果实成熟是果实生长发育历程中重要的阶段,是一种高度协调的、基因程序化的、不可逆的现象,涉及一系列生理生化和感官变化,形成成熟果实特有的风味、质地和色泽等可食用的品质属性[1]。在果实成熟过程中,通常伴随着果实的软化、有机酸减少、淀粉降解、可溶性糖分积累、单宁物质消失、芳香化合物合成、叶绿素降解与类胡萝卜素或花色苷积累等一系列的生理生化变化。
果实成熟是风味和营养品质形成的重要过程,影响果实的商品价值和采后的贮藏性能。果实成熟调控机制的揭示可为提高果实品质分子育种技术提供重要的参考,也是提升果实品质和研发采后保鲜技术的基础。因此,果实成熟的调控一直是国内外研究的热点,研究主要集中在植物激素调控果实成熟的分子机制和转录因子调控果实成熟的分子机制两个方面。
在果实成熟过程中,不同家族的转录因子(transcription factors,TFs)在相关基因的表达中起着转录调控作用,它们通过DNA结合基序与目标基因启动子的特定结构域结合进而对目标基因的表达进行转录激活或抑制,从而调控果实成熟进程[2]。根据目标基因DNA 结合区域的不同,靳进朴等[3]将转录因子分为58 个家族,例如bZIP(basic region-leucine zipper)、MYB(myeloblastosis oncogene)、bHLH(basic helix-loop-helix)、NAC(NAM,ATAF1/2,CUC)、MADS(MCM1-agamous-deficiens-SSRF)、AP2/ERF(apetalous 2/ethylene response factors)、C2H2(cys2/his2-type zinc finger)、DREB(dehydration responsive element binding protein)、SPL(SQUAMOSA promoter binding protein-like)、WRKY(WRKY transcription factors)等。近年来随着对NAC 转录因子家族研究的不断深入,发现其在果实发育成熟中有重要的调控作用,笔者总结了近年来NAC转录因子家族在果实发育及成熟过程中的作用及潜在机制相关研究,以期为果实关键品质性状调控与改良提供参考。
1 NAC基因家族特征
NAC 是广泛存在于植物中的转录因子家族之一,为陆生植物特有,家族成员众多。最初发现的是矮牵牛(Petunia hybrida)NAM(no apical meristem)、拟南芥(Arabidopsis thaliana)ATAF1/2(arabidopsis mestica)转录因子成员最多达253个,其次白梨(Pyrus bretschneideri)和沙梨(P. pyrifolia)两种梨属果树均为185个,而草本果树菠萝(Ananas comosus)和番木瓜(Carica papaya)以及欧亚种葡萄(Vitis vinifera)的NAC 家族成员比较少,分别为73、82 和70个。研究表明这些NAC 转录因子在植物生长发育[12-15]、胁迫应答[16]、果实发育[17]、植株衰老[18]和果实成熟[17,19]等过程中发挥着重要的作用。transcription activation factor)和CUC2(cup shaped cotyledon)3 个基因,因它们都有相似的保守结构,故以它们的首字母命名为NAC 转录因子[4-5]。目前通过全基因组分析NAC 转录因子,发现拟南芥有113 个、矮牵牛有41 个、本氏烟草(Nicotiana benthamiana)有227 个NAC 成员[2,6]。在许多果树中也鉴定了NAC 转录因子(表1),其中苹果(Malus do-
表1 不同果树作物NAC 基因数量
Table 1 Number of NAC genes in different fruit crops
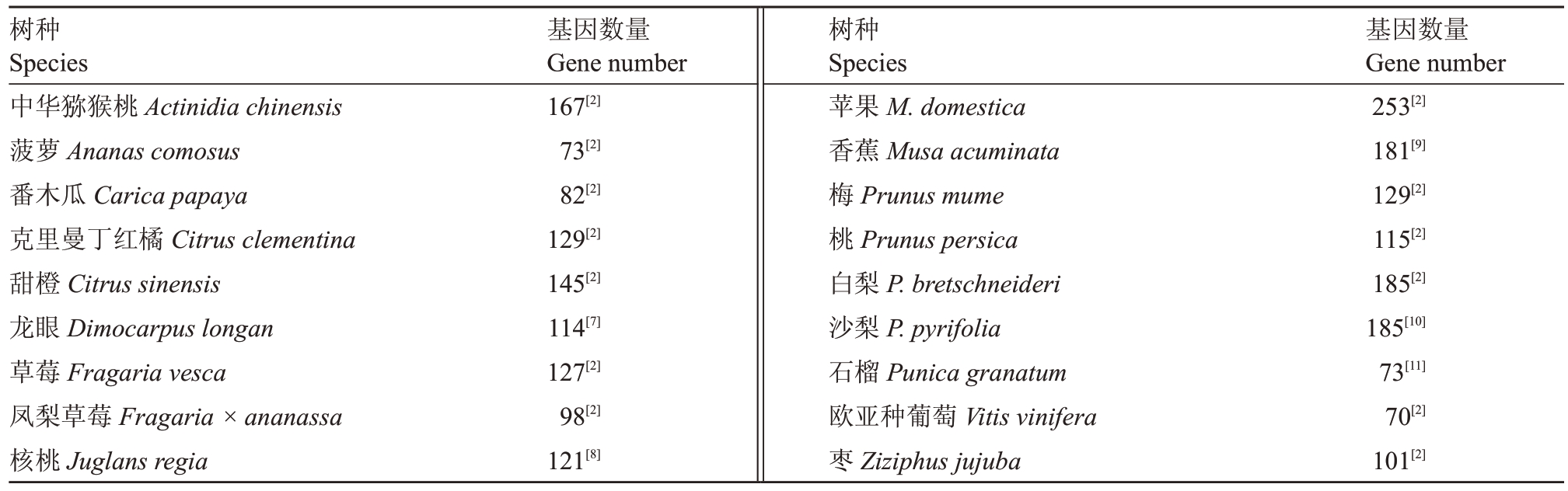
树种Species中华猕猴桃Actinidia chinensis菠萝Ananas comosus番木瓜Carica papaya克里曼丁红橘Citrus clementina甜橙Citrus sinensis龙眼Dimocarpus longan草莓Fragaria vesca凤梨草莓Fragaria×ananassa核桃Juglans regia基因数量Gene number基因数量Gene number 167[2]73[2]82[2]129[2]145[2]114[7]127[2]98[2]121[8]树种Species苹果M.domestica香蕉Musa acuminata梅Prunus mume桃Prunus persica白梨P.bretschneideri沙梨P.pyrifolia石榴Punica granatum欧亚种葡萄Vitis vinifera枣Ziziphus jujuba 253[2]181[9]129[2]115[2]185[2]185[10]73[11]70[2]101[2]
与模式植物拟南芥一样,果树NAC转录因子家族的蛋白序列由约400个氨基酸组成,具有典型(图1-a~i)和非典型两种NAC 结构域[20(]图1-a-ii~iv)。其蛋白结构通过x射线结晶学分析确定不同于经典的螺旋-转角-螺旋结构,而为对称的均二聚体[21](图1-b)。结构特征的多样性使得NAC 转录因子家族具有功能上的多样性,赋予其在植物生长发育各个过程中重要的作用[23]。
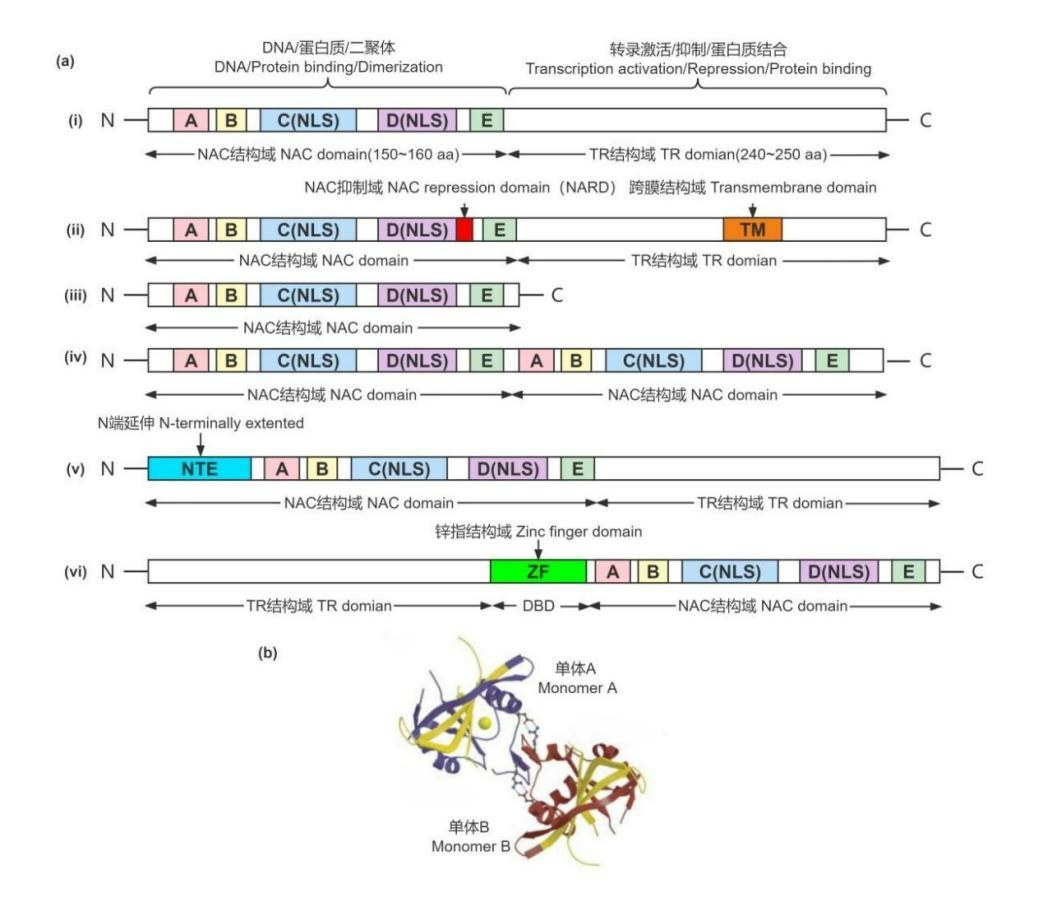
图1 NAC 转录因子结构示意图(a)和蛋白二聚体结构(b)(参考Puranik 等[20]、Ernst 等[21]和赵才美等[22])
