧向扩张;另一方面,RIC1 介导的凹陷区的横向微管又抑制了ROP2/4-RIC4 信号途径[17-18]。此外,Lin 等[19]发现RIC1作为SPK-ROP6的下游效应子,调控根细胞中ROP 信号途径介导的对生长素极性的运输。综上所述,小G 蛋白与上游调控因子和下游效应子之间相互作用共同构成了一系列的信号传递网络。
荧火素酶互补试验(luciferase complementation assay,LCA)主要应用于哺乳动物、植物蛋白质间的相互作用,该原理是以荧火素(luciferin)为底物来检测荧火素酶(luciferase)的活性,利用生物体来源的荧火素酶催化底物荧火素发生氧化反应,发出最强波长在560 nm左右的生物荧光。在试验中,荧火素酶蛋白被分成2 个功能片段:N 端和C 端,即NLuc(2~416 aa)和CLuc(398~550 aa),将2个待检测的目标蛋白分别与NLuc 和CLuc 融合,若2 个目标蛋白有互作,则荧火素酶的NLuc 和CLuc 在空间上能足够靠近并且正确组装,从而就能发挥荧火素酶的活性,即分解荧火素底物产生荧光[20]。目前,检测ROP蛋白活性的普遍方法是利用提前纯化好的MBPRIC1蛋白,从提取的植物总蛋白(或纯化的蛋白)中pull-down 有活性的ROP 蛋白,以此来检测ROP 蛋白的活性变化[21-23]。但是有效的pull-down 试验,需要高纯度的可溶性融合蛋白,找到特定的结合、洗涤和洗脱条件,从而才能实现目标蛋白相互作用的特异性及兼容性。因此,开发一种新的ROP蛋白活性检测方法,已成为研究ROP 蛋白功能的发展趋势。本方法基于RIC1 作为ROP 蛋白的效应子能特异地与活性形式的ROP结合这一特点,通过荧火素酶互补试验,借助烟草(Nicotiana benthamiana)瞬时表达系统就可以成功地检测ROP蛋白的活性变化,为进一步研究ROP蛋白的功能奠定了基础。
1 材料和方法
1.1 试验材料
载 体pCAMBIA1300- NLuc(pNL)和pCAMBIA1300-CLuc(pCL)由南京农业大学梨工程研发中心保存。拟南芥为哥伦比亚野生型(Columbia-0),小叶烟草为本氏烟草(N.benthamiana),分别种植于南京农业大学梨工程研发中心的植物栽培室。大肠杆菌T1(DH5α)购自北京全式金生物技术有限公司。农杆菌GV3101 购自南京百思禾生物科技有限公司。限制性内切酶BamHⅠ、KpnⅠ和SalⅠ购自纽英伦生物技术(北京)有限公司。Phanta Max 高保真DNA聚合酶、2×Rapid Taq master Mix、同源重组连接酶Exnase Ⅱ和定点突变试剂盒(Mut Express® Fast Mutagenesis Kit V2)购自南京诺唯赞生物科技有限公司。胶回收试剂盒、质粒小提试剂盒购自爱思进生物技术(杭州)有限公司。
1.2 RNA提取和cDNA合成
使用成都福际生物技术有限公司生产的多糖多酚植物RNA提取试剂盒提取RNA。反转录试剂盒TransScript® One-Step RT-PCR SuperMix 购自北京全式金生物技术有限公司。
1.3 基因克隆及表达载体构建
从拟南芥信息资源网站(https://www.arabidopsis.org/)中下载目的基因AtROP1、AtRIC1、AtGEF1、AtGDI1和AtGAP1的CDS参考序列,利用软件Primer Premier 5.0设计目的基因的引物(表1)。以拟南芥的花朵cDNA为模板,用F1/R1和F2/R2两对引物分别进行PCR扩增得到目的基因AtROP1片段1和At-RIC1片段2。pCL空载体用BamHⅠ和KpnⅠ进行双酶切,而pNL空载体用BamHⅠ和SalⅠ进行双酶切后,用琼脂糖凝胶回收试剂盒对片段1、2 和双酶切载体进行胶回收,再将片段1与双酶切载体pNL、片段2与双酶切载体pCL在重组酶反应体系下37 ℃、30 min,然后将样品置于冰上终止反应。重组酶反应终止后,立即进行载体转化,从反应体系中取10µL样品在冰上与100µL大肠杆菌T1感受态混合,冰浴30 min,然后在42 ℃水浴锅中热激45 s,立即放入冰上,3 min后在体系中加入700µL无抗LB培养基,在37 ℃摇床中孵育1 h后,将样品涂布于抗性卡那霉素(Kan)固体平板上,在37 ℃培养箱中倒置培养12 h。从上述的转化平板上挑取8个单克隆菌落分别放于装有200 µL Kan 的液体LB 的2 mL 离心管中,在37 ℃摇床中摇菌8~10 h。取其中1µL 菌液为PCR模板,以载体通用引物F3/R3和F4/R4分别对重组载体pNL 和pCL 进行菌落PCR 扩增。菌落PCR 体系为20µL,具体包括模板1µL、正向和反向引物均为1 µL、2×Rapid Taq master Mix 为10 µL,用无菌ddH2O 补至20µL。PCR 产物用琼脂糖凝胶电泳检测,如有目的条带,取100µL 菌液送公司测序。测序结果正确即可得到重组菌株,可以提取质粒进行下一步其他试验。根据诺唯赞公司的点突变试剂盒说明书设计AtROP1 定点突变的引物,再根据说明书步骤进行定点突变试验。试验所需引物见表1。
表1 试验所需引物及用途
Table 1 Primers needed in the experiment and its application

注:小写英文字母为同源臂和酶切位点序列。
Note:The lowercase English letters are homologous arm and enzyme cleavage site sequences.
