莽草酸代谢途径(shikimic acid pathway,SAP)是植物和微生物中的枢纽性代谢[1],具有联系糖代谢和次生代谢的桥梁纽带作用。SAP途径由7个酶促反应组成[2],其中分支酸变位酶(chorismate mutase,CM)是SAP 途径的第七步酶[3],催化分支酸生成预苯酸。在SAP途径的作用下,碳源由莽草酸途径进入苯丙烷代谢途径,形成芳香族氨基酸(色氨酸、苯丙氨酸和酪氨酸)、水杨酸、吲哚乙酸(IAA)等次生代谢物合成的前体。作为莽草酸途径的出口酶,CM控制着SAP途径的碳源流向,其下游途径所合成的次生代谢物与植物生长发育、逆境防御、信号转导等重要过程的关系密切[4-6]。特别是由于哺乳动物中不存在SAP 途径,CM 对动物的氨基酸营养具有重要意义。此外,分支酸变位酶一直以来都是生物化工和制药行业关注的热点,该酶可作为工程菌改造的靶点以提高L-苯丙氨酸和L-酪氨酸的产率[7]。与此同时,病原CM 也是某些疾病(例如肺结核)治疗药物开发的潜在靶点[8]。目前在拟南芥(Arabidopsis thaliana)中共发现3 个CM 编码基因,其中AtCM1和AtCM3定位于叶绿体,表达受苯丙氨酸及酪氨酸的反馈调节;AtCM2 定位于细胞质中,其活性不受芳香族氨基酸的影响[9-10]。CM的表达具有组织特异性,其活性和表达量受逆境、损伤和紫外照射诱导[4],葡萄(Vitis vinifera)的VvCM1 和VvCM2基因在各器官组织中均有表达,VvCM1在果实中表达丰度最高,VvCM2在茎中表达丰度最高,在果实中不同部位随果实发育期表达具有特异性[4]。VvCM1活性不受芳香族氨基酸调控,但VvCM2 的催化活性受色氨酸的影响而显著上调。矮牵牛(Petunia hybrida)中定位于质体中的PhCM1受色氨酸的变构调节,但是不受苯丙氨酸和酪氨酸的反馈调节[11]。
核桃(Juglans regia L.)为胡桃科胡桃属经济林树种,坚果硕大且营养丰富[12]。核桃富含多酚和抗氧化活性物质,因而具有较好的保健功能[13]。目前,莽草酸代谢途径与核桃次生代谢合成的关联研究较少,但就核桃化感关键物质胡桃醌合成途径的研究表明[14],胡桃醌的合成以莽草酸代谢途径产生的物质为前体。鉴于莽草酸途径与核桃次生代谢物合成的密切联系,开展该合成途径关键酶的研究可进一步深化对核桃次生代谢物合成调控的认识。笔者基于核桃的全基因组测序数据[15]开展核桃CM家族基因的筛选鉴定,分析了该家族基因的理化性质、基因结构、染色体定位和进化关系,并对CM家族基因在不同组织中的表达进行qPCR定量。本研究为探究核桃CM家族基因的功能及其在莽草酸途径中的生物学作用奠定了基础。
1 材料和方法
1.1 试验材料
笔者在本研究中以青海省农林科学院引种的优良品种辽宁1 号[16]核桃为研究对象,试验材料采集于2016年嫁接的贵德县河东乡核桃良种嫁接区(东经101°26′48″,北纬36°02′39″)。核桃果实7 月进入硬核期,果实大小已基本定型,青皮成熟,随机选取3 株长势均一植株进行样本采集。采集植株1 年生枝条的成熟叶片、茎、果实青皮及果柄,并进行混样处理,经液氮速冻后保存于-80 ℃冰箱备用。
1.2 核桃CM基因家族成员的鉴定
核桃CM 家族基因的鉴定使用HMM 搜索和BLAST 相结合的方式进行,核桃基因组数据从Ensembl Plants 数据库(https://plants.ensembl.org/index.html)获取。搜索使用的HMM模型CM_1(PF07736)和CM_2(PF01817)从Pfam(http://pfam.xfam.org/)下载。本地BLAST 以拟南芥AtCM1(AT3G29200)为种子序列,E-value阈值设定为1×10-10,经BLASTp搜索后鉴定核桃CM 蛋白。初筛序列经比对后,滤除冗余序列。所有序列经SMART(http://smart.emblheidelberg.de/)和 CDD(https://www.ncbi.nlm.nih.gov/cdd/)数据库进行CM 结构域确认后得到核桃CM家族基因。
1.3 核桃CM基因家族的特征分析