Fig.1 Structural diagram of NAC transcription factor(a)and dimer structure of NAC protein(b)(modified from Puranik et al.[20],Ernst et al.[21]and Zhao et al.[22])
Ooka 等[24]根据结构域中氨基酸序列的相似性预测,将拟南芥和水稻中预测的NAC蛋白分为Ⅰ和Ⅱ两组,Ⅰ组中有14个亚组,Ⅱ组中有4个亚组。笔者在本文中根据氨基酸序列的相似性,构建了荔枝、苹果、香蕉、草莓、甜橙、宽皮柑橘、猕猴桃、葡萄、桃、梨、番木瓜、柿、越橘等果树和水稻、番茄、拟南芥、矮牵牛、烟草、小麦、马铃薯共368个NAC转录因子蛋白的系统进化树。结果如图2 所示,将果树中NAC转录因子蛋白分别聚类到NAP、NAM、SENU5、ONAC003、NAC1、ANAC001、ONAC001、ONAC022、ATAF、NAC2、TERN、AtNAC3、ANAC011、TIP、Os-NAC3、OsNAC7、OsNAC8、ANAC063等18个亚组,其中ANAC063、ONAC003、ANAC001、ONAC001 等4个亚组为Ⅰ组成员,其余的14 个亚组为Ⅱ组成员(图2)。大多数果树NAC 转录因子蛋白聚类到ANAC001、NAM、ANAC063、NAP 亚组中,其中与ANAC001 和NAP 亚组聚类的果树NAC 转录因子数量多。果树与拟南芥及其他大田作物的主要区别是多年生和产品器官果实发育成熟过程中特有的生理生化变化,这两个亚组成员可能在果树的特有性状的形成和调控中发挥重要的功能。ONAC022、ONAC003、ONAC001、SENU5、OsNAC8、OsNAC7、NAC1、ATAF、AtNAC3、TERN、TIP、ANAC011、NAC2亚组只聚类少数果树NAC转录因子蛋白,说明这些亚组在进化关系上与果树NAC 转录因子较远,而没有果树NAC转录因子蛋白聚类在OsNAC3亚组。以上聚类分析结果表明,果树中NAC转录因子在进化上呈现多样性,为其功能的多样化提供一定的遗传基础。
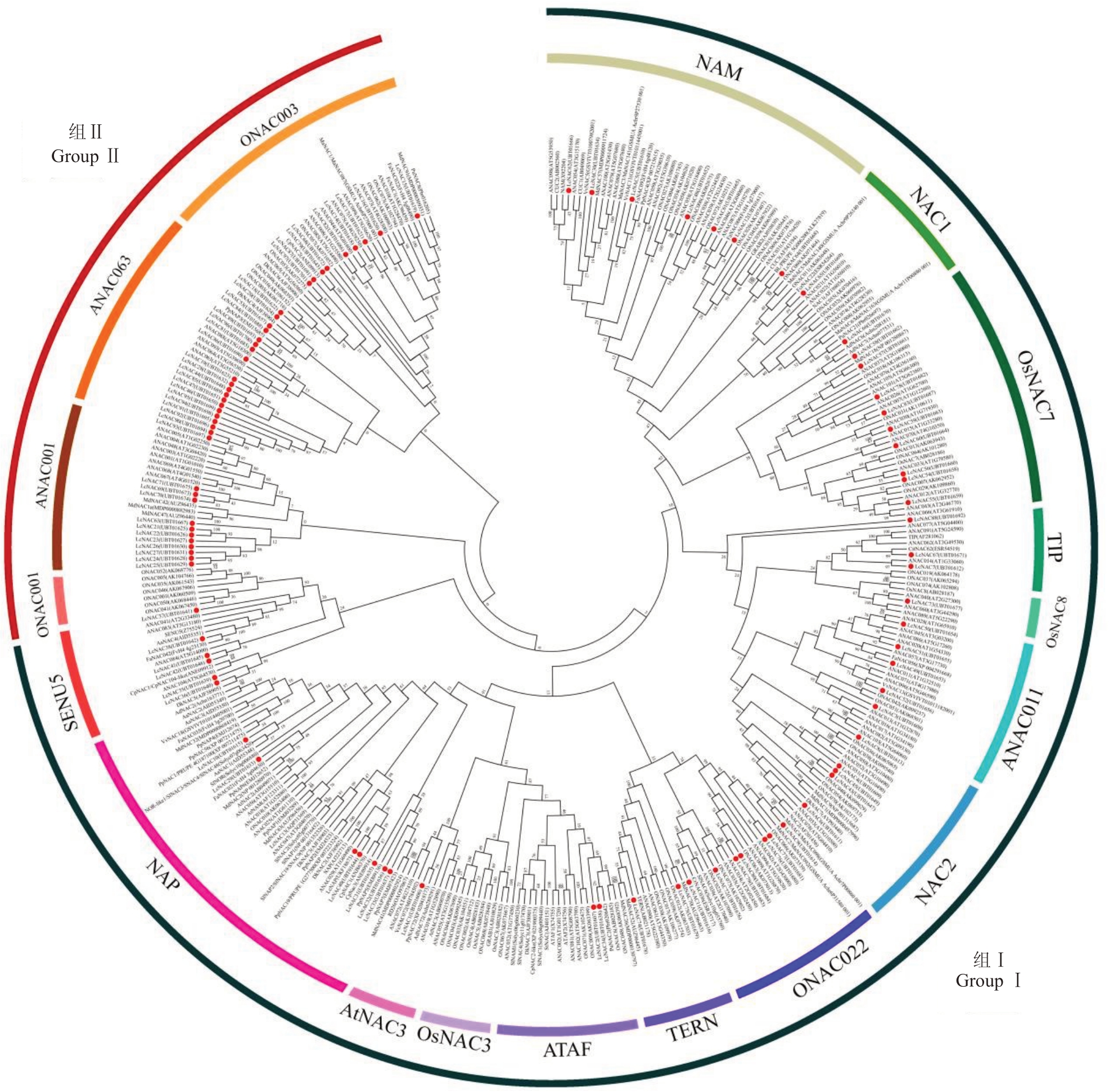
图2 果树NAC 转录因子的系统进化树分析
Fig.2 Phylogenetic tree of NAC transcription factors in fruit trees
该图构建了荔枝(LcNAC)、苹果(MdNAC)、香蕉(MaNAC)、草莓(FaNAC,FcNAC)、甜橙(CitNAC,RD26)、宽皮柑橘(CrNAC)、金橘(FcrNAC)、猕猴桃(AdNAC,AaNAC)、葡萄(VvNAC)、桃(PpNAC,BL)、梨(PuNAC)、番木瓜(CpNAC)、柿(DkNAC)、越橘(VcNAC)等果树和水稻(ONAC,OsNAC3/4/5/6/7/8)、番茄(SlNAC,SlNOR,SENU5)、拟南芥(ANAC,ATAF1/2,CUC1/2/3,NAC1/2,AtNAM,TIP)、矮牵牛(NAM)、烟草(TERN)、小麦(GRAB1/2)、马铃薯(StNAC)共368 个NAC 转录因子蛋白的系统进化树,氨基酸序列从NCBI(https://www.ncbi.nlm.nih.gov/),GDR(https://www.rosaceae.org/),PlantTFDB(http://planttfdb.gao-lab.org/)等在线网站获取。使用TBtools 软件“One Step Build a ML Tree”功能以极大似然法构建系统发育树[25],分支长度表示遗传距离。
The figure constructs the phylogenetic tree of 368 NAC transcription factor proteins: litchi (LcNAC), apple (MdNAC), banana (MaNAC), strawberry (FaNAC, FcNAC), sweet orange (CitNAC, RD26), mandarin citrus (CrNAC), kumquat (FcrNAC), kiwifruit (AdNAC, AaNAC), grape(VvNAC),peach(PpNAC,BL),pear(PuNAC),papaya(CpNAC),persimmon(DkNAC),cranberry(VcNAC)and other fruit trees and rice(ONAC,OsNAC3/4/5/6/7/8),tomato(SlNAC,SlNOR,SENU5),arabidopsis(ANAC,ATAF1/2,CUC1/2/3,NAC1/2,AtNAM,TIP),petunia(NAM),tobacco(TERN),wheat(GRAB 1/2),potato(StNAC).Amino acid sequence was obtained from NCBI(https://www.ncbi.nlm.nih.gov/),GDR(https://www.rosaceae.org/), PlantTFDB (http://planttfdb.gao-lab.org/) and other online websites to access. The phylogenetic trees[25] was constructed by maximum likelihood using the TBtools software“One Step Build a ML Tree”function,with branch lengths representing genetic distance.