引物名称Primer name AtRIC1-F1 AtRIC1-R1 AtROP1-F2 AtROP1-R2 AtGEF1-GFP-F AtGEF1-GFP-R AtGDI1-GFP-F AtGDI1-GFP-R AtGAP1-GFP-F AtGAP1-GFP-R pNL-F3 pNL-R3 pCL-F4 pCL-R4 F-35S GFP-R AtROP1-CA-F AtROP1-CA-R AtROP1-DN-F AtROP1-DN-R引物序列(5’-3’)Primer sequence(5’-3’)gcgtcccggggcggtaccATGGCGACGACAATGAAG agtccatttgttggatccGATAATATCGTTACAGGTTGTATCA ctcggtacccggggatccATGAGCGCTTCGAGGTTCGT gtacgagatctggtcgacTAGAATGGAGCATGCCTTCTGC gagaacacgggggactctagaATGGGGAGCT TATCTTCTGA gcccttgctcaccatggatccATCTCTTTCCGGCGTCA gagaacacgggggactctagaATGGATGAAG AGTACGAAGTCATA gcccttgctcaccatggatccTTCCTCCTCTGCAGCA gagaacacgggggactctagaATGACTGAAGTTCTTCACTTTCC gcccttgctcaccatggatccCCGCCAAGCTTCAGTAC ATCCTTCGCAAGACCCTTCCTC TGCTCTCCAGCGGTTCCATC CCTCATAAAGGCCAAGAAGGGCG GATTCTGGTGTGTGCGCAATG TCCACTGACGTAAGGGATG CGTCGTCCTTGAAGAAGATG TGCAGGGCTAGAGGATTACAATAGATTAAGACCACTGAGTTAC TAATCCTCTAGCCCTGCAGTGTCCCACAATCCAAGATTC AAAGCTTGCTCTTCGAGATAAACAGTTCTTTATCGACC CTCGAAGAGCAAGCTTTGTTCCAACAAGGACGATG用途Application基因克隆Gene cloning重组质粒PCR验证PCR verification of recombinant plasmid点突变基因克隆Point mutation gene cloning
1.4 荧火素酶互补试验
将目的基因AtROP1和AtRIC1分别构建到载体pNL和pCL上,并将AtROP1-pNL和AtRIC1-pCL的质粒转入农杆菌(GV3101),经PCR 鉴定为阳性后放在28 ℃、200 r·min-1摇床中培养1~2 d。采用张皓等[24]的方法,将得到的农杆菌液按体积比为1∶50扩繁后5000 r·min-1离心10 min收集菌液,并将菌液悬浮在含有10 mmol·L-1 MES、10 mmol·L-1 MgCl2和100 μmol·L-1 乙酰丁香酮的诱导剂中,调OD600=1.0时放置于室温50 r·min-1的摇床中孵育4 h,即得到侵染液。将含有目标基因表达载体的侵染液(At-ROP1-pNL 和AtRIC1-pCL)按体积比1∶1 混合并设置好对照组,然后将等体积的混合侵染液用1 mL注射器注入小叶烟草背面的相应位置。最后将小叶烟草放置于正常培养环境中继续培养40~48 h。荧火素酶互补试验参照赵燕等[20]的方法,稍作修改具体如下:
植物活体成像仪定性检测荧火素酶活性:(1)取已注射农杆菌40~48 h 后的小叶烟草叶片,用含1 mmol·L-1荧火素底物反应液D-luciferin 将叶片背面喷湿,在黑暗中静置烟草叶片5~10 min。(2)将烟草叶片背面向上放置于培养皿中,移入植物活体成像系统中检测发光情况。如果有荧光,即说明激活态的AtROP1 可以与AtRIC1 存在互作,同时也说明该方法可以检测到AtROP1蛋白的活性。为了确保试验的准确性和一致性,同一株烟草至少选取3 个重复进行荧光检测。(3)根据试验结果调整曝光强度并拍照。
酶标仪定量检测荧光强度:(1)取已注射农杆菌40~48 h 后的小叶烟草叶片,用打孔器(直径约0.6 cm)打孔,将烟草小圆片转移到含有200 μL无菌ddH2O的白色96孔酶标板中。为了保证试验的准确性,同一烟草注射部位至少设8 个重复。(2)用移液枪去除96孔酶标板内的无菌ddH2O后,加入200 μL含有1 mmol·L-1荧火素底物反应液D-luciferin,孵育10 min。在试验过程中枪头尽量不要触碰叶片,避免应激反应。(3)使用酶标仪进行荧光扫描。(4)根据结果的数值绘制柱形图,以显示荧火素酶分解底物后产生荧光的强度来判断目标蛋白AtROP1活性强弱的程度。
2 结果与分析
2.1 基因AtROP1和AtRIC1的克隆及融合载体构建
利用F1/R1 和F2/R2 引物分别进行PCR 扩增得到目的基因AtROP1片段1和AtRIC1片段2。凝胶电泳结果显示,片段1 和片段2 位于500~750 bp 之间,与预期的591 bp和672 bp基本相符(图1-A)。将片段1 与双酶切载体pNL 片段、片段2 与双酶切载体pCL 片段分别在重组酶Exnase Ⅱ的作用下进行连接,转化和测序。菌液PCR结果与目的基因条带基本一致(图1-B~C),测序结果显示目的基因AtROP1和AtRIC1已经连接到载体上。

图1 基因克隆(A)及阳性克隆的PCR 检测(B、C)
Fig.1 Gene cloning(A)and PCR detection of positive clones(B,C)
A.1~2 分别为AtROP1 和AtRIC1 的基因克隆,M 为DNA Marker;B.1~8 分别为重组质粒AtRIC1-pCL 转化大肠杆菌的PCR 检测;C.1~8 分别为重组质粒AtROP1-pNL 转化大肠杆菌的PCR 检测。
A.1-2.Gene clones of AtROP1 and AtRIC1,respectively.M.DNA Marker.B.1-8.PCR detection of recombinant plasmid AtRIC1-pCL was transformed into E.coli,respectively.C.1-8.PCR detection of recombinant plasmid AtROP1-pNL transformed into E.coli,respectively.