通过在线软件ProtParam 预测CM 家族蛋白结构,分析目的基因编码蛋白质的氨基酸组成、蛋白质的相对分子质量、理论等电点及稳定性等参数。采用ExPASy 中的ProtScale 工具对核桃CM 基因编码蛋白的疏水性和亲水性进行预测。根据从基因组注释文件中获取的CM基因家族在染色体上的位置数据,对其染色体定位进行可视化分析。利用在线软件(http://www.csbio.sjtu.edu.cn/bioinf/plant- multi/)进行蛋白的亚细胞定位预测。蛋白跨膜结构的预测使用TMHMM Server v.2.0 在线工具进行。蛋白质三级结构的预测使用SWISS-MODEL进行。
1.4 核桃CM基因结构、启动子及结构域的分析
从基因组注释文件中提取基因结构信息,并使用TBtools 对基因外显子-内含子结构进行可视化分析。使用TBtools 软件提取7 个核桃CM 基因的启动子序列(ATG 起始密码上游2 kb),并将序列提交至PlantCARE 数据库(http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)[17]进行顺式作用元件预测,结果的可视化分析使用TBtools[18]进行。
1.5 CM蛋白序列比对及系统进化树构建
利用CLUSTALW 软件对CM 基因编码蛋白序列进行比对,使用ESPript 3 expript 查看比对结果。其余CM家族蛋白序列从PLAZA数据库(https://bioinformatics.psb.ugent.be/plaza/versions/plaza_v4_dicots/)获取。使用MEGA11 软件进行系统进化分析,构建采用邻接法(neighbor-joining,NJ),Poisson model 模型,Bootstrap 值设为1000。进化树的美化采用Evolview(http://www.evolgenius.info/evolview/#/treeview)进行。
1.6 核桃CM基因的组织表达特异性分析
使用Oligo Architect Online 在线工具设计CM基因荧光定量引物,内参基因选择Walnut 18S,引物序列详细信息见表1。使用RNAprep Pure 试剂盒(天根)提取组织总RNA,实时定量反应以反转录的cDNA 为模板,使用赛默飞7500 实时荧光定量PCR 仪进行。实时定量PCR 的反应程序为:95 ℃10 min;95 ℃15 s;55 ℃30 s;72 ℃15 s,40 个循环。反应结束后,使用2-ΔΔCt法[19]计算CM基因的相对表达量。
表1 基因荧光定量引物信息
Table 1 Sequence information of the primers used for qRT-PCR

基因Gene JrCM1 JrCM2 JrCM3 JrCM4 JrCM5 JrCM6 JrCM7 Walnut 18S上游引物Forward primer(5’-3’)GAGCCATTGTTGCCACCTT TGAAGAAGAGAGTTGCGAAGAAG TTCTTCATCCTCCTCGTCTCC AAGATTGGTTGAGGAAGGAGATG CCTGGTGATGATGGCAATTATG AGGCTGCCATTAGAGTACAGGA AGGCTGCCATTAGAGTACAGGA GGTCAATCTTCTCGTTCCCTT下游引物Reverse primer(5’-3’)ATCCGCAGTTGCCATCATC AATCAAGACGGTGTAGGAGGTA AGAGTGCCTTATGCCGTCTA TCTGATGGCGAGTTCATAGGTA CTCCTCCACACTCTCAACTG CGATTGCTGCTTCCACTGTT TTCGCAGCAAGTATTCAACCT TCGCATTTCGCTACGTTCTT
2 结果与分析
2.1 核桃CM基因家族鉴定及特征分析