2 NAC 转录因子在果实成熟中的作用
近年的研究表明,NAC转录因子家族在果实成熟中发挥着重要的调控作用,参与果实软化、风味物质代谢、色素的合成以及成熟激素合成和信号传导(表2)。
表2 一些NAC 转录因子成员在果实成熟中的作用
Table 2 The roles of NAC transcription factor members in the ripening of some fruit species

树种Fruit species软枣猕猴桃Actinidia arguta中华猕猴桃A.chinensis NAC基因成员NAC gene member AaNAC2/3/4 AcNAC2/3/4美味猕猴桃A.deliciosa AdNAC2/3/6/7/72宽皮柑橘Citrus reticulata甜橙C.sinensis甜橙C.sinensis番木瓜Carica papaya番木瓜C.papaya柿Diospyros kaki枇杷Eriobotrya japonica草莓Fragaria×ananassa草莓F.×ananassa CrNAC036 CitNAC62 RD26 CpNAC1/2 CpNAC3 DkNAC1/3/5/6/7/13/16 EjNAC82 FaNAC035/42/92 FaNAC035智利草莓F.chiloensis FcNAC1金橘Fortunella crassifolia荔枝Litchi chinensis香蕉Musa acuminata香蕉M.acuminata香蕉M.×paradisiaca苹果Malus domestica主要功能Main function单萜合成[26]Monoterpene synthesis单萜合成、乙烯合成[26-27]Monoterpene synthesis,ethylene synthesis乙烯和萜类合成、果实软化[28-30]Ethylene and terpenoid synthesis,fruit softening ABA合成[31]Abscisic acid synthesis降低柠檬酸含量[32]Reduce citric acid content葡萄糖、果糖积累[33]Accumulation of glucose and fructose类胡萝卜素合成[34-36]Carotenoid synthesis乙烯信号转导[37]Ethylene signal transduction果实脱涩[38-40]Fruit de-astringency类胡萝卜素合成[41]Carotenoid synthesis果实软化[42]Fruit softening色素、糖和香气合成[42]Synthesis of Color,sugar and aroma细胞壁重塑、ABA信号响应[43]Cell wall remodeling,ABA signaling response类胡萝卜素合成[44]Carotenoid synthesis花色素生物合成[45]Anthocyanin biosynthesis香气成分合成[46]Synthesis of aroma components乙烯信号转导[8,46-47]Ethylene signal transduction叶绿素降解[48]Chlorophyll degradation乙烯生物合成[49,50-52]Ethylene biosynthesis苹果M.domestica苹果M.domestica FcrNAC22 LcNAC13 MaNAC1/5/87/140 MaNAC1/2/3/4/6/16/83/87/92/94/95 MpSNAC67 MdNAC1/1a/2/16/18.1/26/32/41/47/5 7/63/78/80/91/119/141 MdNAC18.1 MdNAC9/42/52桃Prunus persica BL(BLOOD),PpNAC1/PpNAC2桃P.persica桃P.persica果实软化[49]Fruit softening黄酮和花色素积累[53-55]Flavonoid and anthocyanin accumulation花色素、挥发性酯、脂肪酸合成[56-59]Synthesis of anthocyanins,volatile esters,fatty acid synthesis类胡萝卜素合成[60]Carotenoid synthesis乙烯合成、ABA响应[61-64]Ethylene synthesis,ABA response秋子梨Pyrus ussuriensis PpNAC19 PpNAC1/PpNAC4/PpNAC6,PpNAC.A59,Purpe.4G181700 PuNAC2/PuNAC8/PuNAC21沙梨P.pyrifolia越橘Vaccinium corymbosum葡萄Vitis vinifera PpNAC61/70/172/176/23 VcNAC072 VvNAC03/11/13/33/60乙烯合成及信号转导[65]Ethylene synthesis and signal transduction果实着色[10]Fruit coloration花色素积累[66]Anthocyanin accumulation浆果发育[67]Berry development
2.1 果实软化
果实软化是成熟的重要特征之一,直接影响采后物流、货架寿命和商品性。在草莓中,通过研究112 个NAC 基因的表达模式,发现FaNAC006/021/022/035/042/096 在果实软化过程中表达上调,其中FaNAC035 转录水平最高,可能参与草莓果实软化过程的调控[42]。在智利草莓(F.chiloensis)果实成熟过程中也发现FcNAC1 可以调节细胞壁果胶代谢,导致果实软化[43]。在美味猕猴桃(A. deliciosa)中,AdNAC6 和AdNAC7 可以促进猕猴桃果实成熟过程中的软化[31]。另外,在对苹果的研究中发现,与番茄成熟调控关键转录因子NON-RIPENING(NOR)的直系同源基因MdNAC18.1 是果实硬度多态性的决定因子,可以作为成熟采收时和冷藏3 个月后果实硬度的预测基因,通过调节乙烯的产生以一种保守的方式控制苹果果实的成熟,是决定苹果果实硬度和收获期的决定因子[49,68]。
2.2 果实着色
在果实成熟过程中往往伴随着叶绿素的降解、不同类型花色苷的合成和在液泡中的积累以及类胡萝卜素(如β-胡萝卜素、叶黄素酯、叶黄素、番茄红素等)的积累[69],在外观上表现为果实着色。果实的色泽是决定果实品质的关键因素之一,均匀且鲜艳的果实着色吸引动物采食,继而为种子的传播提供可能,而对于生产者来说,着色良好的果实意味着具有更好的商品价值与市场竞争力。
2.2.1 叶绿素降解和类胡萝卜素积累 叶绿素降解是果实底色消退和良好着色的前提条件。大蕉(Musa×paradisiaca)中,过表达MpSNAC67会降低叶片叶绿素含量,促进其脱绿和黄化,说明该基因可能正向调控叶绿素降解过程[48]。虽然目前叶绿素降解途径比较清楚,但关于NAC转录因子在果实成熟过程中对叶绿素降解所起的具体作用研究报道很少。
研究者更多关注于NAC 转录因子在果实成熟过程中表色如花色苷和类胡萝卜素生物合成的作用。番木瓜中,CpNAC1 是果实成熟过程中类胡萝卜素生物合成的正调控因子[34,36]。CpNAC2 和乙烯不敏感3a(ethylene-insensitive 3a)CpEIN3a 相互作用,共同参与番木瓜果实采后成熟过程中的类胡萝卜素合成[35-36]。金橘果实中的FcrNAC22 转录水平受红光诱导,并随果实颜色变化而表达上调,可以加速果实脱色和类胡萝卜素积累促进果实着色[44]。类胡萝卜素的积累是生物合成和降解代谢的综合体现。黄肉桃果实中类胡萝卜素含量显著高于白肉桃果实,黄肉桃果实中PpNAC19 表达量较高,而类胡萝卜素降解关键基因类胡萝卜素裂解双加氧酶4(carotenoid-cleaving dioxygenase 4)PpCCD4 的表达量却显著低于白肉桃果实,PpNAC19能够转录抑制PpCCD4 启动子活性,从而促进桃果实成熟过程中果肉类胡萝卜素的积累[60]。在红肉枇杷大红袍中,EjNAC82 表达量也显著高于白肉枇杷白沙,在枇杷果实类胡萝卜素合成中起着积极调控作用[41]。
2.2.2 类黄酮和花色苷合成 对苹果的研究发现,NAC转录因子MdNAC52转录水平在果实着色期间增加,过表达MdNAC52 能够促进苹果愈伤组织花色苷的积累[53]。在红肉苹果中,MdNAC42表达量高于白肉苹果,与果实成熟时花色苷含量积累呈正相关,在红肉苹果花色苷生物合成中起着重要的作用[54]。另一NAC 成员MdNAC9 则与红肉苹果的黄酮醇生物合成密切相关,该基因转录水平与黄酮醇积累呈正相关,在苹果愈伤组织中过表达MdNAC9后黄酮醇含量显著增加[55]。在荔枝果实成熟过程中,转录组分析发现6个NAC转录因子在果皮成熟过程中有着很高的转录丰度,可能在荔枝果皮成熟中扮演着积极的角色[70]。后续研究中发现,LcNAC13 能够负调控果皮花色苷生物合成[45]。对红色和白色草莓果实比较转录组分析发现,NAC家族成员可能参与草莓果实成熟过程中花色苷积累的调控[71]。进一步的研究发现沉默FaNAC035 的草莓株系花色苷生物合成相关酶基因表达下调,果实着色延迟,而过表达FaNAC035 则会使草莓果实着色提前[42]。在越橘中克隆出1个NAC基因VcNAC072,该基因的表达随着果实成熟着色而上调,可能在越橘果实花色苷的积累中起正向调节作用[66]。在血桃中研究者鉴定出属于NAC转录因子家族的基因BL(BLOOD),该基因的表达蛋白在果实发育后期能与PpNAC1转录因子形成异源二聚体激活PpMYB10.1的表达,进而促进花色苷生物合成途径结构基因的表达,最终导致果肉大量积累花色苷而呈血红色[56,59]。
2.3 糖分积累
可溶性糖积累是果实成熟的重要生化变化,与果实风味品质密切相关。果实成熟过程中甜度增加主要是可溶性糖持续输入或多糖淀粉等降解的结果,少数果实还存在葡糖异生现象,有机酸的持续下降也是果实甜度体系的一个重要方面,糖酸比是判断果实成熟度的重要指标[69]。荔枝果肉糖的组分主要决定于蔗糖水解酶特别是酸性转化酶,而蔗糖转运载体LcSUT4 的表达则是糖分积累量的重要决定因子[72-74],但关于糖积累的上游调控因子则尚无相关报道。近年研究发现NAC 转录因子家族在果实成熟过程中对糖分积累起着积极作用。在草莓成熟过程中,FaNAC035上调表达,促进蔗糖在草莓果实成熟早期的积累[42]。在甜橙中鉴定出在晚熟突变体和正常熟期品种之间存在差异表达NAC 基因RD26(responsive to desiccation 26),在果实成熟过程中该基因的表达与果糖、葡萄糖的积累具有高度的相关性[33]。
2.4 芳香化合物的合成
果实成熟过程中香气的增加得益于挥发性芳香化合物包括醛、酯、醇、酮、酸、萜类、内酯和硫化物等的合成与积累,通常只有几种挥发性化合物组成特定水果的整体香气和独特味道[75-77]。挥发性化合物是决定水果风味的重要因素,浓厚的香气能够使果树吸引动物采食继而使其种子得到传播,同时赋予水果更好的商品价值。在桃的研究中发现转录因子PpNAC1 能够调控挥发性酯类化合物的合成[56]。最新的研究发现,过表达PpNAC1 可以促进桃果实挥发性化合物(E)-2-己烯醛和(Z)-3-己烯醇合成前体ω-3 脂肪酸亚麻酸(18∶3)及其衍生挥发物的产生,说明PpNAC1在调节脂肪酸通路产生果实风味相关挥发性有机物中也起着重要作用[57]。此外,Pp-NAC1也可以与PpNAC2相互作用调控酯类化合物合成[58]。草莓中,转录因子FaNAC035 在控制芳香化合物的生物合成中发挥重要作用[42]。AaNAC2、AaNAC3、AaNAC4 在软枣猕猴桃果实成熟过程中正调控单萜芳香化合物的形成[26]。同样在美味猕猴桃中发现AdNAC6 和AdNAC7 促进猕猴桃成熟过程中萜类香气化合物的形成[28]。