2.2 定性和定量检测激活态的AtROP1与AtRIC1的互作
为了检测AtRIC1 是AtROP1 蛋白下游的效应子,将构建好的表达载体(AtROP1-pNL 和AtRIC1-pCL)转化到农杆菌感受态GV3101,得到含有目标基因的农杆菌,通过农杆菌瞬时转化体系来验证AtROP1 与AtRIC1 的互作。如图2 所示,将烟草叶片背面向上放置于培养皿中喷洒1 mmol·L-1荧火素底物反应液D-luciferin,移入植物活体成像系统中检测发光情况以及利用酶标仪检测荧光强度。如果有荧光,即说明激活态的AtROP1 可以与AtRIC1 存在互作,同时也说明该方法可以检测到AtROP1 蛋白的活性。
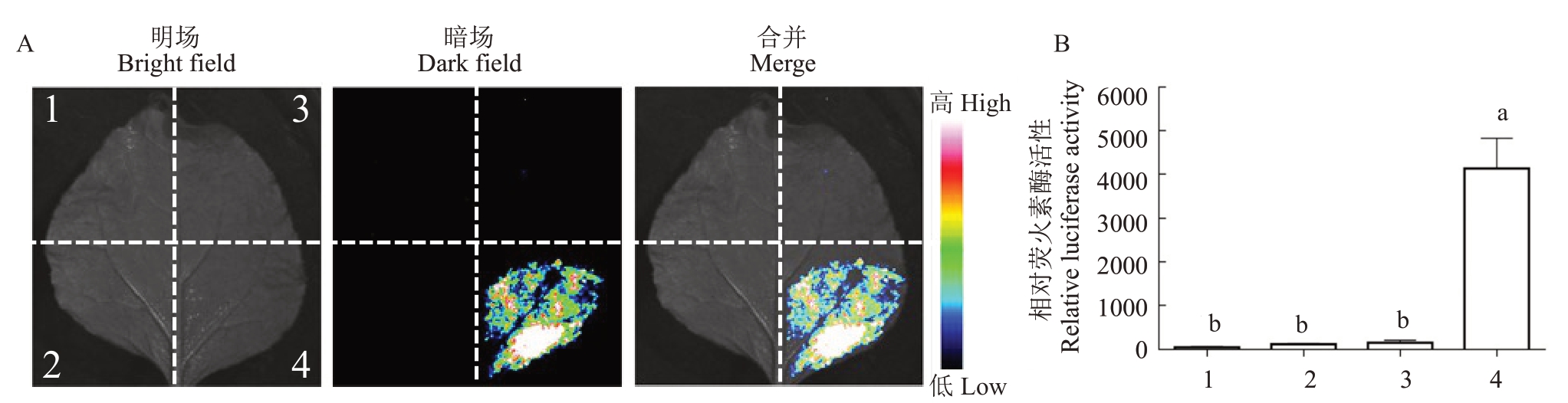
图2 定性和定量检测AtROP1 与AtRIC1 的互作
Fig.2 Qualitative and quantitative detection of the interaction between AtROP1 and AtRIC1
A.AtROP1 与AtRIC1 互作检测;B.AtROP1 与AtRIC1 互作的荧火素酶活性检测。1.NLuc+CLuc;2.NLuc+AtRIC1-CLuc;3.AtROP1-NLuc+CLuc;4.AtROP1-NLuc+AtRIC1-CLuc。
A.Detection of AtROP1 and AtRIC1 interactions; B.Detection of luciferase activity of of AtROP1 interacting with AtRIC1.1.NLuc+CLuc;2.NLuc+AtRIC1-CLuc;3.AtROP1-NLuc+CLuc;4.AtROP1-NLuc+AtRIC1-CLuc.