通过HMM搜索、序列比对和结构域确认,最终选定7条核桃CM家族基因序列,7条序列的理化性质见表2。核桃CM 家族基因对应编码的氨基酸数为258~367 个,蛋白分子质量为29.26~41.74 ku,理论等电点为5.72~8.64,不稳定系数为47.67~57.64(>40),表明7个CM蛋白均为不稳定性蛋白;其平均亲/疏水性均小于0,故均为亲水性蛋白;除JrCM5外其余成员均无信号肽,JrCM5 的信号肽切割位点位于第24 和25 个氨基酸残基之间。染色体定位分析(表2、图1)显示7个成员分布在6条染色体上,其中JrCM1、JrCM2、JrCM3、JrCM4、JrCM5 五个基因均分布于不同的染色体上;JrCM6 和JrCM7 两个基因均位于16号染色体上。亚细胞定位预测显示,核桃CM蛋白均定位于叶绿体中。
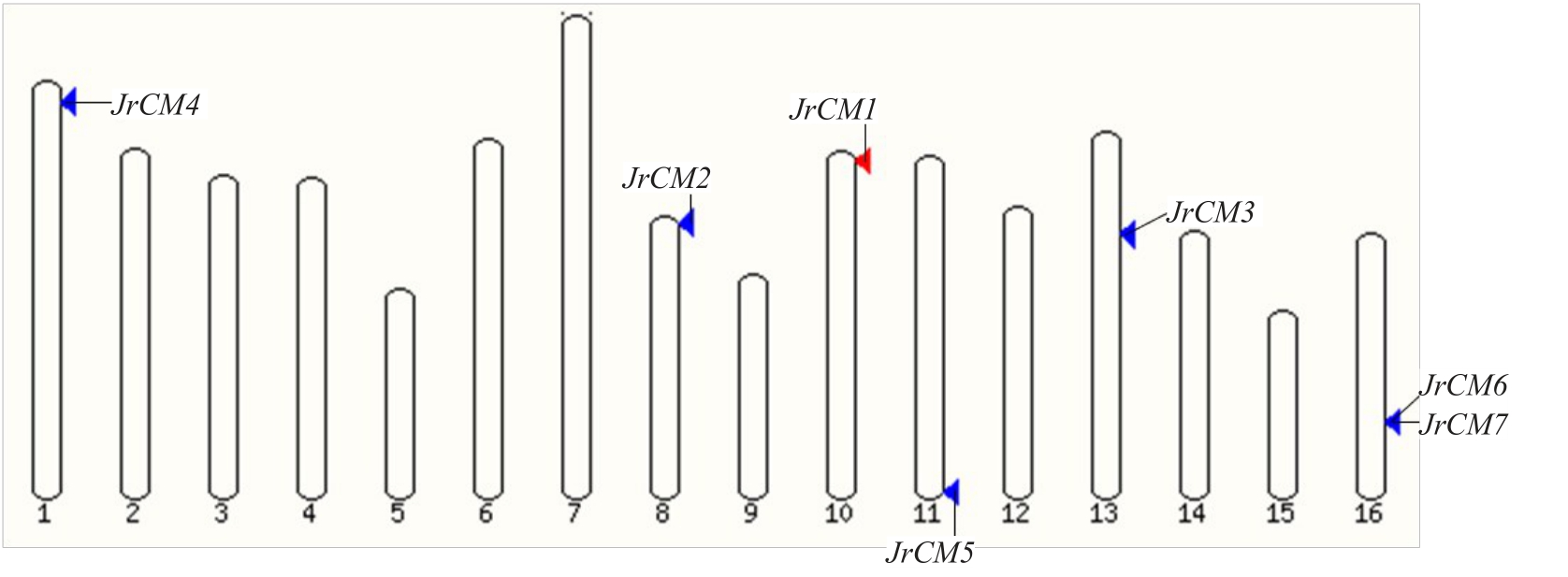
图1 核桃CM 家族基因的染色体定位
Fig.1 Chromosome location of CM family genes in walnut
表2 核桃CM 家族蛋白的理化性质
Table 2 Physicochemical properties of walnut CM family proteins

基因名称Gene name JrCM1基因ID Gene ID染色体定位Chromosome location等电点Isoelectric point Jr10_01790基因长度Gene Length/bp 1327编码区长度Length of CDS/bp 954分子质量Molecular mass/ku 36.06 5.64氨基酸数量Number of amino acids/aa 318不稳定系数Coefficient of instability 47.67亲/疏水性Hydrophilicity/hydrophobicity-0.305信号肽Signal peptide无No JrCM2 Jr08_00830 Chr.10(+):1 068 413-1 072 381 Chr.8(-):718 168-720 7221279 774 29.26 5.82 258 49.71-0.250无No JrCM3 Jr13_14800 1290 960 36.36 7.62 320 57.64-0.348无No JrCM4 Jr01_03110 1561 1101 41.74 5.83 367 51.19-0.213无No JrCM5 Jr11_30250 1228 840 31.64 8.45 280 52.93-0.163 JrCM6 Jr16_12450 972 969 36.77 8.4 323 51.86-0.378 AMALN无No JrCM7 Jr16_12460 Chr.13(+):11 172 097-11 176 153 Chr.1(+):2 236 541-2 240 708 Chr.11(+):36 459 446-36 461 883 Chr.16(-):20 451 797-20 456 245 Chr.16(-):20 451 797-20 456 245 930 900 34.05 8.16 300 51.43-0.408无No亚细胞定位预测Subcellular localization predicted叶绿体Chloroplast叶绿体Chloroplast叶绿体Chloroplast叶绿体Chloroplast叶绿体Chloroplast叶绿体Chloroplast叶绿体Chloroplast
对跨膜结构的预测显示,核桃CM 家族蛋白中仅JrCM4 蛋白N 端含有1 个跨膜结构域,其余各成员均无跨膜结构。蛋白质三级结构预测结果显示(图2),CM 家族成员具有与模板蛋白(拟南芥AtCM1蛋白4ppu.1.A)相似的结构,核桃CM家族蛋白与模板蛋白的序列相似度总体较高。其中JrCM5和JrCM2与AtCM1的相似度最低,分别为53.10%和55.05%;其余序列相似度均在68%以上。

图2 核桃CM 家族蛋白的三级结构
Fig.2 Tertiary structure of walnut CM family proteins
2.2 核桃CM基因启动子顺势作用原件、基因结构及结构域分析