2.5 参与果实成熟其他过程
在柿果实脱涩过程中,DkNAC1/3/5/6/7/13/16表达上调,可能在柿果实成熟脱涩过程中起重要作用[38- 40]。在柑橘成熟过程中,CitNAC62 和Cit-WRKY1可以相互作用激活CitAco3的启动子,促进其表达,从而降低果实的柠檬酸含量[33]。
3 NAC调节果实成熟的机制
3.1 NAC 通过调控结构基因或与其他转录因子互作影响果实成熟
3.1.1 NAC 影响果实软化的机制 果实的软化主要是由细胞壁组成和结构的变化引起的,包括果胶溶解、解聚以及果胶侧链上中性糖损失、纤维素和半纤维素解聚、纤维素-半纤维素网络松弛、细胞壁膨胀等[78-81]。这些变化是细胞壁的降解与重塑的结果,其中细胞壁降解酶与重塑酶发挥着关键的作用。在智利草莓果实成熟过程中发现FcNAC1与果胶裂解酶(pectate lyase,PL)基因FcPL 的启动子结合并激活其转录,调节果实软化过程中的细胞壁果胶代谢[43]。在凤梨草莓中,FaNAC035 通过促进β-木糖苷酶(β-xylosidase,XYL)基因FaXYL3、果胶裂解酶(pectate lyase,PL)基因FaPL3-4、内切-1,4-葡聚糖酶(endo-1,4-beta-glucanase,GH9B)基因FaGH9B15、松 弛 蛋 白(expansin,EXP)基 因FaEXP1-3、果胶甲基酯酶(pectin methylesterase,PME)基因FaPME39、阿拉伯半乳糖蛋白(arabino galactan-proteins,AGPs)基因FaAGPs 等细胞壁降解与重塑相关酶基因的表达,参与草莓果实软化过程的调控。
3.1.2 NAC 影响果实着色的机制 在果实着色方面,过表达MdNAC52 通过增强与MdMYB9、Md-MYB11 的启动子相互作用促进苹果愈伤组织花色苷的积累,MdNAC52 还可以与MdLAR 启动子结合增强其表达从而促进原花青素的合成[53]。在红肉苹果中,MdNAC42 可与MdMYB10 相互作用,过表达MdNAC42 能够上调MdCHS、MdCHI、MdF3H、MdDFR、MdANS 和MdUFGT 等结构基因的表达而增加花色苷的积累[54]。另外,在苹果愈伤组织中过表达MdNAC9 后黄酮醇合成酶基因MdFLS 转录水平和黄酮醇含量显著升高,进一步研究发现MdNAC9 通过激活MdFLS表达正向调控黄酮醇[55]。
BL(BLOOD)在血桃果实发育后期通过与Pp-NAC1 形成的异源二聚体激活PpMYB10.1 的表达,促进血桃花色苷生物合成途径的结构基因表达[56]。荔枝LcNAC13 能负调控果皮花色苷生物合成相关基因LcCHS1/2、LcCHI、LcF3H、LcF3’H、LcDFR 和关键转录因子基因LcMYB1 的表达,而荔枝中R1MYB 成员LcR1MYB1 可与LcNAC13 发生物理相互作用并消除其对上述基因表达的抑制作用[45,82-83]。与此相反,越橘中VcNAC072 可结合于MYB 转录因子AtPAP1 启动子并激活其表达,在拟南芥中过表达VcNAC072 可以促进AtPAP1 及AtDFR、AtANS表达而获得花色苷积累显著增加的种子,揭示VcNAC072在越橘果实中正调控花色苷的积累的机制[67]。
番木瓜CpNAC1可结合类胡萝卜素生物合成关键基因八氢番茄红素去饱和酶2/4(phytoene desaturase 2/4)CpPDS2/4启动子的NAC结合位点基序,是果实成熟过程中类胡萝卜素生物合成的正调控因子[34,36]。除了CpPDS2/4 外,进一步研究还发现Cp-NAC1 可以通过激活类胡萝卜素合成相关基因ζ-胡萝卜素去饱和酶(ζ-carotene desaturase)CpZDS、番茄红素环化酶-e(lycopene cyclase-e)CpLCY-e 和胡萝卜素羟化酶-b(carotene hydroxylase-b)CpCHY-b 的表达来调控番木瓜类胡萝卜素代谢[35-36]。在金橘果实中,FcrNAC22能够激活编码类胡萝卜素生物合成途径中三个关键酶基因的启动子,包括番茄红素β-环化酶(lycopene β-cyclase)FcrLCYB1、β-胡萝卜素羟化酶2(β-carotene Hydroxylase 2)FcrBCH2和9-顺式-环氧类胡萝卜素双加氧酶5(9-cis-epoxycarotenoid dioxygenases 5)FcrNCED5,在柑橘和番茄愈伤组织中过表达FcrNAC22能够增强类胡萝卜素生物合成途径中多数基因的表达以促进色素积累,而敲除FcrNAC22则减弱组织着色,表明FcrNAC22通过激活类胡萝卜素代谢途径关键基因的表达促进柑橘果实的着色[44]。
3.1.3 NAC 影响果实香气物质合成的机制 除了影响果实软化,草莓FaNAC035被沉默后,丁香酚合成酶2(eugenol synthase 2)基因FaEGS2和苯丙烷丁香酚生物合成关键基因FaEOBII、FaDOF2 表达下调,参与萜烯类挥发性化合物芳樟醇和橙花醇的生物合成的橙花醇合成酶(nerolidol synthase 1)FaNES1 表达也同时下调。说明FaNAC035 在共同调控草莓果实的软化和芳香化合物的生物合成中发挥重要的功能。相同的,美味猕猴桃AdNAC6和Ad-NAC7 可以结合细胞壁降解途径中β-甘露聚糖酶(endo-β-mannanase,MAN)基因AdMAN1 和高松油烯形成关键酶萜烯合成酶1(terpene synthase 1)AaTPS1的启动子区域并激活其表达,同时促进猕猴桃果实成熟过程中的软化和芳香物质的形成[28]。在软枣猕猴桃中,AaNAC2、AaNAC3、AaNAC4可结合萜烯合成酶1(terpene synthase 1)AaTPS1 启动子而促进其表达,而在中华猕猴桃中,因为NAC 结合位点 突 变 使AcNAC2、AcNAC3、AcNAC4 不 能 与AcTPS1 启动子结合,导致萜类化合物生物合成受阻,通过引入12 bp 的NAC 核心结合区域可以恢复对AcTPS1 启动子的激活作用,进一步揭示NAC 转录因子在猕猴桃成熟过程中调控单萜芳香化合物形成的机制[26]。
桃PpNAC1通过直接结合挥发性酯类合成的重要酶乙醇酰基转移酶(alcohol acyltransferases,AATs)基因PpAAT1 的启动子促进其表达而调控酯类化合物的合成,在番茄SlAAT1突变体植株中过表达PpNAC1 可以恢复SlAAT1 的表达及成熟果实挥发性酯类的形成,另外,苹果中PpNAC1的同源基因MdNAC5的蛋白也能通过结合MdAAT1的启动子激活其表达,调控挥发性酯类的合成[57]。最新的研究发现,过表达PpNAC1 可以促进桃果实挥发性化合物(E)-2-己烯醛和(Z)-3-己烯醇合成前体ω-3 脂肪酸亚麻酸(18∶3)及其衍生挥发物的产生,进一步的实验证明PpNAC1 可以结合亚麻酸(18:3)合成基因PpFAD3-1启动子并激活其表达,说明PpNAC1在调节脂肪酸通路产生果实风味相关挥发性有机物中也起着重要作用[58]。此外,PpNAC1 可以与PpNAC2形成二聚体,后者也可以结合PpAAT1 启动子激活转录[59]。
3.1.4 NAC 影响果实脱涩和糖积累的机制 柿果实中,DkNAC7 不仅可以反式靶向激活脱涩关键调控基因乙烯反应因子DkERF9 的表达,还可以激活柿果实丙酮酸脱羧酶2(pyruvate decarboxylase)基因DkPDC2 的表达,从而调控柿果实的脱涩[39]。DkNAC13 和DkNAC16 可以分别结合脱涩相关基因DkPDC2 的激活因子DkERF9 和乙醇脱氢酶(alcohol dehydrogenase)基因DkADH1的启动子区域并激活其表达,从而促进柿果实脱涩[40]。
草莓FaNAC035除了正调控果实软化和香气合成,还通过上调表达抑制糖酵解、发酵等消耗糖的过程,并调节蔗糖合酶(sucrose synthase 1)FaSUS和蔗糖磷酸合成酶(sucrose phosphate synthase)FaSPS1的表达而在草莓果实成熟早期的蔗糖积累中起重要调控作用[42]。
3.2 NAC 通过调控激素生物合成和信号转导影响果实成熟
植物激素对果实发育和成熟的调节是不可或缺的,而乙烯和ABA(abscisic acid,ABA)被认为是调控果实成熟的核心激素,乙烯在呼吸跃变型果实中起着明显的作用,而呼吸非跃变型果实通常与ABA有关,但越来越多的研究表明果实成熟涉及多种内源激素的交互[84]。对荔枝的研究结果显示ABA 在果皮花色苷合成中而乙烯在叶绿素降解中发挥重要的调控作用[85]。近年来随着对NAC 转录因子研究的不断深入,许多的研究发现NAC通过参与乙烯和ABA生物合成和信号转导过程,在调控果实成熟中发挥重要的作用。
3.2.1 通过乙烯通路调控 乙烯是一种重要的果实成熟植物激素,微量的乙烯可以触发许多细胞代谢事件,特别是在呼吸跃变型果实中[1]。近年来大量的研究表明NAC 转录因子参与乙烯生物合成和乙烯信号转导的调控,在果实成熟调控中起着积极的作用。An等[51]研究发现MdNAC47能够直接与乙烯生物合成的正调控因子MdERF3启动子区域结合并激活其表达,说明MdNAC47可以通过乙烯依赖途径参与果实发育的调控。在金冠(Golden Delicious)苹果中,MdNAC2可能通过响应乙烯和与恢复乙烯敏感性基因(reversion to ethylene sensitivity,RTE)MdRTE1b相互作用,参与乙烯信号介导的果实成熟调控[52]。
在香蕉中,MaNAC1 和MaNAC2 能与乙烯信号通路下游元件MaEIL5(ethylene insensitive 3-like protein)相互作用并下调其表达。在后续研究中发现,MdNAC1和MdNAC2可以直接与乙烯生物合成的负调控因子MaERF11的启动子区结合,抑制其转录,从而激活MaERF11 的下游靶基因MaACS1 和MaACO1 的表达,促进乙烯的生物合成。乙烯反应抑制因子RTE1-HOMOLOG 1(RTH1)是果实成熟的负调控因子,也作为MaNAC2 的下游靶标,并受其转录抑制,因此,MaNAC1 和MaNAC2 可能通过介导乙烯促进的一系列事件而参与调节香蕉果实的成熟[46,86-87]。
在美味猕猴桃果实软化等成熟变化过程中,Ad-NAC2 和AdNAC3 两个基因可以直接结合乙烯生物合成关键酶1-氨基环丙烷-1-羧酸合成酶(1-aminocyclopropane- 1- carboxylic acid synthase)基 因AdACS1 的启动子区域,激活AdACS1 表达,促进内源乙烯产生。此后的研究也发现,AdNAC6 和Ad-NAC7 可以直接结合AdACS1 和AdACO1 的启动子区域并激活它们的表达[28],从而促进乙烯生物合成和果实成熟。AdNAC2和AdNAC72的表达被乙烯上调,它们的表达蛋白可以结合于甲硫氨酸亚砜还原酶(methionine sulfoxide reductase B)基因AdMsrB1启动子上促进该基因的表达,过表达AdNAC72不仅上调AdMsrB1的表达,还提高游离甲硫氨酸(methionine,Met)和乙烯合成直接前体ACC 的含量和乙烯生成速率,这些结果揭示了NAC转录因子在促进乙烯合成进而调控猕猴桃果实成熟中的重要作用[30]。Nieuwenhuizen 等[27]在中华猕猴桃中也发现AcNAC2/3/4 通过控制AcACS1 的表达水平来调控果实成熟早期系统Ⅰ乙烯和成熟后期系统Ⅱ乙烯的生物合成,进而影响果实成熟与品质形成。