2.3 定性和定量检测AtROP1-CA 和AtROP1-DN的蛋白活性
利用点突变试剂盒对AtROP1 基因进行定点突变(图3),从而获得持续激活态的AtROP1(At-ROP1-CA)和持续失活态的AtROP1(AtROP1-DN)。将构建好的表达载体(AtROP1-CA-pNL 和AtROP1-DN-pCL)转化到农杆菌感受态GV3101 得到含有目标基因的农杆菌,通过农杆菌瞬时转化体系检测点突变后AtROP1蛋白活性的变化。试验结果表明AtROP1-CA 蛋白的荧火素酶荧光强度和活性明显比AtROP1 的高(图4),而AtROP1-DN 蛋白的荧火素酶荧光强度和活性明显比AtROP1 的低(图5)。
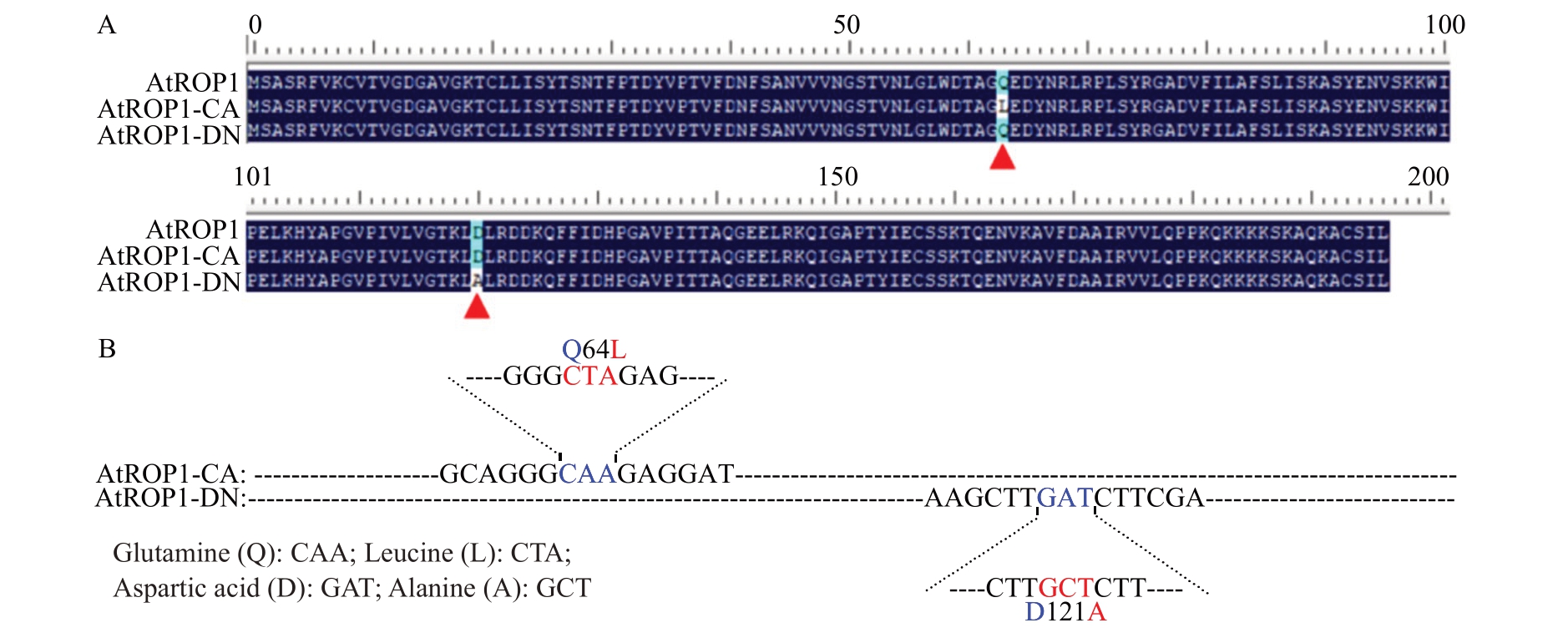
图3 ROP 蛋白序列比对与点突变位点图解
Fig.3 Sequence alignment and point mutation site diagram
A.AtROP1、AtROP1-CA 和AtROP1-DN 的蛋白序列比对;B.AtROP1-CA 和AtROP1-DN 点突变位点的图解。
A.Protein sequence alignment of AtROP1,AtROP1-CA and AtROP1-DN;B.Graphics of point mutation sites in AtROP1-CA and AtROP1-DN.