通过分析7 个CM 家族基因的启动子区域(图3),发现除典型的CAAT-box 和TATA-box 外,还包含光响应(AT1-motif、G-box、GA-motif、Gap-box、AE-box 等)、激素响应(ABRE、CGTCA-motif、TGA-element、TGACG-motif、as-1 等)、胁迫响应(AAGAA- motif、LTR、ARE、STRE、WUN- motif等)、转录因子结合位点(AC-I、MYB、W box、MYC、AP-1 等)、生长和发育响应(CAT-box、MSAlike、GCN4-motif、RY-element、AT-rich element 等)、昼夜节律调控(circadian)等7 大类,296 个顺式作用元件,部分元件的功能不明(如A-box 和BoxⅢ)。以7 个JrCM 基因的元件总数计,MYB 转录因子结合位点的数量最多(表3),其次为ERE 乙烯响应原件,但同一元件在不同JrCM 基因启动子中的数量不一。此外,供MYC 转录因子结合(MYC)、光响应调节(Box 4、G-box)、激素响应(ABRE、O2-site、TCA-element)和胁迫响应(ARE、STRE)的相关元件数量也较多。

图3 核桃CM 家族基因的启动子中的响应元件
Fig.3 Response elements in the promoter of walnut CM family genes
表3 核桃CM 基因启动子优势顺势作用原件统计
Table 3 Number of cis-acting motifs that predicted to be abundant in CM promoters
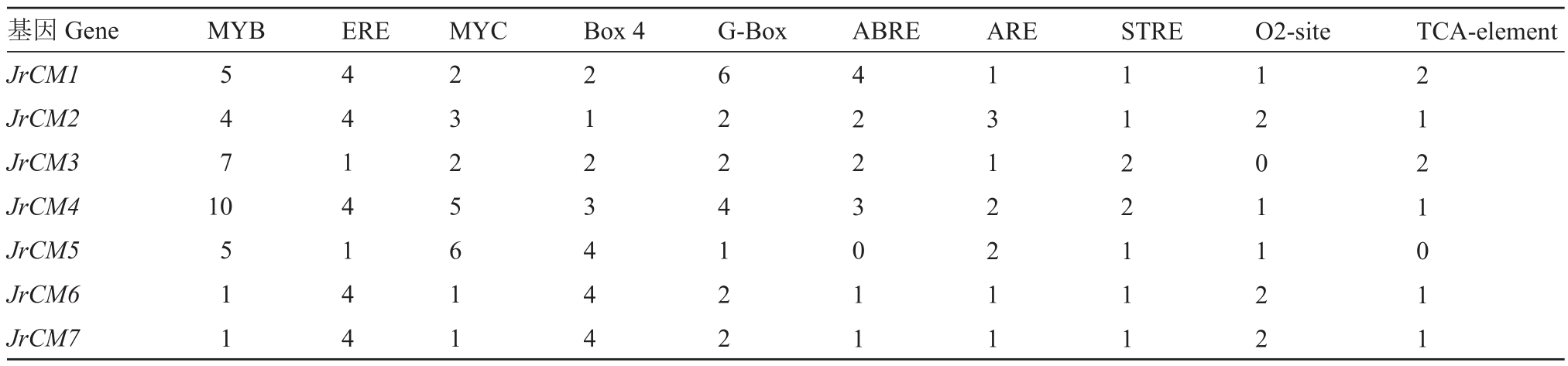
基因Gene JrCM1 JrCM2 JrCM3 JrCM4 JrCM5 JrCM6 JrCM7 MYB ERE MYC Box 4 G-Box ABRE ARE STRE O2-site TCA-element 5 4 7 10 5 1 1 4 4 1 4 1 4 4 2 3 2 5 6 1 1 2 1 2 3 4 4 4 6 2 2 4 1 2 2 4 2 2 3 0 1 1 1 3 1 2 2 1 1 1 1 2 2 1 1 1 1 2 0 1 1 2 2 2 1 2 1 0 1 1
基因结构分析显示(图4),核桃CM家族基因的外显子数量变化不大,均为5~6 个。各基因长度差异主要是内含子的长度差异较大导致的。
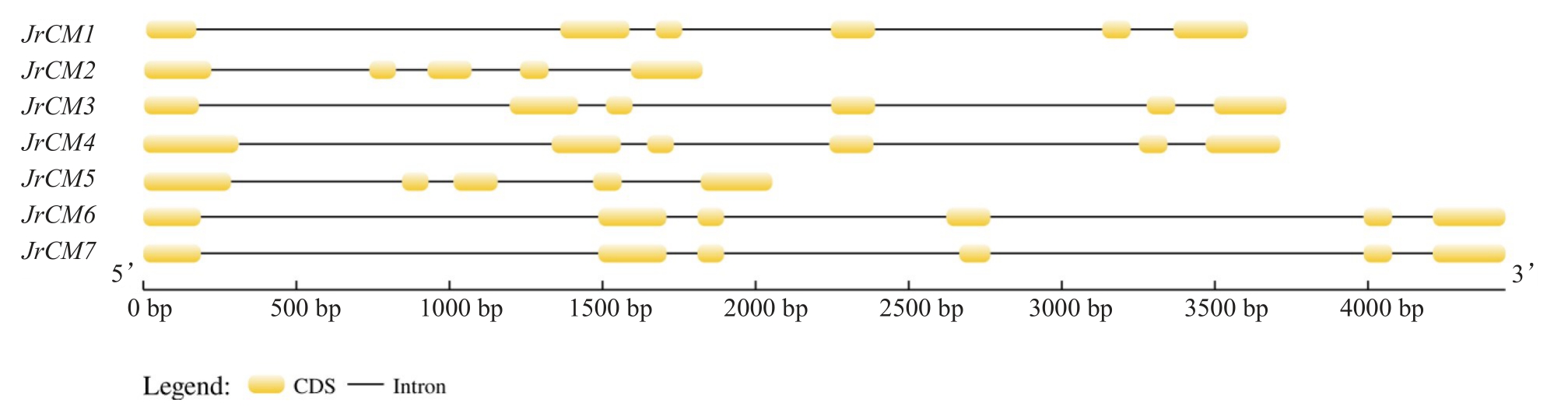
图4 核桃CM 家族基因结构分析
Fig.4 Gene structure analysis of walnut CM family genes
分析7个核桃CM的保守基序和保守结构域,结果显示核桃CM家族基因中共得到10种保守基序;7个核桃CM 蛋白均含有该家族特有的CM_2 结构域,且PLN02344 和CM-plyst 亚结构域完整(图5)。结合基序分析结果可以看出,CM_2 的特征性结构域主要由Motif 1、2、3、4 所构成(图6),特别是Jr-CM2仅含有上述4个基序,且上述4个基序在所有7个核桃CM家族蛋白中都是保守的。部分基序具有一定的特异性,如Motif 8仅存在于JrCM3和JrCM5中;Motif 9 仅存在于JrCM3、JrCM1 和JrCM4 中,而Motif 10(289~293 bp)仅存在于JrCM7、JrCM6和Jr-CM3中。

图5 核桃CM 家族蛋白的结构域
Fig.5 Structural domain of walnut CM family proteins

图6 核桃CM 家族蛋白的保守基序
Fig.6 Conserved motifs of walnut CM family proteins
A.CM 蛋白序列的保守基序;B.保守结构域的序列保守性。
A.Conserved motifs of CM protein sequences were analyzed.B.The conservation of the sequences for each conserved domain was also presented.
