塞外红苹果又名鸡心果,是由通辽市林业和草原科学研究所王宝侠研究员团队历经20多年自主选育出的小苹果新品种,是内蒙古自治区首个通过中国森林认证的经济林树种。因为塞外红苹果具有适应性强、丰产性好、抗逆性强、果实色泽艳丽、口味香甜、耐贮藏等特性,已经发展成为内蒙古自治区种植面积最大的苹果品种之一。目前,在内蒙古自治区通辽市种植面积约为2 万hm2,成为当地果农的主要收入来源。另外,在辽宁、河北、山东、甘肃等省也有引种。
苹果褪绿叶斑病毒(apple chlorotic leaf spot virus,ACLSV)、苹果茎痘病毒(apple stem pitting virus,ASPV)和苹果茎沟病毒(apple stem grooving virus,ASGV)是苹果上发生率最高的3 种病毒,在世界范围内广泛发生[1]。1959 年,Mink 等[2]等首次报道了苹果上发生ACLSV。随后发现ACLSV还侵染梨、桃、山楂等果树[3-5]。1954年,Smith[6]在出现茎痘斑和坏死症状的苹果砧木上发现了ASPV,近年发现ASPV 在梨、山楂、樱桃、枇杷等果树上都会发生[7-9]。Lister 等[10]于1965 年最先在苹果上发现ASGV,伴随着RT-PCR 和siRNA 高通量测序技术的应用,ASGV的寄主范围从蔷薇科果树上扩展到百合、月季、竹等观赏植物[11-14]。这3种苹果病毒单独侵染时,在大多数商业苹果品种上不引起明显症状,但复合侵染时可以引起苹果树“高接病”、树势衰退、果实畸形、皱缩等,是苹果无病毒苗木生产中需要脱除的对象。伴随着塞外红苹果种植面积的不断扩大,病毒侵染导致苹果品质下降已成为制约塞外红苹果产业发展的重要因素,开展3 种病毒的检测和分子变异研究,对塞外红苹果产业发展有重要意义,也将为无病毒苗木生产提供指导数据。
笔者在本研究中采用RT-PCR对采自内蒙古自治区通辽市8个乡镇的96份塞外红苹果样品进行了检测及分析,明确了通辽市塞外红苹果中3 种苹果潜隐病毒的发生情况和遗传多样性。
1 材料和方法
1.1 供试材料
2021 年2 月25 日在开鲁县的开鲁镇、大榆树镇、建华镇、小街基镇、东风镇、东来镇、麦新镇、吉日嘎郎吐镇等8个乡镇采集了96份塞外红苹果枝条样品(每个乡镇随机采集12份),选择15~20 cm长的一年生枝条,用石蜡封口,置于4 ℃冰箱保存备用。
1.2 主要试剂及实验仪器
1.2.1 主要试剂 TaKaRa MiniBEST plant RNA Extraction Kit[宝日医生物技术(北京)有限公司]、PrimeScript™Ⅱ1st Strand cDNA Synthesis Kit[宝日医生物技术(北京)有限公司]、M5 超光速mix(北京聚合美生物科技有限公司)、琼脂糖凝胶DNA 回收试剂盒[天根生化科技(北京)有限公司]、零背景pTOPO-TA 克隆试剂盒(北京艾德莱生物科技有限公司)、E.coli JM109 感受态细胞[宝日医生物技术(北京)有限公司]等;具体实验操作步骤根据试剂盒说明书完成。
1.2.2 主要实验仪器 生工Sangon Biotech 台式高速微量离心机(Hico21)、电子天平、海尔医用低温保存箱、Scllogex sloloe离心机、涡轮振荡机、卧式冷藏冷冻转换柜、恒温水浴锅、微波炉、Allsheng 微量分光光度计(Nano-300)、微型涡旋混合仪、流水式制冰机、电热恒温培养箱、BIO-RAD Thermal Cycler、移液枪、DLAB数显恒温金属浴。
1.3 方法
1.3.1 植物总RNA 提取 按照TaKaRa MiniBEST plant RNA Extraction Kit说明书步骤,从植物枝条中提取总RNA。得到的总RNA 使用Allsheng 微量分光光度计(Nano-300)进行浓度和纯度的测定。存放于-20 ℃冰箱备用。
1.3.2 反转录合成cDNA 第一链 以提取的样品总RNA 为模板,使用反转录试剂盒PrimeScript™Ⅱ1st Strand cDNA Synthesis Kit 进行反转录,过程如下:1 μL 随机引物(50 μmoL·L-1)、1 μL dNTP(10 mmoL·L-1)、1.5 μL模板RNA、6.5 μL无酶水,将反应液放入PCR 仪内65 ℃5 min;冰上迅速冷却后继续加入4 μL 5×PrimeScript ⅡBuffer、0.5 μL RNase Inhibitor(40 U·μL- 1)、1 μL PrimeScript ⅡRTase(200 U·μL-1)、4.5 μL无酶水。将上述20 μL反应液轻轻混匀,放入PCR 仪内42 ℃30 min、70 ℃15 min后,取出冷却。反转录产物可以在-20 ℃冰箱保存。