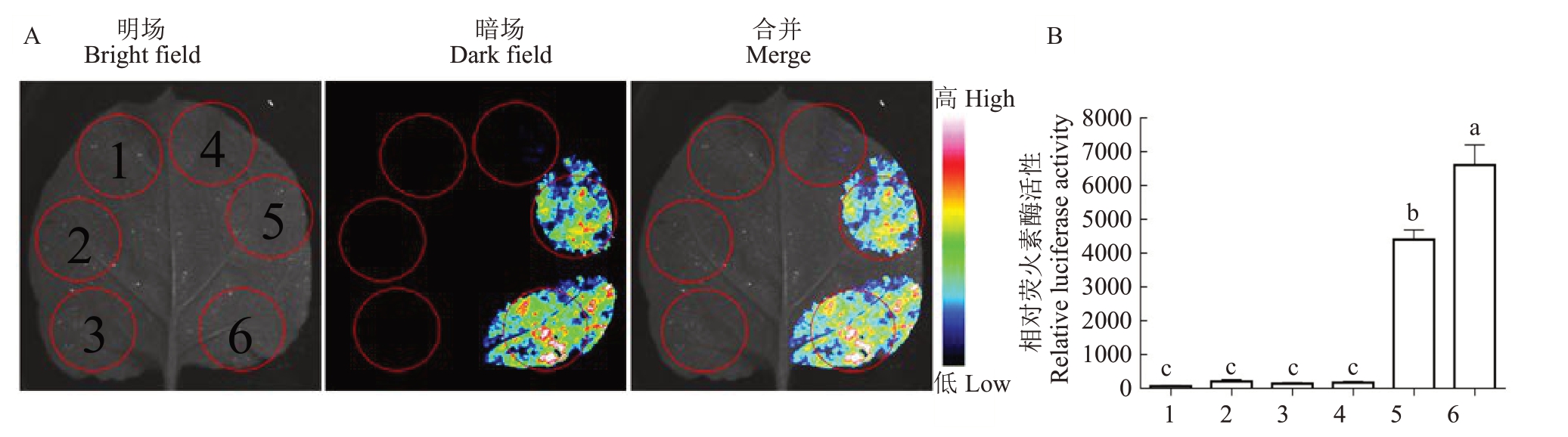
图4 定性和定量检测AtROP1-CA 的活性
Fig.4 Qualitative and quantitative detection of AtROP1-CA activity
A.AtROP1-CA 与AtRIC1 互作检测;B.AtROP1-CA 与AtRIC1 互作的荧火素酶活性检测。1.NLuc+CLuc;2.NLuc+AtRIC1-CLuc;3.At-ROP1-NLuc+CLuc;4.AtROP1-CA-NLuc+CLuc;5.AtROP1-NLuc+AtRIC1-CLuc;6.AtROP1-CA-NLuc+AtRIC1-CLuc。
A.Detection of AtROP1-CA and AtRIC1 interactions;B.Detection of luciferase activity of AtROP1-CA interacting with AtRIC1.1.NLuc+CLuc;2.NLuc+AtRIC1-CLuc;3.AtROP1-NLuc+CLuc;4.AtROP1-CA-NLuc+CLuc;5.AtROP1-NLuc+AtRIC1-CLuc;6.AtROP1-CA-NLuc+AtRIC1-CLuc.

图5 定性和定量检测AtROP1-DN 的活性
Fig.5 Qualitative and quantitative detection of AtROP1-DN activity
A.AtROP1-DN 与AtRIC1 互作检测;B.AtROP1-DN 与AtRIC1 互作的荧火素酶活性检测。1.NLuc+CLuc;2.NLuc+AtRIC1-CLuc;3.At-ROP1-NLuc+CLuc;4.AtROP1-DN-NLuc+CLuc;5.AtROP1-NLuc+AtRIC1-CLuc;6.AtROP1-DN-NLuc+AtRIC1-CLuc。
A.Detection of AtROP1-DN and AtRIC1 interactions;B.Detection of luciferase activity of AtROP1-DN interacting with AtRIC1.1.NLuc+CLuc;2.NLuc+AtRIC1-CLuc; 3.AtROP1-NLuc+CLuc; 4.AtROP1-DN-NLuc+CLuc;5.AtROP1-NLuc+AtRIC1-CLuc; 6.AtROP1-DN-NLuc+AtRIC1-CLuc.
2.4 AtGEF1、AtGAP1 和AtGDI1 蛋白对AtROP1蛋白活性的影响
为了进一步验证该试验方法的可靠性,成功构建了AtGAP1-GFP 和AtGDI1-GFP 载体,通过该试验体系验证抑制AtROP1 活性的上游调控因子(At-GAP1-GFP 和AtGDI1-GFP)是否同样抑制AtROP1蛋白活性。试验结果表明,与对照和加入GFP空载的处理组相比,AtGAP1-GFP处理组的AtROP1蛋白的荧火素酶荧光强度和活性都出现了明显的下降(图6-A~B);并检测到GFP 和AtGAP1-GFP 蛋白在该体系中确实有表达(图6-C),而AtGAP1-GFP 的表达明显降低AtROP1活性。同样,AtGDI1-GFP处理组与对照和加入GFP 空载的处理组相比,At-ROP1 蛋白的荧火素酶荧光强度和活性也出现明显的下降(图7-A~B);也检测到GFP 和AtGDI1-GFP蛋白在该体系中确实有表达(图7-C),而AtGDI1-GFP的表达明显降低AtROP1活性。

图6 定性和定量检测AtGAP1 对AtROP1 活性的影响
Fig.6 Qualitative and quantitative detection of the effect of AtGAP1 on AtROP1 protein activity
A.AtGAP1 对AtROP1 与AtRIC1 互作检测;B.AtGAP1 对AtROP1 与AtRIC1 互作的荧火素酶活性检测;C.AtGAP1、GFP 蛋白在烟草细胞中稳定表达,标尺为20 μm。1.NLuc+CLuc;2.NLuc+AtRIC1-CLuc;3.AtROP1-NLuc+CLuc;4.AtROP1-NLuc+AtRIC1-CLuc;5.AtROP1-NLuc+AtRIC1-CLuc+GFP;6.AtROP1-NLuc+AtRIC1-CLuc+AtGAP1-GFP。
A.Detection from AtGAP1 on the interaction between AtROP1 and AtRIC1;B.Detection of luciferase activity of AtGAP1 on AtROP1 interacting with AtRIC1;C.Stable expression of AtGAP1 and GFP proteins in tobacco cells.Bar=20 μm.1.NLuc+CLuc;2.NLuc+AtRIC1-CLuc;3.AtROP1-NLuc+CLuc;4.AtROP1-NLuc+AtRIC1-CLuc;5.AtROP1-NLuc+AtRIC1-CLuc+GFP;6.AtROP1-NLuc+AtRIC1-CLuc+AtGAP1-GFP.