2.3 核桃CM 家族蛋白的多序列比对及系统进化分析
对7 个核桃CM 家族蛋白进行多序列比对发现,该家族成员间具有较高的序列相似度和同源性(图7)。系统进化分析表明(图8)CM 家族蛋白可分为4 个亚组,源自核桃的7 个CM 蛋白主要分布在Ⅱ和Ⅳ两个较大的支系上。其中JrCM2 和Jr-CM5 聚集在第Ⅱ支系,与胡杨(Populus trichocar-pa)CM 同源蛋白Potri.T174200 的亲缘关系较近;JrCM6、JrCM7、JrCM4、JrCM1 及JrCM3 聚集在第Ⅳ支系上,其中JrCM6、JrCM7 和JrCM4 与葡萄CM 同源蛋白GSVIVG01010091001 的亲缘关系最近;而JrCM1 和JrCM3 则与蝴蝶兰(Phalaenopsis aphrodite)CM 同源蛋白PEQU04269 的亲缘关系最近。
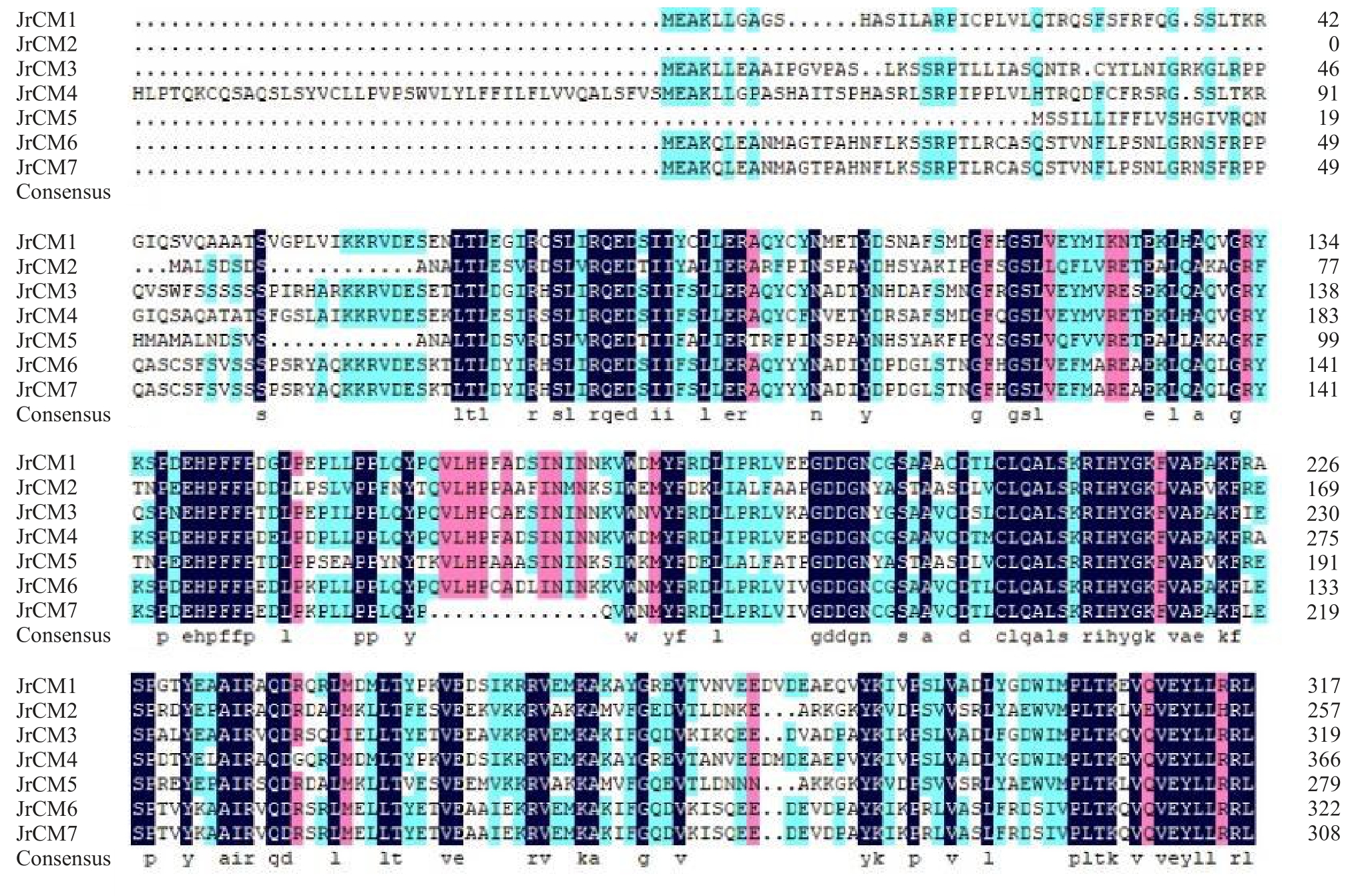
图7 核桃CM 家族蛋白的多序列比对
Fig.7 Multi sequence alignment of walnut CM family proteins

图8 核桃CM 家族基因的系统进化关系
Fig.8 Phylogenetic relationship of walnut CM family genes
2.4 CM家族基因的组织特异性表达分析
笔者分析了该家族7 个基因在不同组织(叶、茎、果柄、青皮)中的表达量(图9),结果显示:该家族基因在各组织中均有表达,但表达模式存在差异。JrCM4 在果柄中的表达量最高,显著高于叶和青皮;JrCM2在果柄中的转录水平较高,显著高于其他3个组织,其他各组织间不存在显著差异;JrCM1在茎和果柄中的表达量较高,显著高于叶和青皮;JrCM5在叶片中的表达量最高,显著高于茎,茎显著高于果柄和青皮;JrCM3 的表达量在各组织间不存在显著差异;JrCM6 在叶和茎中的表达量显著高于果柄和青皮;JrCM7在茎中的表达量最高,显著高于其他3个组织。上述结果说明:核桃CM基因在各个组织中均有不同程度的表达,预示各成员在功能上可能存在分化和差异。

图9 CM 家族基因在核桃不同组织中的表达量
Fig.9 Expression of CM family genes in different tissues of Walnut
3 讨 论
莽草酸途径具有联系植物初级代谢和次生代谢的枢纽作用,对次生代谢物丰富的核桃(如青皮)来说具有重要意义。多酚类物质的生物合成是莽草酸途径下游的一个分支,迄今大量的研究只关注与多酚生物合成紧密相连的苯丙烷代谢途径和类黄酮代谢途径,对其碳供应链上游的莽草酸途径报道较少[4]。核桃作为经济林树种,坚果富含多种氨基酸、多酚及高抗氧化性的次生代谢物,开展莽草酸途径关键酶的研究对提升核桃品质具有重要意义。目前关于CM的研究多集中于微生物和植物中,如大肠杆菌(Escherichia coli)[20]、结核分枝杆菌(Mycobacterium tuberculosis)[21]、葡 萄[4]、玉 米(Zea mays)[22]、拟南芥[10]、龙须菜(Gracilariopsis lemaneiformis)[23]等。较为有趣的是,CM 不仅是动物病原的有效靶点,在近期关于植物病原与宿主的相关作用研究中发现,核盘菌[Sclerotinia sclerotiorum(Lib.)de Bary]分支酸变位酶SsCm1 可直接与大豆(Glycine max)CM 蛋白相互作用,抑制水杨酸的合成[24]。与此类似,黑粉菌(Ustilago maydis)的分支酸变位酶可通过与宿主CM蛋白间形成二聚体降低胞内水杨酸水平,进而达到抑制宿主免疫的目的[25]。目前,关于核桃CM 基因的研究尚属空白,笔者在本研究中基于核桃的全基因组测序数据,共鉴定出7个核桃CM家族基因,并对其进行了生物信息学分析。7个CM家族蛋白中,仅JrCM5 含有信号肽,这与矮牵牛PhCM1、丹参(Salvia miltiorrhiza)SmCM1 的结构特征一致[6],表明JrCM5 可能为外泌蛋白。亚细胞定位预测结果显示,核桃CM蛋白均定位于叶绿体中,这与其他报道中分支酸变位酶发挥作用的部位较为一致。结合现有报道所阐释的CM 同源蛋白的功能,可以推测出核桃CM 蛋白不仅直接参与了重要次生代谢产物的合成,且极有可能也是病原-宿主相互作用中的重要靶点。病原可能通过操纵核桃分支酸变位酶干扰植株水杨酸合成,进而影响其防御反应[26-27]。