在呼吸跃变型果实桃中,NAC转录因子亚家族NAP转录因子PpNAP1、PpNAP4、PpNAP6在果实软化过程中表达上调,并伴随着乙烯的快速生成,参与桃果实成熟的调控[61,63]。与番茄NOR[68]高度相似的Purpe.4G181700 在早熟桃果实成熟阶段表达量很高,可能通过一个以依赖乙烯的保守方式控制果实的成熟时间和过程[62]。PpNAC.A59 能够结合于乙烯 响 应 因 子A16(ethylene response factor A16)PpERF.A16启动子促进其表达,并通过NAC-ERF信号级联间接介导乙烯的生物合成,诱导PpACS1 和ACC氧化酶(ACC oxidase)PpACO1的表达[63]。
在番木瓜果实中,CpNAC3 可以单独或与Cp-MADS4 相互作用特异结合于乙烯信号转导的关键基因CpERF9和CpEIL5的启动子并激活它们表达,说明CpNAC3 可以独立或与CpMADS4 协同激活乙烯信号转导通路而在果实成熟调控过程中发挥作用[37]。在秋子梨果实成熟过程中,PuNAC2 和Pu-NAC8 表达水平在呼吸高峰后显著上调,可能参与果实成熟过程中乙烯生物合成和信号转导,而Pu-NAC21 表达显著下调,说明它可能是梨果实成熟的负调节因子[65]。
3.2.2 通过ABA 通路调控 ABA 是参与果实成熟调控的重要激素,在果实的软化、淀粉和糖分积累、色泽发育等过程中起着重要的作用[88]。近年的研究表明NAC 转录因子家族在ABA 调控果实成熟分子网络扮演重要的角色。在草莓果实成熟过程中,沉默FaNAC035 会下调ABA 生物合成关键基因FaNCED2/3/5 和玉米黄质环氧化酶(zeaxanthin epoxidase)FaZEP 的表达,使果实ABA 含量减少,同时ABA 信号转导基因表达也有显著的差异,着色等成熟进程明显延迟,表明FaNAC035 通过调控ABA 生物合成和信号转导参与草莓果实成熟的调控[42]。Zhu 等[31]在宽皮柑橘中获得了一个由CrNAC036 异常高表达引起的ABA 缺失突变体,CrNAC036 可以直接或与CrMYB68 互作结合于CrNCED5 启动子区域并抑制其表达,说明CrNAC036 在抑制ABA 积累和调控柑橘成熟过程中发挥积极作用。
3.2.3 其他激素 除了乙烯和ABA,近年研究证明其他激素在果实成熟过程中也具有重要作用,NAC通过参与这些激素的生物合成或信号转导途径,对果实成熟进行调控。在草莓成熟过程中沉默Fa-NAC035 后,与生长素和多胺生物合成及信号转导的相关基因表达有显著差异,说明FaNAC035 可能也通过生长素和多胺信号途径参与果实成熟的调控[42]。
4 影响NAC基因表达的因素与机制
4.1 响应乙烯、ABA等激素信号
Zhang 等[50]在182 个苹果NAC 基因中筛选出13个在果实生长和成熟阶段具有差异表达和组织特异性表达的成员,其中MdNAC1a 和MdNAC78 的表达在采后储藏过程中受到乙烯的抑制和1-MCP 的诱导,而MdNAC2/26/41/57/80/91/119/141 则被乙烯上调表达,其转录与乙烯的产生速率一致,说明NAC转录因子可能通过乙烯依赖性和非依赖性机制参与苹果发育和成熟过程的调控。乙烯处理能够明显上调香蕉果皮和果肉中MaNAC1 和MaNAC2 的表达,并与乙烯产生量的增加保持一致,MaNAC2 启动子在乙烯处理后被激活,证实其受乙烯诱导[46]。Li等[9]在香蕉成熟过程中进行乙烯处理,发现有10 个MaNACs 基因表达上调,其中MaNAC016/083/094/095 基因在果肉中被乙烯显著上调,而MaNAC094被认为是香蕉乙烯信号转导通路的关键调控因子[47]。外源茉莉酸甲酯(methyl jasmonate,MeJA)可以协同加强外源乙烯对美味猕猴桃成熟的诱导,同时诱导AdNAC2和AdNAC3表达[28]。用ABA处理桃果实后,PpNAC1、PpNAC4、PpNAC6(文献中称为PpNAP1、PpNAP4、PpNAP6)表达水平升高,果实硬度显著下降,说明PpNAC1、PpNAC4、PpNAC6 能够响应ABA 信号,参与果实成熟的调控[61-62]。外源施加ABA还可以反馈促进FaNAC035的表达,进而增加ABA生物合成量[42]。FcNAC1在智利草莓果实成熟过程中表达增加,其启动子序列中含有ABA和生长素等多种激素的顺式反应元件,表明FcNAC1 可能响应ABA等激素信号参与果实成熟的调控[43]。
许田等[89]也发现草莓果实成熟过程中Fa-NAC56 表达量急剧增加,其启动子区含有ABA、赤霉素、生长素、乙烯等激素响应元件,该基因的表达水平受这些激素诱导,说明多种激素通过调控Fa-NAC56影响草莓果实的发育和成熟。
4.2 受转录或翻译后水平修饰调控
香蕉中,RING E3 连接酶XA21 结合蛋白3(MaXB3)可以与MaNAC2、MaACS1和MaACO1相互作用,并通过泛素化途径促进其降解,从而在转录或翻译后水平上抑制乙烯生物合成和下游反应,在香蕉果实和番茄中的瞬时和异源过表达MaXB3,可以抑制乙烯生物合成、推迟果实成熟,证实了MaXB3 的 作 用。同 时MaXB3 也 是MaNAC1 和MaNAC2的下游目标基因,并受到它们的直接转录抑制。MaXB3 和MaNAC1、MaNAC2、MaACS1 和MaACO1 在乙烯形成中依赖反馈调节机制的多层次调控级联[86]。研究发现AdNAC6 和AdNAC7 转录后被miR164靶向降解,这一降解途径位于乙烯信号的下游并受乙烯抑制,亚细胞定位分析表明,Ad-NAC6 和AdNAC7 在细胞核和细胞质中都有分布,AdNAC6 和AdNAC7 可以形成同源二聚体或异源二聚体,只在细胞核中定位。然而,miR164 的存在会阻止AdNAC6 和AdNAC7 进入细胞核,并导致它们留在细胞质中。这种miR164-NAC转录后水平调控途径保守存在于苹果、香蕉、草莓、桃、柑橘、葡萄等果实中并与水果成熟调控有关[28]。此外,功能性或调节性蛋白的氧化还原修饰已成为翻译后修饰的一个重要机制,在香蕉中,MaNAC42 在氧化胁迫下直接与氧化应激和成熟相关基因的启动子结合,MaNAC42中的蛋氨酸氧化会导致其DNA结合能力和转录活性下降,而蛋氨酸亚砜还原酶B 可以靶向MaNAC42,在氧化胁迫下,MaMsrB2 可以部分修复氧化的MaNAC42 并恢复其DNA 结合能力,这些研究结果揭示了一个涉及MaMsrB2 介导的成熟相关转录因子MaNAC42 的氧化还原调节机制[90]。在血桃果实发育早期,BL-PpNAC1异源二聚体的反式激活活性被上游转录因子PpSPL1(SQUAMOSA promoter-binding-like 1)所抑制,阻遏下游靶标基因PpMYB10.1的转录,从而限制了花色苷的合成,而在发育后期,PpSPL1表达下调,BL-PpNAC1异源二聚体转录激活活性升高,同时可能涉及bHLH-WD40复合体的协同作用,增强PpMYB10.1 的转录,从而促进后期花色苷的形成[59]。除了转录控制外,表观遗传学分析表明,桃果实成熟期间,PpNAC1 和PpAAT1表达的增加与表观遗传标记H3K4me3的增加有关,在未成熟的果实中观察到PpNAC1 和PpAAT1 位点的抑制性组蛋白标记hyper-H3K27me3,但在成熟的果实中没有,揭示了表观遗传因素的变化在调节NAC 基因表达和随后的挥发性酯类合成的保守机制[57]。
4.3 响应环境因子及非生物胁迫
大蕉中MpSNAC67 能够响应干旱、高盐和过氧化物等胁迫,过量表达MpSNAC67 的转基因香蕉品系会加速叶片脱绿和黄化,进一步研究发现Mp-SNAC67 可以激活叶绿素降解基因PAO-like(Pheophorbide-a-oxygenase,脱镁叶绿酸a 加氧酶)、HCAR-like(hydroxymethyl chlorophyll-a-reductase,羟甲基叶绿素加氧酶)、NYC/NOL-like(Chlorophyll-b-reductase,叶绿素b 还原酶)以及ORS1-like的表达,正向调节叶绿素的降解[48]。金橘果实中的FcrNAC22转录水平在红光照射下被显著诱导,并随果实颜色变化而表达上调[44]。在苹果着色期间,Md-NAC52 受光诱导表达上调,并且受编码光调节器(light-regulator)的MdHY5 靶向而对光信号做出反应,进一步的实验证明MdHY5 可以结合MdNAC52启动子中的G-box 顺式作用元件,通过激发Md-NAC52 的启动子活性参与着色[52]。通过RNA-Seq和实时定量PCR 分析,PpNAC61/70/172/176/23 可能参与蓝光诱导下梨果实的着色[10]。在香蕉果实中,低温条件可以抑制MaNAC67-like 的表达,抑制其对β-淀粉酶基因MaBAM6、磷酸葡聚糖磷酸酶基因MaSEX4、麦芽糖转运体MaMEX1 淀粉降解关键酶基因的转录激活和乙烯信号转导因子MaEBF1(EIN3 binding F-box-1)的相互作用,有助于延缓香蕉果实成熟[91]。在柿果实脱涩过程中,DkNAC1/3/5/6/7/13/16 能够响应CO2处理并表达上调,促进果实脱涩[38~40]。
5 总结与展望
NAC 转录因子是植物中最大的转录因子家族之一,其结构特征、表达特性及功能已在几十种植物中得到描述和鉴定。NAC 转录因子在植物的生长发育和响应生物、非生物逆境以及果实发育和成熟过程中发挥着关键的调控作用。随着现代分子生物学技术的飞速发展,NAC转录因子在果实发育及成熟中的具体功能逐渐得到揭示,NAC 与MYB、乙烯、ABA 相关转录因子协同作用形成调控网络,响应内源和环境信号,调节果实成熟过程的不同方面。但对于NAC 转录因子发挥功能的更多具体分子机制,调控网络及不同蛋白的结构与功能的内在联系,仍有待进一步深入研究,NAC 上游的调控因子和更多的下游靶标基因等仍急需深入挖掘。相关的研究一方面将有助于NAC 转录因子家族在果实成熟和品质形成中功能和调控机制的深入揭示,另一方面自高效的成簇规律间隔短回文重复序列(clustered regularly interspaced short palindromic repeats,CRISPR)/CRISPR相关蛋白9(Cas9)基因组编辑技术出现及高速发展以来,已经在香蕉[92]、草莓[93]、猕猴桃[94]、葡萄[95-97]及苹果[95]等果树中实现决定果实品质关键性状的改良[98],通过基因编辑精准调控NAC基因表达水平或NAC蛋白活性从而加强其对果实成熟和品质形成的调控在未来的研究或会得到更多的应用。研究将为利用改良果实特定品质性状的果树分子育种方法,以及在果实成熟调控技术研发和理论创新方面奠定坚实的基础。
[1] PRASANNA V,PRABHA T N,THARANATHAN R N. Fruit ripening phenomena:An overview[J]. Critical Reviews in Food Science and Nutrition,2007,47(1):1-19.