图7 定性和定量检测AtGDI1 对AtROP1 活性的影响
Fig.7 Qualitative and quantitative detection of the effect of AtGDI1 on AtROP1 protein activity
A.AtGDI1 对AtROP1 与AtRIC1 互作检测;B.AtGDI1 对AtROP1 与AtRIC1 互作的荧火素酶活性检测;C.AtGDI1、GFP 蛋白在烟草细胞中稳定表达,标尺为20 μm。1.NLuc+CLuc;2.NLuc+AtRIC1-CLuc;3.AtROP1-NLuc+CLuc;4.AtROP1-NLuc+AtRIC1-CLuc;5.AtROP1-NLuc+AtRIC1-CLuc+GFP;6.AtROP1-NLuc+AtRIC1-CLuc+AtGDI1-GFP。
A.Detection from AtGDI1 on the interaction between AtROP1 and AtRIC1; B.Detection of luciferase activity of AtGDI1 on AtROP1 interacting with AtRIC1;C.Stable expression of AtGDI1 and GFP proteins in tobacco cells.Bar=20 μm.1.NLuc+CLuc;2.NLuc+AtRIC1-CLuc;3.AtROP1-NLuc+CLuc;4.AtROP1-NLuc+AtRIC1-CLuc;5.AtROP1-NLuc+AtRIC1-CLuc+GFP;6.AtROP1-NLuc+AtRIC1-CLuc+AtGDI1-GFP.
AtROP1 活性上游的调控因子AtGEF1 能够激活ROP 蛋白的活性,为证实该方法的适用性,成功构建了AtGEF1-GFP载体。试验结果表明,AtGEF1-GFP处理组比任何对照组的AtROP1蛋白的荧火素酶荧光强度和活性都明显增强(图8-A~B);并检测到GFP和AtGEF1-GFP蛋白在该体系中确实有表达(图8-C),而AtGEF1-GFP 的表达明显增强AtROP1活性。

图8 定性和定量检测AtGEF1 对AtROP1 活性的影响
Fig.8 Qualitative and quantitative detection of the effect of AtGEF1 on AtROP1 protein activity
A.AtGEF1 对AtROP1 与AtRIC1 互作检测;B.AtGEF1 对AtROP1 与AtRIC1 互作的荧火素酶活性检测;C.AtGEF1、GFP 蛋白在烟草细胞中稳定表达,标尺为20 μm。1.NLuc+CLuc;2.NLuc+AtRIC1-CLuc;3.AtROP1-NLuc+CLuc;4.AtROP1-NLuc+AtRIC1-CLuc;5.AtROP1-NLuc+AtRIC1-CLuc+GFP;6.AtROP1-NLuc+AtRIC1-CLuc+AtGEF1-GFP。
A.Detection from AtGEF1 on the interaction between AtROP1 and AtRIC1;B.Detection of luciferase activity of AtGEF1 on AtROP1 interacting with AtRIC1;C.Stable expression of AtGEF1 and GFP proteins in tobacco cells.Bar=20 μm.1.NLuc+CLuc;2.NLuc+AtRIC1-CLuc;3.AtROP1-NLuc+CLuc;4.AtROP1-NLuc+AtRIC1-CLuc;5.AtROP1-NLuc+AtRIC1-CLuc+GFP;6.AtROP1-NLuc+AtRIC1-CLuc+AtGEF1-GFP.