核桃CM基因启动子区域含有光、激素、逆境胁迫以及生长和发育、转录因子结合、昼夜节律调控等多种顺式作用元件,说明影响核桃CM 家族基因转录的因素是多样的,但转录因子(MYB、MYC)、光照、逆境胁迫(缺氧、干旱)、ABA(ABRE)、赤霉素(O2-site)、水杨酸(TCA-element)可能是调控JrCM表达的主要因素,这与其他植物中已报道的CM 基因易受逆境、光照度、紫外照射等影响和调控的结论较为一致[28-29]。李小溪[4]的研究证实环境因子可诱导MYB 转录因子表达,促进其与莽草酸途径相关基因启动子顺式作用原件的结合,从而调控转录和莽草酸-类黄酮途径代谢产物的积累。
核桃青皮富含多酚类物质,具有较强的抑菌、抗氧化活性,核桃1 年生枝条和叶片的酚类物质含量在6—7月份呈现上升状态[13],笔者在本研究中选择7 月份核桃生长关键时期对核桃各组织进行采样,研究JrCM 家族基因的组织特异性表达,结果显示该家族基因在各组织中均有表达,但表达模式存在差异。拟南芥的AtCM1 和AtCM3 基因在花器中表达最高,但在根和茎中表达量较低,活性受芳香族氨基酸的调节;AtCM2 基因在根中的表达量最高,其次是茎和叶,其活性基本不受芳香族氨基酸的影响[9]。丹参SmCM1在茎和叶中的表达量分别为根中1.8 和2.8 倍,并且可受酵母和银离子的联合诱导而上调表达[6],进而大量合成苯丙氨酸和酪氨酸,最终提高体内酚酸类化合物的含量。葡萄VvCM1 和VvCM2 在果实中不同部位随发育期表达具有特异性,VvCM1 在果实中丰度最高,而VvCM2 在茎中表达量最高,葡萄CM 同源基因的差异表达可能与光合同化产物通过莽草酸途径大量流向原花色素、酚酸和木质素的合成有关[30]。
核桃CM家族基因在不同组织中的表达具有普遍性,这与CM 的“枢纽”作用有关。核桃中含有丰富的酚类物质,是苯丙烷代谢和类黄酮代谢途径的产物或衍生物,均与莽草酸代谢途径的关系密切。了解分支酸变位酶家族基因及其表达特性,有助于更好地理解核桃代谢产物(各类氨基酸、酚类和抗氧化活性物质)的生成,但次生代谢产物对CM活性的影响及其调节机制仍需进一步深入研究。鉴于CM在植物-病原相互作用中的重要作用,分析CM在核桃抗病中的功能也是值得探索的方向。
4 结 论
笔者在本研究中鉴定得到了7 个核桃CM 家族基因,分别为JrCM1~7,均为不稳定的亲水性蛋白。其中JrCM4、JrCM2 和JrCM1 的pH 小于7,显酸性,其余4 个显碱性。7 个基因分布在6 条染色体上,其中JrCM6 和JrCM7 定位在同一条染色体上,且两者的理化性质、亲缘关系及相似度均较高,故这2 个基因可能具有更为相似的生物学功能。分析各基因的启动子发现,核桃CM 的转录受控于7 个大类多种不同类型的顺式作用原件,其中转录因子(MYB、MYC)、光照、激素(ABA、赤霉素、水杨酸)和逆境胁迫(缺氧、干旱)可能是调控JrCM 表达的关键因素。表达分析结果显示,JrCM 在核桃的不同组织中表达量各不相同,显示具有组织特异性。系统进化分析结果表明,植物CM 蛋白可分为4 个大组,其中核桃CM 蛋白主要聚集在第Ⅱ和Ⅳ分支上。本研究结果丰富和深化了对核桃CM 家族基因的认识,为进一步开展基因功能研究奠定了基础。
[1]RUAN J,HAERDTER R,GERENDÁS J.Impact of nitrogen supply on carbon/nitrogen allocation:A case study on amino acids and catechins in green tea [Camellia sinensis (L.) O.Kuntze]plants[J].Plant Biology,2010,12(5):724-734.