[2] MOHANTA T K,YADAV D,KHAN A,HASHEM A,TABASSUM B,KHAN A L,ABD ALLAH E F,AL-HARRASI A. Genomics,molecular and evolutionary perspective of NAC transcription factors[J].PLoS One,2020,15(4):e0231425.
[3] 靳进朴,郭安源,何坤,张禾,朱其慧,陈新,高歌,罗静初.植物转录因子分类、预测和数据库构建[J].生物技术通报,2015,31(11):68-77.JIN Jinpu,GUO Anyuan,HE Kun,ZHANG He,ZHU Qihui,CHEN Xin,GAO Ge,LUO Jingchu. Classification,prediction and database construction of plant transcription factors[J]. Biotechnology Bulletin,2015,31(11):68-77.
[4] SOUER E,VAN HOUWELINGEN A,KLOOS D,MOL J,KOES R. The No apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries[J]. Cell,1996,85(2):159-170.
[5] AIDA M,ISHIDA T,TASAKA M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis:interaction among the CUP- SHAPED COTYLEDON and SHOOT MERISTEMLESS genes[J]. Development,1999,126(8):1563-1570.
[6] TRUPKIN S A,ASTIGUETA F H,BAIGORRIA A H,GARCÍA M N,DELFOSSE V C,GONZÁLEZ S A,DE LA TORRE M C P,MOSCHEN S,LÍA V V,FERNÁNDEZ P,HEINZ R A.Identification and expression analysis of NAC transcription factors potentially involved in leaf and petal senescence in Petunia hybrida[J].Plant Science,2019,287:110195.
[7] MUNIR N,CHEN Y K,CHEN X H,NAWAZ M A,IFTIKHAR J,RIZWAN H M,SHEN X,LIN Y L,XU X H,LAI Z X. Genome-wide identification and comprehensive analyses of NAC transcription factor gene family and expression patterns during somatic embryogenesis in Dimocarpus longan Lour.[J]. Plant Physiology and Biochemistry,2020,157:169-184.
[8] 亢超,郭彩华,张雪蒙,刘金明,袁星,全绍文,牛建新. 核桃NAC 基因家族的全基因组鉴定与分析[J].果树学报,2021,38(9):1444-1458.KANG Chao,GUO Caihua,ZHANG Xuemeng,LIU Jinming,YUAN Xing,QUAN Shaowen,NIU Jianxin. Genome- wide identification and analysis of NAC gene family in walnut (Juglans regia L. )[J]. Journal of Fruit Science,2021,38(9):1444-1458.
[9] LI B,FAN R Y,YANG Q S,HU C H,SHENG O,DENG G M,DONG T,LI C Y,PENG X X,BI F C,YI G J. Genome-wide identification and characterization of the NAC transcription factor family in Musa acuminata and expression analysis during fruit ripening[J]. International Journal of Molecular Sciences,2020,21(2):634.
[10] AHMAD M,YAN X H,LI J Z,YANG Q S,JAMIL W,TENG Y W,BAI S L. Genome wide identification and predicted functional analyses of NAC transcription factors in Asian pears[J].BMC Plant Biology,2018,18(1):214.
[11] 李圣龙,王传铭,李晓静.石榴NAC 转录因子家族的生物信息学分析[J].分子植物育种,2021,19(1):88-99.LI Shenglong,WANG Chuanming,LI Xiaojing. Bioinformatic analysis of the NAC transcription factor family in Punica granatum L.[J].Molecular Plant Breeding,2021,19(1):88-99.
[12] NAKANO Y,YAMAGUCHI M,ENDO H,REJAB N A,OHTANI M.NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants[J].Frontiers in Plant Science,2015,6:288.
[13] XIE Q,FRUGIS G,COLGAN D,CHUA N H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development[J]. Genes & Development,2000,14(23):3024-3036.
[14] HE X J,MU R L,CAO W H,ZHANG Z G,ZHANG J S,CHEN S Y.AtNAC2,a transcription factor downstream of ethylene and auxin signaling pathways,is involved in salt stress response and lateral root development[J].The Plant Journal,2005,44(6):903-916.
[15] KO J H,YANG S H,PARK A H,LEROUXEL O,HAN K H.ANAC012,a member of the plant-specific NAC transcription factor family,negatively regulates xylary fiber development in Arabidopsis thaliana[J]. The Plant Journal,2007,50(6):1035-1048.
[16] NURUZZAMAN M,SHARONI A M,KIKUCHI S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants[J].Frontiers in Microbiology,2013,4:248.
[17] FORLANI S,MIZZOTTI C,MASIERO S.The NAC side of the fruit:Tuning of fruit development and maturation[J]. BMC Plant Biology,2021,21(1):238.
[18] PODZIMSKA-SROKA D,O’SHEA C,GREGERSEN P L,SKRIVER K. NAC transcription factors in senescence:From molecular structure to function in crops[J]. Plants,2015,4(3):412-448.
[19] KOU X H,ZHOU J Q,WU C E,YANG S,LIU Y F,CHAI L P,XUE Z H. The interplay between ABA/ethylene and NAC TFs in tomato fruit ripening:A review[J]. Plant Molecular Biology,2021,106(3):223-238.
[20] PURANIK S,SAHU P P,SRIVASTAVA P S,PRASAD M.NAC proteins:regulation and role in stress tolerance[J]. Trends in Plant Science,2012,17(6):369-381.
[21] ERNST H A,OLSEN A N,SKRIVER K,LARSEN S,LO LEGGIO L. Structure of the conserved domain of ANAC,a member of the NAC family of transcription factors[J]. EMBO Reports,2004,5(3):297-303.
[22] 赵才美,黄兴奇,殷富有,李定琴,陈越,陈玲,程在全. 水稻NAC 转录因子家族的研究进展[J]. 植物科学学报,2020,38(2):278-287.ZHAO Caimei,HUANG Xingqi,YIN Fuyou,LI Dingqin,CHEN Yue,CHEN Ling,CHENG Zaiquan. Research progress on NAC transcription factor family in Oryza sativa L.[J]. Plant Science Journal,2020,38(2):278-287.
[23] DIAO P F,CHEN C,ZHANG Y Z,MENG Q W,LÜ W,MA N N.The role of NAC transcription factor in plant cold response[J].Plant Signaling&Behavior,2020,15(9):1785668.
[24] OOKA H,SATOH K,DOI K,NAGATA T,OTOMO Y,MURAKAMI K,MATSUBARA K,OSATO N,KAWAI J,CARNINCI P,HAYASHIZAKI Y,SUZUKI K,KOJIMA K,TAKAHARA Y,YAMAMOTO K,KIKUCHI S.Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana[J].DNA Research,2003,10(6):239-247.
[25] CHEN C J,CHEN H,ZHANG Y,THOMAS H R,FRANK M H,HE Y H,XIA R. TBtools:An integrative toolkit developed for interactive analyses of big biological data[J]. Molecular Plant,2020,13(8):1194-1202.
[26] NIEUWENHUIZEN N J,CHEN X Y,WANG M Y,MATICH A J,PEREZ R L,ALLAN A C,GREEN S A,ATKINSON R G.Natural variation in monoterpene synthesis in kiwifruit:Transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-like transcription factors[J].Plant Physiology,2015,167(4):1243-1258.
[27] NIEUWENHUIZEN N J,CHEN X Y,PELLAN M,ZHANG L,GOU L,LAING W A,SCHAFFER R J,ATKINSON R G,ALLAN A C. Regulation of wound ethylene biosynthesis by NAC transcription factors in kiwifruit[J]. BMC Plant Biology,2021,21(1):411.
[28] WANG W Q,WANG J,WU Y Y,LI D W,ALLAN A C,YIN X R. Genome-wide analysis of coding and non-coding RNA reveals a conserved miR164-NAC regulatory pathway for fruit ripening[J].New Phytologist,2020,225(4):1618-1634.
[29] WU Y Y,LIU X F,FU B L,ZHANG Q Y,TONG Y,WANG J,WANG W Q,GRIERSON D,YIN X R. Methyl jasmonate enhances ethylene synthesis in kiwifruit by inducing NAC genes that activate ACS1[J]. Journal of Agricultural and Food Chemistry,2020,68(10):3267-3276.
[30] FU B L,WANG W Q,LIU X F,DUAN X W,ALLAN A C,GRIERSON D,YIN X R. An ethylene-hypersensitive methionine sulfoxide reductase regulated by NAC transcription factors increases methionine pool size and ethylene production during kiwifruit ripening[J].New Phytologist,2021,232(1):237-251.
[31] ZHU F,LUO T,LIU C Y,WANG Y,ZHENG L,XIAO X,ZHANG M F,YANG H B,YANG W,XU R W,ZENG Y L,YE J L,XU J,XU J G,LARKIN R M,WANG P W,WEN W W,DENG X X,FERNIE A R,CHENG Y J. A NAC transcription factor and its interaction protein hinder abscisic acid biosynthesis by synergistically repressing NCED5 in Citrus reticulata[J].Journal of Experimental Botany,2020,71(12):3613-3625.