3 讨 论
ROP/RAC 蛋白是植物中重要的信号转导调节因子,参与植物细胞形态发育[17- 18,25- 26]、极性生长[8,15-16,27-28]、激素信号转导[9-10,19,29]、抗病[2,30-32]以及抗逆[3,21,33]等各种重要生命过程。最近,Lin等[34]还发现ROP 蛋白参与调控植物细胞自噬来应对逆境和营养的缺失。尽管通过pull-down 技术检测有活性的ROP 蛋白的方法已得到普遍使用[21-23],但其仍然存在一些缺点。如需要较高纯度的融合蛋白,合适的结合、洗涤和洗脱条件才能实现目标蛋白相互作用的特异性及兼容性。因此,笔者在本研究中建立一种操作简单且高效的方法,其对ROP蛋白的活性检测和功能验证具有重要意义。
荧火素酶互补试验作为一种具有简单、灵敏、可靠、高效和低背景等优点的方法,被广泛应用于植物学和动物学蛋白质互作研究[20]。与其他蛋白互作研究技术相比,利用烟草瞬时表达系统的荧火素酶互补试验可以定性、定量分析生物发光或发光强度,从而检测植物目标蛋白之间是否存在相互作用及互作强度[20]。通过该方法成功地检测到AtROP1与AtRIC1 有荧光,二者存在互作,同时还通过对AtROP1 基因进行定点突变试验获得了AtROP1-CA和AtROP1-DN 蛋白表达载体,通过检测发现RIC1蛋白可以检测突变ROP蛋白的活性变化。此外,通过进一步验证发现AtGEF1 蛋白可以明显地激发AtROP1 蛋白活性,同时AtGAP1 和AtGDI1 蛋白显著地抑制AtROP1 蛋白活性。目前利用纯化好的RIC1-MBP融合蛋白来检测ROP蛋白活性是一种常见的技术手段[21-23],但传统的pull-down 方法存在试验环节较多,即需要表达纯化多个蛋白,检测蛋白是否表达以及蛋白的纯度,操作繁杂且重复,往往还导致目的蛋白表达量偏低等问题。针对这些问题,该方法基于RIC1 作为ROP 的效应子能特异地与活性形式的ROP 结合这一特点,通过荧火素酶互补试验,借助烟草瞬时表达系统,将含有融合蛋白的表达载体转化到农杆菌后注射小叶烟草叶片。利用植物活体成像仪或酶标仪来实现定性定量检测荧光强度,以判定ROP蛋白活性的强弱程度。同时还可以添加其他目的蛋白来检测其对ROP 蛋白活性的影响。该方法具有高灵敏度、可视化、可定量化和操作简单高效等特点,为进一步研究ROP蛋白的功能提供了一定的技术支持。
4 结 论
ROP/RAC 蛋白是植物中多个信号转导途径的中枢调节因子。该方法通过荧火素酶互补实验,借助烟草瞬时表达系统就可以成功地检测ROP 蛋白活性的变化。与其他方法相比,该方法具有高灵敏度、可视化、可定量化和操作简单等特点,并且适用于检测其他蛋白对ROP蛋白活性的影响,所以该方法是一种可靠的体内检测ROP蛋白活性的新方法。
[1]TAKAI Y,SASAKI T,MATOZAKI T.Small GTP-binding proteins[J].Physiological Reviews,2001,81(1):153-208.
[2]ONO E,WONG H L,KAWASAKI T,HASEGAWA M,KODAMA O,SHIMAMOTO K.Essential role of the small GTPase Rac in disease resistance of rice[J].Proceedings of the National Academy of Sciences of the United States of America,2001,98(2):759-764.
[3]MIAO H X,SUN PG,LIU J H,WANG JY,XU BY,JIN Z Q.Overexpression of a novel ROP gene from the banana (MaROP5g)confers increased salt stress tolerance[J].International Journal of Molecular Sciences,2018,19(10):3108.
[4]BERKEN A.ROPs in the spotlight of plant signal transduction[J].Cellular and Molecular Life Sciences,2006,63(21):2446-2459.
[5]CRADDOCK C,LAVAGI I,YANG Z B.New insights into Rho signaling from plant ROP/Rac GTPases[J].Trends in Cell Biology,2012,22(9):492-501.
[6]ZHENG Z L,YANG Z B.The Rop GTPase:an emerging signaling switch in plants[J].Plant Molecular Biology,2000,44(1):1-9.
[7]YANG Z B.Small GTPases:Versatile signaling switches in plants[J].The Plant Cell,2002,14(S):375-388.
[8]MOLENDIJK A J,BISCHOFF F,RAJENDRAKUMAR C S,FRIML J,BRAUN M,GILROY S,PALME K. Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth[J].The EMBO Journal,2001,20(11):2779-2788.
[9]ZHENG Z L,NAFISI M,TAM A,LI H,CROWELL D N,CHARY S N,SCHROEDER J I,SHEN J J,YANG Z B.Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis[J].The Plant Cell,2002,14(11):2787-2797.
[10]LI Z X,KANG J,SUI N,LIU D.ROP11 GTPase is a negative regulator of multiple ABA responses in Arabidopsis[J].Journal of Integrative Plant Biology,2012,54(3):169-179.
[11]VETTER I R,WITTINGHOFER A.The guanine nucleotidebinding switch in three dimensions[J].Science,2001,294(5545):1299-1304.
[12]BERKEN A.ROPs in the spotlight of plant signal transduction[J].Cellular and Molecular Life Sciences,2006,63(21):2446-2459.
[13]NIBAU C,WU H M,CHEUNG A Y.RAC/ROP GTPases:‘Hubs’for signal integration and diversification in plants[J].Trends in Plant Science,2006,11(6):309-315.
[14]WU G,GU Y,LI S D,YANG Z B.A genome-wide analysis of Arabidopsis Rop- interactive CRIB motif- containing proteins that act as Rop GTPase targets[J].The Plant Cell,2001,13(12):2841-2856.
[15]GU Y,FU Y,DOWD P,LI S D,VERNOUD V,GILROY S,YANG Z B.A Rho family GTPase controls actin dynamics and tip growth via two counteracting downstream pathways in pollen tubes[J].The Journal of Cell Biology,2005,169(1):127-138.
[16]LEE Y J,SZUMLANSKI A,NIELSEN E,YANG Z B.Rho-GTPase-dependent filamentous actin dynamics coordinate vesicle targeting and exocytosis during tip growth[J].The Journal of Cell Biology,2008,181(7):1155-1168.
[17]FU Y,GU Y,ZHENG Z L,WASTENEYS G,YANG Z B.Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis[J].Cell,2005,120(5):687-700.
[18]FU Y,XU T D,ZHU L,WEN M Z,YANG Z B.A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis[J].Current Biology,2009,19(21):1827-1832.