[2]CHEN K,DOU J,TANG S R,YANG Y S,WANG H,FANG H Q,ZHOU C L.Deletion of the aroK gene is essential for high shikimic acid accumulation through the shikimate pathway in E.coli[J].Bioresource Technology,2012,119:141-147.
[3]OSBORNE A,THORNELEY R N F,ABELL C,BORNEMANN S.Studies with substrate and cofactor analogues provide evidence for a radical mechanism in the chorismate synthase reaction[J].Journal of Biological Chemistry,2000,275(46):35825-35830.
[4]李小溪.葡萄果实莽草酸途径和类黄酮代谢协同调节机制的研究[D].北京:中国农业大学,2016.LI Xiaoxi.Collaborative expression mechanism between shikimate pathway and flavonoid metabolism[D].Beijing:China Agricultural University,2016.
[5]ZHANG Z Z,LI X X,CHU Y N,ZHANG M X,WEN Y Q,DUAN C Q,PAN Q H.Three types of ultraviolet irradiation differentially promote expression of shikimate pathway genes and production of anthocyanins in grape berries[J].Plant Physiology and Biochemistry,2012,57:74-83.
[6]王亚君,黄璐琦,蒋超,申业.丹参分支酸变位酶基因的克隆和生物信息学分析[J].中国中药杂志,2013,38(11):1697-1702.WANG Yajun,HUANG Luqi,JIANG Chao,SHEN Ye.Cloning and bioinformatics analysis of chorismate mutase gene from Salvia miltiorrhiza[J].China Journal of Chinese Materia Medica,2013,38(11):1697-1702.
[7]CHÁVEZ-BÉJAR M I,LARA A R,LÓPEZ H,HERNÁNDEZCHÁVEZ G,MARTINEZ A,RAMÍREZ O T,BOLÍVAR F,GOSSET G.Metabolic engineering of Escherichia coli for L-tyrosine production by expression of genes coding for the chorismate mutase domain of the native chorismate mutase- prephenate dehydratase and a cyclohexadienyl dehydrogenase from Zymomonas mobilis[J].Applied and Environmental Microbiology,2008,74(10):3284-3290.
[8]SCHNEIDER C Z,PARISH T,BASSO L A,SANTOS D S.The two chorismate mutases from both Mycobacterium tuberculosis and Mycobacterium smegmatis:Biochemical analysis and limited regulation of promoter activity by aromatic amino acids[J].Journal of Bacteriology,2008,190(1):122-134.
[9]MOBLEY E M,KUNKEL B N,KEITH B.Identification,characterization and comparative analysis of a novel chorismate mutase gene in Arabidopsis thaliana[J].Gene,1999,240(1):115-123.
[10]WESTFALL C S,XU A,JEZ J M.Structural evolution of differential amino acid effector regulation in plant chorismate mutases[J].The Journal of Biological Chemistry,2014,289(41):28619-28628.
[11]COLQUHOUN T A,SCHIMMEL B C J,KIM J Y,REINHARDT D,CLINE K,CLARK D G.A petunia chorismate mutase specialized for the production of floral volatiles[J].The Plant Journal,2010,61(1):145-155.
[12]刘畅,周家春.植物多酚抗氧化性研究[J].粮食与油脂,2011,24(2):43-46.LIU Chang,ZHOU Jiachun.Research on antioxidation of plant polyphenols[J].Cereals&Oils,2011,24(2):43-46.
[13]周晔,王伟,王成章,裴东.核桃属(Juglans)植物多酚类物质研究进展[J].南京林业大学学报(自然科学版),2013,37(5):146-152.ZHOU Ye,WANG Wei,WANG Chengzhang,PEI Dong.Research progress on polyphenols from Juglans plants[J].Journal of Nanjing Forestry University (Natural Sciences Edition),2013,37(5):146-152.
[14]MCCOY R M,UTTURKAR S M,CROOK J W,THIMMAPURAM J,WIDHALM J R.The origin and biosynthesis of the naphthalenoid moiety of juglone in black walnut[J].Horticul-ture Research,2018,5:67.
[15]MARTÍNEZ-GARCÍA P J,CREPEAU M W,PUIU D,GONZALEZ-IBEAS D,WHALEN J,STEVENS KA,PAUL R,BUTTERFIELD T S,BRITTON M T,REAGAN R L,CHAKRABORTY S,WALAWAGE S L,VASQUEZ-GROSS H A,CARDENO C,FAMULA R A,PRATT K,KURUGANTI S,ARADHYA M K,LESLIE C A,DANDEKAR A M,SALZBERG S L,WEGRZYN J L,LANGLEY C H,NEALE D B.The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols[J].The Plant Journal,2016,87(5):507-532.