[32] LI S J,YIN X R,WANG W L,LIU X F,ZHANG B,CHEN K S. Citrus CitNAC62 cooperates with CitWRKY1 to participate in citric acid degradation via up-regulation of CitAco3[J]. Journal of Experimental Botany,2017,68(13):3419-3426.
[33] WU J X,FU L L,YI H L. Genome-wide identification of the transcription factors involved in Citrus fruit ripening from the transcriptomes of a late-ripening sweet orange mutant and its wild type[J].PLoS One,2016,11(4):e0154330.
[34] FU C C,HAN Y C,FAN Z Q,CHEN J Y,CHEN W X,LU W J,KUANG J F. The Papaya transcription factor CpNAC1 modulates carotenoid biosynthesis through activating phytoene desaturase genes CpPDS2/4 during fruit ripening[J]. Journal of Agricultural and Food Chemistry,2016,64(27):5454-5463.
[35] FU C C,HAN Y C,KUANG J F,CHEN J Y,LU W J. Papaya CpEIN3a and CpNAC2 co-operatively regulate carotenoid biosynthesis-related genes CpPDS2/4,CpLCY-e and CpCHY-b during fruit ripening[J]. Plant and Cell Physiology,2017,58(12):2155-2165.
[36] 付长春.NAC 类转录因子参与调控番木瓜果实后熟过程中类胡萝卜素代谢的机制研究[D].广州:华南农业大学,2017.FU Changchun. Mechanism analysis of NAC transcription factors in regulation of carotenoid biosynthesis during Papaya fruit ripening[D]. Guangzhou:South China Agricultural University,2017.
[37] FU C C,CHEN H J,GAO H Y,WANG S L,WANG N,JIN J C,LU Y,YU Z L,MA Q,HAN Y C. Papaya CpMADS4 and Cp-NAC3 co-operatively regulate ethylene signal genes CpERF9 and CpEIL5 during fruit ripening[J]. Postharvest Biology and Technology,2021,175:111485.
[38] MIN T,WANG M M,WANG H X,LIU X F,FANG F,GRIERSON D,YIN X R,CHEN K S. Isolation and expression of NAC genes during persimmon fruit postharvest astringency removal[J]. International Journal of Molecular Sciences,2015,16(1):1894-1906.
[39] JIN R,ZHU Q G,SHEN X Y,WANG M M,JAMIL W,GRIERSON D,YIN X R,CHEN K S. DkNAC7,a novel high-CO2/hypoxia-induced NAC transcription factor,regulates persimmon fruit de-astringency[J].PLoS One,2018,13(3):e0194326.
[40] JAMIL W,WU W,AHMAD M,ZHU Q G,LIU X F,JIN R,YIN X R. High-CO2/hypoxia-modulated NAC transcription factors involved in de-astringency of persimmon fruit[J]. Scientia Horticulturae,2019,252:201-207.
[41] 柴吉钏,王康,杨民杰,董婉琪,曹士锋,陈伟,杨震峰,施丽愉.枇杷EjNAC82 的克隆及其对类胡萝卜素合成基因EjPSY、EjBCH 的转录激活分析[J].核农学报,2022,36(6):1115-1126.CHAI Jichuan,WANG Kang,YANG Minjie,DONG Wanqi,CAO Shifeng,CHEN Wei,YANG Zhenfeng,SHI Liyu.Cloning of EjNAC82 in loquat and its transcriptional regulation on carotenoid biosynthetic genes EjPSY and EjBCH[J]. Journal of Nuclear Agricultural Sciences,2022,36(6):1115-1126.
[42] MARTÍN-PIZARRO C,VALLARINO J G,OSORIO S,MECO V,URRUTIA M,PILLET J,CASAÑAL A,MERCHANTE C,AMAYA I,WILLMITZER L,FERNIE A R,GIOVANNONI J J,BOTELLA M A,VALPUESTA V,POSÉ D.The NAC transcription factor FaRIF controls fruit ripening in strawberry[J]. The Plant Cell,2021,33(5):1574-1593.
[43] CARRASCO- ORELLANA C,STAPPUNG Y,MENDEZYAÑEZ A,ALLAN A C,ESPLEY R V,PLUNKETT B J,MOYA-LEON M A,HERRERA R. Characterization of a ripening-related transcription factor FcNAC1 from Fragaria chiloensis fruit[J].Scientific Reports,2018,8(1):10524.
[44] GONG J L,ZENG Y L,MENG Q N,GUAN Y J,LI C Y,YANG H B,ZHANG Y Z,AMPOMAH-DWAMENA C,LIU P,CHEN C W,DENG X X,CHENG Y J,WANG P W. Red lightinduced kumquat fruit coloration is attributable to increased carotenoid metabolism regulated by FcrNAC22[J]. Journal of Experimental Botany,2021,72(18):6274-6290.
[45] JIANG G X,LI Z W,SONG Y B,ZHU H,LIN S,HUANG R M,JIANG Y M,DUAN X W. LcNAC13 physically interacts with LcR1MYB1 to coregulate anthocyanin biosynthesis-related genes during Litchi fruit ripening[J]. Biomolecules,2019,9(4):135.
[46] SHAN W,KUANG J F,CHEN L,XIE H,PENG H H,XIAO Y Y,LI X P,CHEN W X,HE Q G,CHEN J Y,LU W J. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening[J]. Journal of Experimental Botany,2012,63(14):5171-5187.
[47] LÜ P T,YU S,ZHU N,CHEN Y R,ZHOU B Y,PAN Y,TZENG D,FABI J P,ARGYRIS J,GARCIA-MAS J,YE N H,ZHANG J H,GRIERSON D,XIANG J,FEI Z J,GIOVANNONI J,ZHONG S L. Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening[J]. Nature Plants,2018,4(10):784-791.
[48] TAK H,NEGI S,GUPTA A,GANAPATHI T R.A stress associated NAC transcription factor MpSNAC67 from banana(Musa×paradisiaca) is involved in regulation of chlorophyll catabolic pathway[J]. Plant Physiology and Biochemistry,2018,132:61-71.
[49] MIGICOVSKY Z,YEATS T H,WATTS S,SONG J,FORNEY C F,BURGHER-MACLELLAN K,SOMERS D J,GONG Y H,ZHANG Z Q,VREBALOV J,VAN VELZEN R,GIOVANNONI J G,ROSE J K C,MYLES S.Apple ripening is controlled by a NAC transcription factor[J]. Frontiers in Genetics,2021,12:671300.
[50] ZHANG Q J,LI T,ZHANG L J,DONG W X,WANG A D.Expression analysis of NAC genes during the growth and ripening of apples[J].Horticultural Science,2018,45(1):1-10.
[51] AN J P,YAO J F,XU R R,YOU C X,WANG X F,HAO Y J.An apple NAC transcription factor enhances salt stress tolerance by modulating the ethylene response[J]. Physiologia Plantarum,2018,164(3):279-289.
[52] WANG A D,XU K N. Characterization of two orthologs of REVERSION-TO-ETHYLENE SENSITIVITY1 in apple[J]. Journal of Molecular Biology Research,2012,2(1):24-41.
[53] SUN Q G,JIANG S H,ZHANG T L,XU H F,FANG H C,ZHANG J,SU M Y,WANG Y C,ZHANG Z Y,WANG N,CHEN X S. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11[J].Plant Science,2019,289:110286.
[54] ZHANG S Y,CHEN Y X,ZHAO L L,LI C Q,YU J Y,LI T T,YANG W Y,ZHANG S N,SU H Y,WANG L. A novel NAC transcription factor,MdNAC42,regulates anthocyanin accumulation in red-fleshed apple by interacting with MdMYB10[J].Tree Physiology,2020,40(3):413-423.
[55] 孙庆国,姜生辉,房鸿成,张天亮,王楠,陈学森. 苹果Md-NAC9 的克隆及其调控黄酮醇合成功能的鉴定[J].园艺学报,2019,46(11):2073-2081.SUN Qingguo,JIANG Shenghui,FANG Hongcheng,ZHANG Tianliang,WANG Nan,CHEN Xuesen. Cloning of MdNAC9 and functional of its regulation on flavonol synthesis[J]. Acta Horticulturae Sinica,2019,46(11):2073-2081.
[56] 周晖.桃花青苷着色及原花青素合成的调控机制研究[D].北京:中国科学院大学(中国科学院武汉植物园),2015.ZHOU Hui. Mechanisms underlving the regulation of anthocyanincoloration and proanthocyanidin synthesis in peach[D].Beijing:University of Chinese Academy of Sciences(Wuhan Botanical Garden,Chinese Academy of Sciences),2015.
[57] CAO X M,WEI C Y,DUAN W Y,GAO Y,KUANG J F,LIU M C,CHEN K S,KLEE H,ZHANG B.Transcriptional and epigenetic analysis reveals that NAC transcription factors regulate fruit flavor ester biosynthesis[J]. The Plant Journal,2021,106(3):785-800.
[58] JIN Z N,WANG J J,CAO X M,WEI C Y,KUANG J F,CHEN K S,ZHANG B.Peach fruit PpNAC1 activates PpFAD3-1 transcription to provide ω-3 fatty acids for the synthesis of shortchain flavor volatiles[J]. Horticulture Research,2022,9:uhac085.
[59] 曹香梅.桃果实酯类芳香物质的代谢与调控研究[D].杭州:浙江大学,2019.CAO Xiangmei. Metabolism and regulation of aromatic esters in peach fruit[D].Hangzhou:Zhejiang University,2019.
[60] 秦娟,余凡,刘璐,朱婷婷,陈伟,曹士锋,杨震峰,施丽愉.桃PpNAC19 的克隆及其对PpCCD4 启动子活性的调节分析[J].核农学报,2021,35(6):1273-1280.QIN Juan,YU Fan,LIU Lu,ZHU Tingting,CHEN Wei,CAO Shifeng,YANG Zhenfeng,SHI Liyu. Cloning of peach Pp-NAC19 and its regulation on PpCCD4 promoter activity[J].Journal of Nuclear Agricultural Sciences,2021,35(6):1273-1280.
[61] LI F,LI J J,QIAN M,HAN M Y,CAO L J,LIU H K,ZHANG D,ZHAO C P. Identification of peach NAP transcription factor genes and characterization of their expression in vegetative and reproductive organs during development and senescence[J].Frontiers in Plant Science,2016,7:147.
[62] 李芳.NAP 家族成员克隆及其在桃果实发育与成熟期间的表达特性研究[D].杨凌:西北农林科技大学,2016.LI Fang. Cloning and expression characterization of NAP familymembers during development and ripening of peach[D]. Yangling:Northwest A&F University,2016.