[19]LIN D S,NAGAWA S,CHEN J S,CAO L Y,CHEN X,XU T D,LI H J,DHONUKSHE P,YAMAMURO C,FRIML J,SCHERES B,FU Y,YANG Z B.A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots[J].Current Biology,2012,22(14):1319-1325.
[20]赵燕,周俭民.萤火素酶互补实验检测蛋白互作[J].植物学报,2020,55(1):69-75.ZHAO Yan,ZHOU Jianmin.Luciferase complementation assay for detecting protein interactions[J].Chinese Bulletin of Botany,2020,55(1):69-75.
[21]LI C J,LU H M,LI W,YUAN M,FU Y.A ROP2-RIC1 pathway fine-tunes microtubule reorganization for salt tolerance in Arabidopsis[J].Plant,Cell & Environment,2017,40(7):1127-1142.
[22]KANG E F,ZHENG M Z,ZHANG Y,YUAN M,YALOVSKY S,ZHU L,FU Y.The microtubule-associated protein MAP18 affects ROP2 GTPase activity during root hair growth[J].Plant Physiology,2017,174(1):202-222.
[23]杨波.拟南芥SnRK1 通过调节ROP 活性参与铺板细胞形态建成的机制研究[D].北京:中国农业大学,2015.YANG Bo.SnRK1 is involved in leaf pavement cell morphogenesis through regulation of ROP GTPase activity in Arabidopsis[D].Beijing:China Agricultural University,2015.
[24]张皓,刘雪莹,钱铭,高鸿儒,汤超,张华,王鹏,张绍铃,吴巨友.一种快速添加或替换蛋白标签的新方法及应用[J].南京农业大学学报,2021,44(6):1046-1053.ZHANG Hao,LIU Xueying,QIAN Ming,GAO Hongru,TANG Chao,ZHANG Hua,WANG Peng,ZHANG Shaoling,WU Juyou.A new method of rapid protein tag addition or replacement and its application[J].Journal of Nanjing Agricultural University,2021,44(6):1046-1053.
[25]LIN W W,TANG W X,PAN X,HUANG A B,GAO X Q,ANDERSON C T,YANG Z B.Arabidopsis pavement cell morphogenesis requires FERONIA binding to pectin for activation of ROP GTPase signaling[J].Current Biology,2022,32(3):497-507.
[26]LIN D S,REN H B,FU Y.ROP GTPase-mediated auxin signaling regulates pavement cell interdigitation in Arabidopsis thaliana[J].Journal of Integrative Plant Biology,2015,57(1):31-39.
[27]BURKART G M,BASKIN T I,BEZANILLA M.A family of ROP proteins that suppresses actin dynamics,and is essential for polarized growth and cell adhesion[J].Journal of Cell Science,2015,128(14):2553-2564.
[28]LI E,ZHANG Y L,SHI X L,LI H,YUAN X F,LI S,ZHANG Y.A positive feedback circuit for ROP-mediated polar growth[J].Molecular Plant,2021,14(3):395-410.
[29]HUANG J B,LIU H L,CHEN M,LI X J,WANG M Y,YANG Y L,WANG C L,HUANG J Q,LIU G L,LIU Y T,XU J,CHEUNG A Y,TAO L Z.ROP3 GTPase contributes to polar auxin transport and auxin responses and is important for embryogenesis and seedling growth in Arabidopsis[J].The Plant Cell,2014,26(9):3501-3518.
[30]KAWANO Y,AKAMATSU A,HAYASHI K,HOUSEN Y,OKUDA J,YAO A,NAKASHIMA A,TAKAHASHI H,YOSHIDA H,WONG H L,KAWASAKI T,SHIMAMOTO K.Activation of a rac GTPase by the NLR family disease resistance protein pit plays a critical role in rice innate immunity[J].Cell Host&Microbe,2010,7(5):362-375.
[31]王爱荣,陈新,张冬梅,陈惠红,鲁国东,王宗华.拟南芥不同ROP 蛋白对病原细菌增殖的影响[J].福建农林大学学报(自然科学版),2008,37(6):610-613.WANG Airong,CHEN Xin,ZHANG Dongmei,CHEN Huihong,LU Guodong,WANG Zonghua.Effects of different Arabidopsis ROPs on multiplication of Pseudomonas syringae pv.tomato DC3000[J].Journal of Fujian Agriculture and Forestry University(Natural Science Edition),2008,37(6):610-613.
[32]ZHANG Z W,ZHANG X L,NA R,YANG S Q,TIAN Z M,ZHAO Y,ZHAO J.StRac1 plays an important role in potato resistance against Phytophthora infestans via regulating H2O2 production[J].Journal of Plant Physiology,2020,253:153249.
[33]金微微,徐昌杰,李鲜,王平,张波,孙崇德,陈昆松.采后玉露桃果实冷害发生与ROP 基因的表达调控[J].果树学报,2009,26(5):608-613.JIN Weiwei,XU Changjie,LI Xian,WANG Ping,ZHANG Bo,SUN Chongde,CHEN Kunsong.Occurrence of chilling injury and regulation of ROP gene expression in Yulu peach fruit during storage[J].Journal of Fruit Science,2009,26(5):608-613.
[34]LIN Y S,ZENG Y L,ZHU Y,SHEN J B,YE H,JIANG L W.Plant Rho GTPase signaling promotes autophagy[J].Molecular Plant,2021,14(6):905-920.