[16]刘小利,魏海斌,顾文毅,廖东.五个早实薄皮核桃品种的区域引种试验[J].北方园艺,2017(22):67-70.LIU Xiaoli,WEI Haibin,GU Wenyi,LIAO Dong.Regional introduction experiment of five early walnut varieties[J].Northern Horticulture,2017(22):67-70.
[17]LESCOT M,DÉHAIS P,THIJS G,MARCHAL K,MOREAU Y,VAN DE PEER Y,ROUZÉ P,ROMBAUTS S.PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J].Nucleic Acids Research,2002,30(1):325-327.
[18]CHEN C J,CHEN H,ZHANG Y,THOMAS H R,FRANK M H,HE Y H,XIA R.TBtools:An integrative toolkit developed for interactive analyses of big biological data[J].Molecular Plant,2020,13(8):1194-1202.
[19]VANDESOMPELE J,DE PRETER K,PATTYN F,POPPE B,VAN ROY N,DE PAEPE A,SPELEMAN F.Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes[J].Genome Biology,2002,3(7):0034.
[20]钱晋.大肠杆菌莽草酸代谢途径的相关研究[D].上海:华东理工大学,2011.QIAN Jin.The study of shikimic acid pathway of Escherichia coli[D].Shanghai:East China University of Science and Technology,2011.
[21]FONSECA I O,SILVA R G,FERNANDES C L,DE SOUZA O N,BASSO L A,SANTOS D S.Kinetic and chemical mechanisms of shikimate dehydrogenase from Mycobacterium tuberculosis[J].Archives of Biochemistry and Biophysics,2007,457(2):123-133.
[22]SODERLUND C,DESCOUR A,KUDRNA D,BOMHOFF M,BOYD L,CURRIE J,ANGELOVA A,COLLURA K,WISSOTSKI M,ASHLEY E,MORROW D,FERNANDES J,WALBOT V,YU Y.Sequencing,mapping,and analysis of 27,455 maize full- length cDNAs[J].PLoS Genetics,2009,5(11):e1000740.
[23]林丽春,吕燕,孙鹏,孙雪,徐年军.龙须菜(Gracilariopsis lemaneiformis)中两种分支酸代谢酶对温度和水杨酸的响应及其原核表达研究[J].海洋与湖沼,2019,50(1):220-227.LIN Lichun,LÜ Yan,SUN Peng,SUN Xue,XU Nianjun.Responses of two chotismate metabolic enzymes to temperature and salicylic acid in gracilariopsis lemaneiformis and their prokaryotic expression analysis[J].Oceanologia et Limnologia Sinica,2019,50(1):220-227.
[24]杨凤.核盘菌分支酸变位酶SsCm1 的作用机制研究[D].长春:吉林大学,2021.YANG Feng.Pathogenic mechanism of chorismate mutase Ss-Cm1 in the Sclerotinia sclerotiorum[D].Changchun:Jilin University,2021.
[25]HAN X W,ALTEGOER F,STEINCHEN W,BINNEBESEL L,SCHUHMACHER J,GLATTER T,GIAMMARINARO P I,DJAMEI A,RENSING S A,REISSMANN S,KAHMANN R,BANGE G.A kiwellin disarms the metabolic activity of a secreted fungal virulence factor[J].Nature,2019,565(7741):650-653.
[26]刘耀,何其光,廖小淼,徐良向,刘文波,林春花,李潇,郑服丛,缪卫国.橡胶树白粉菌分支酸变位酶基因克隆及蛋白表达纯化[J].基因组学与应用生物学,2020,39(3):1120-1127.LIU Yao,HE Qiguang,LIAO Xiaomiao,XU Liangxiang,LIU Wenbo,LIN Chunhua,LI Xiao,ZHENG Fucong,MIAO Weiguo.Cloning,prokaryotic expression and purification of chorismate mutase from Oidium heveae[J].Genomics and Applied Biology,2020,39(3):1120-1127.
[27]盛寅生,任爱芝,常雪,赵培宝.核盘菌一分支酸变位酶同源效应蛋白的亚细胞定位[J].植物保护,2018,44(5):176-180.SHENG Yinsheng,REN Aizhi,CHANG Xue,ZHAO Peibao.Subcellular localization of an effector homologous with chorismate mutase in Sclerotinia sclerotiorum[J].Plant Protection,2018,44(5):176-180.
[28]李小溪,问亚琴,李春兰,潘秋红.低温对葡萄果实莽草酸途径入口酶及后分支酸途径关键酶基因表达的影响[J].热带生物学报,2011,2(3):250-255.LI Xiaoxi,WEN Yaqin,LI Chunlan,PAN Qiuhong.Effects of low temperature on expression of genes encoding entrance enzyme of grape shikimate pathway and key enzymes of post chorimate pathway[J].Journal of Tropical Organisms,2011,2(3):250-255.
[29]初英娜,张珍珍,潘秋红.紫外照射对葡萄果实莽草酸途径相关基因表达的影响[J].植物生理学通讯,2010,46(9):902-908.CHU Yingna,ZHANG Zhenzhen,PAN Qiuhong.Effects of ultraviolet radiation on expression of shikimate pathway-related genes in grape (Vitis vinifera L.) berries[J].Plant Physiology Communications,2010,46(9):902-908.
[30]张晨,陈灵婧,李小溪,王军.葡萄果实分支酸变位酶基因的克隆与表达分析[J].中外葡萄与葡萄酒,2014(1):6-11.ZHANG Chen,CHEN Lingjing,LI Xiaoxi,WANG Jun.Cloning and expression of gene encoding chorismate mutase in grape berries[J].Sino-Overseas Grapevine&Wine,2014(1):6-11.