[63] TAN Q P,LI S,ZHANG Y Z,CHEN M,WEN B B,JIANG S,CHEN X D,FU X L,LI D M,WU H Y,WANG Y,XIAO W,LI L.Chromosome-level genome assemblies of five Prunus species and genome-wide association studies for key agronomic traits in peach[J].Horticulture Research,2021,8(1):213.
[64] GUO Z H,ZHANG Y J,YAO J L,XIE Z H,ZHANG Y Y,ZHANG S L,GU C. The NAM/ATAF1/2/CUC2 transcription factor PpNAC. A59 enhances PpERF. A16 expression to promote ethylene biosynthesis during peach fruit ripening[J].Horticulture Research,2021,8(1):209.
[65] HUANG G H,LI T,LI X Y,TAN D M,JIANG Z Y,WEI Y,LI J C,WANG A D.Comparative transcriptome analysis of climacteric fruit of Chinese pear (Pyrus ussuriensis) reveals new insights into fruit ripening[J].PLoS One,2014,9(9):e107562.
[66] 宋杨,刘红弟,王海波,张红军,刘凤之.越橘VcNAC072 克隆及其促进花青素积累的功能分析[J].中国农业科学,2019,52(3):503-511.SONG Yang,LIU Hongdi,WANG Haibo,ZHANG Hongjun,LIU Fengzhi. Molecular cloning and functional characterization of vc NAC072 reveals its involvement in anthocyanin accumulation in blueberry[J]. Scientia Agricultura Sinica,2019,52(3):503-511.
[67] ZENONI S,D’INCÀ E,TORNIELLI G B. Genetic dissection of grape berry ripening control:defining a role for NAC transcription factors[J].Acta Horticulturae,2019(1248):387-402.
[68] GIOVANNONI J J. Genetic regulation of fruit development and ripening[J].The Plant Cell Online,2004,16(S1):S170-S180.
[69] BRADY C J.Fruit ripening[J].Annual Review of Plant Physiology,1987,38(1):155-178.
[70] LAI B,HU B,QIN Y H,ZHAO J T,WANG H C,HU G B.Transcriptomic analysis of Litchi chinensis pericarp during maturation with a focus on chlorophyll degradation and flavonoid biosynthesis[J].BMC Genomics,2015,16(1):225.
[71] LIN Y X,JIANG L Y,CHEN Q,LI Y L,ZHANG Y T,LUO Y,ZHANG Y,SUN B,WANG X R,TANG H R.Comparative transcriptome profiling analysis of red- and white-fleshed strawberry (Fragaria × ananassa) provides new insight into the regulation of the anthocyanin pathway[J]. Plant and Cell Physiology,2018,59(9):1844-1859.
[72] 王惠聪,黄辉白,黄旭明.荔枝果实的糖积累与相关酶活性[J].园艺学报,2003,30(1):1-5.WANG Huicong,HUANG Huibai,HUANG Xuming. Sugar accumulation and related enzyme activities in the Litchi fruit of‘Nuomici’and‘Feizixiao’[J]. Acta Horticulturae Sinica,2003,30(1):1-5.
[73] YANG Z Y,WANG T D,WANG H C,HUANG X M,QIN Y H,HU G B. Patterns of enzyme activities and gene expressions in sucrose metabolism in relation to sugar accumulation and composition in the aril of Litchi chinensis Sonn.[J]. Journal of Plant Physiology,2013,170(8):731-740.
[74] WANG T D,ZHANG H F,WU Z C,LI J G,HUANG X M,WANG H C. Sugar uptake in the aril of Litchi fruit depends on the apoplasmic post-phloem transport and the activity of proton pumps and the putative transporter LcSUT4[J]. Plant and Cell Physiology,2015,56(2):377-387.
[75] SONG J,FORNEY C F. Flavour volatile production and regulation in fruit[J]. Canadian Journal of Plant Science,2008,88(3):537-550.
[76] DEFILIPPI B G,MANRÍQUEZ D,LUENGWILAI K,GONZÁLEZ-AGÜERO M. Aroma volatiles:Biosynthesis and mechanisms of modulation during fruit ripening[J].Advances in Botanical Research,2009,50:1-37.
[77] WANG L B,BALDWIN E A,BAI J H. Recent advance in aromatic volatile research in tomato fruit:The metabolisms and regulations[J]. Food and Bioprocess Technology,2016,9(2):203-216.
[78] BRUMMELL D A. Cell wall disassembly in ripening fruit[J].Functional Plant Biology,2006,33(2):103-119.
[79] HAYAMA H,TATSUKI M,ITO A,KASHIMURA Y. Ethylene and fruit softening in the stony hard mutation in peach[J]. Postharvest Biology and Technology,2006,41(1):16-21.
[80] GOULAO L F,OLIVEIRA C M.Cell wall modifications during fruit ripening:When a fruit is not the fruit[J]. Trends in Food Science&Technology,2008,19(1):4-25.
[81] POSÉ S,PANIAGUA C,MATAS A J,GUNNING A P,MORRIS V J,QUESADA M A,MERCADO J A. A nanostructural view of the cell wall disassembly process during fruit ripening and postharvest storage by atomic force microscopy[J]. Trends in Food Science&Technology,2019,87:47-58.
[82] LAI B,LI X J,HU B,QIN Y H,HUANG X M,WANG H C,HU G B. LcMYB1 is a key determinant of differential anthocyanin accumulation among genotypes,tissues,developmental phases and ABA and light stimuli in Litchi chinensis[J]. PLoS One,2014,9(1):e86293
[83] LAI B,DU L N,LIU R,HU B,SU W B,QIN Y H,ZHAO J T,WANG H C,HU G B.Two LcbHLH transcription factors interacting with LcMYB1 in regulating late structural genes of anthocyanin biosynthesis in Nicotiana and Litchi chinensis during anthocyanin accumulation[J].Frontiers in Plant Science,2016,7:166.
[84] FENN M A,GIOVANNONI J J. Phytohormones in fruit development and maturation[J].The Plant Journal,2021,105(2):446-458.
[85] WANG H C,HUANG H B,HUANG X M. Differential effects of abscisic acid and ethylene on the fruit maturation of Litchi chinensis Sonn. [J]. Plant Growth Regulation,2007,52(3):189-198.
[86] SHAN W,KUANG J F,WEI W,FAN Z Q,DENG W,LI Z G,BOUZAYEN M,PIRRELLO J,LU W J,CHEN J Y. MaXB3 modulates MaNAC2,MaACS1,and MaACO1 stability to repress ethylene biosynthesis during banana fruit ripening[J].Plant Physiology,2020,184(2):1153-1171.
[87] WEI W,YANG Y Y,SU X G,KUANG J F,CHEN J Y,LU W J,SHAN W. MaRTH1 suppression of ethylene response during banana fruit ripening and is controlled by MaXB3-MaNAC2 regulatory module[J]. Postharvest Biology and Technology,2021,182:111707.
[88] 王鹏洋,曲姗姗.ABA 对果实品质的影响研究进展[J].现代农业科技,2019(7):198.WANG Pengyang,QU Shanshan. Research progress on the effect of ABA on fruit quality[J]. Modern Agricultural Science and Technology,2019(7):198.
[89] 许田,吴敏,陈雪雪,黄芸,沈元月.转录因子FaNAC56 在草莓果实成熟中的功能分析[J]. 果树学报,2021,38(12):2072-2081.XU Tian,WU Min,CHEN Xuexue,HUANG Yun,SHEN Yuanyue. Functional analysis of transcription factor FaNAC56 in strawberry fruit ripening[J]. Journal of Fruit Science,2021,38(12):2072-2081.
[90] YAN H L,JIANG G X,WU F W,LI Z W,XIAO L,JIANG Y M,DUAN X W. Sulfoxidation regulation of transcription factor NAC42 influences its functions in relation to stress-induced fruit ripening in banana[J]. Journal of Experimental Botany,2021,72(2):682-699.
[91] SONG Z Y,QIN J J,ZHENG Q L,DING X C,CHEN W X,LU W J,LI X P,ZHU X Y. The involvement of the banana F-box protein MaEBF1 in regulating chilling-inhibited starch degradation through interaction with a MaNAC67-like protein[J]. Biomolecules,2019,9(10):552.
[92] KAUR N,ALOK A,SHIVANI,KUMAR P,KAUR N,AWASTHI P,CHATURVEDI S,PANDEY P,PANDEY A,PANDEY A K,TIWARI S. CRISPR/Cas9 directed editing of lycopene epsilon-cyclase modulates metabolic flux for β-carotene biosynthesis in banana fruit[J]. Metabolic Engineering,2020,59:76-86.
[93] 陈文君. FvCO4 在草莓果实早期发育中的功能鉴定[D]. 沈阳:沈阳农业大学,2019.CHEN Wenjun. Function analysis of FvCO4 in the early development of strawberry fruit[D].Shenyang:Shenyang Agricultural University,2019.
[94] 李创.猕猴桃表皮毛发育基因的筛选及其在海沃德猕猴桃中的基因编辑研究[D].杨凌:西北农林科技大学,2019.LI Chuang. Screening the genes regulating trichome development in Actinidia and their genome editing in‘Hayward’kiwifruit[D].Yangling:Northwest A&F University,2019.
[95] OSAKABE Y,LIANG Z C,REN C,NISHITANI C,OSAKABE K,WADA M,KOMORI S,MALNOY M,VELASCO R,POLI M,JUNG M H,KOO O J,VIOLA R,KANCHISWAMY C N. CRISPR- Cas9- mediated genome editing in apple and grapevine[J].Nature Protocols,2018,13(12):2844-2863.
[96] REN C,LIU X J,ZHANG Z,WANG Y,DUAN W,LI S H,LIANG Z C. CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay(Vitis vinifera L.)[J].Scientific Reports,2016,6:32289.
[97] REN C,LIU Y F,GUO Y C,DUAN W,FAN P G,LI S H,LIANG Z C.Optimizing the CRISPR/Cas9 system for genome editing in grape by using grape promoters[J]. Horticulture Research,2021,8:52.
[98] 何玉坤,欧阳嫣惟,张秀梅,林文秋,潘晓璐,张红娜.CRISPR/Cas9 基因编辑技术在果树作物中的应用研究进展[J].果树学报,2022,39(5):870-881.HE Yukun,OUYANG Yanwei,ZHANG Xiumei,LIN Wenqiu,PAN Xiaolu,ZHANG Hongna. Application of CRISPR/Cas9 gene editing technology in fruit trees[J]. Journal of Fruit Science,2022,39(5):870-881.