1.3.3 病毒基因组克隆(1)引物设计。根据NCBI的GenBank 数据库中存储的ACLSV、ASGV、ASPV基因序列的保守区域,设计了3对病毒检测引物(表1)。ACLSV1-F/R 用于扩增ACLSV 的部分复制蛋白和部分移动蛋白基因;ASPV1-F/R用于扩增ASPV的部分外壳蛋白基因和3’非编码区;ASGV1-F/R用于扩增ASGV的部分移动蛋白和外壳蛋白基因。
表1 检测塞外红样品中ACLSV、ASGV、ASPV 所用引物
Table 1 Primers used for detection of ACLSV,ASGV and ASPV in Saiwaihong

注:Y.C/T;K.G/T;N.A/G/C/T。
引物名称Primers ACLSV1 F ACLSV1 R ASPV1 F ASPV1 R ASGV1 F ASGV1 R引物序列(5’→3’)Sequences(5’→3’)ATCGCAGAAGGGGATATTCCAAT TGTCKAGCAAACTGAAAYTCTCT GACTTCTTYTTYGGAGTNGARAG GAAAATCTAGTTAAAACAAAAATAAG ATGAGTTTGGAAGACGTGCTTCA CAAAGCTTYCKGAACGTACATTC产物大小Size/bp 1016 344 449退火温度Tm/℃61 61 58 58 58 58
(2)病毒基因片段扩增。以反转录合成的cDNA 为模板,使用2×M5 Hiper 超光速mix 分别扩增ACLSV、ASPV、ASGV 基因片段。扩增体系如下:10 μL 2×M5 Hipor 超光速mix、1 μL 上游引物、1 μL下游引物、5 μL cDNA、3 μL 无酶水。反应条件:95 ℃预变性3 min,35 次循环:94 ℃变性25 s、退火25 s(ACLSV 61 ℃,ASPV 58 ℃,ASGV 58 ℃)、72 ℃延伸(ACLSV 60 s,ASPV 30 s,ASGV 30 s),72 ℃补平5 min。
(3)PCR 产物的纯化、克隆及序列测定。将检测到的ACLSV、ASGV、ASPV 目标片段纯化回收,纯化后的PCR 产物用pTOPO-TA 试剂盒进行连接,连接产物转化大肠杆菌JM109 感受态细胞并涂板。对培养出来的单个菌落进行PCR 菌液鉴定。每个样品随机挑选3个阳性克隆菌液送华大基因公司进行测序。
1.3.4 序列一致性分析及系统发育分析。运用软件Vector NTI Advance 11(Invitrogen Corporation,CA,USA)对测序结果进行分析和拼接;使用SDT 软件CLE序列比对程序将测序基因序列与参考基因序列进行两两比对,参考序列均从NCBI GenBank数据库下载[15]。用MEGA-X 软件构建系统发育进化树,处理序列采用ClustalW 算法进行比对,选用邻接法(neighbor-joining method,NJ)构建系统发育树,自展校正值为1000,其他均为软件默认值[16]。
2 结果与分析
2.1 病毒的检出率
RT-PCR 检测结果显示,96 份塞外红苹果枝条样品中,有29份样品携带ACLSV,检出率为30.2%;有12份样品携带ASPV,检出率为12.5%;有21份样品携带ASGV,检出率为22.0%。通过对每个乡镇的病毒检出率比较,ACLSV检出率最高的地区在开鲁县开鲁镇,为66.7%;ASPV 检出率最高的地区在开鲁县东风镇,为41.7%;ASGV 检出率最高的地区在开鲁县开鲁镇,为58.3%(表2)。在所采集的96份塞外红苹果枝条样品中,有4份样品同时携带ACLSV、ASPV 和ASGV,检出率为4.2%;有4 份样品同时携带ACLSV 和ASPV,检出率为4.2%;有13 份样品同时携带ACLSV和ASGV,检出率为13.5%;有3份样品同时携带ASPV和ASGV,检出率为3.1%(表3)。
表2 开鲁塞外红苹果受单一病毒侵染的情况
Table 2 Occurrence of infection with a single virus in Saiwaihong in Kailu

地点Location开鲁镇Kailu大榆树镇Dayushu建华镇Jianhua小街基镇Xiaojieji东风镇Dongfeng东来镇Donglai麦新镇Maixin吉日嘎郎吐镇Jirigalangtu阳性株数/检测总量Number of diseased plants/total number of samples ACLSV 8/12 4/12 1/12 4/12 3/12 3/12 4/12 2/12 ASPV 1/12 1/12 1/12 0/12 5/12 1/12 2/12 1/12 ASGV 7/12 3/12 3/12 2/12 2/12 1/12 2/12 1/12
表3 开鲁塞外红苹果受复合病毒侵染的情况
Table 3 Occurrence of mixed infection with viruses in Saiwaihong in Kailu
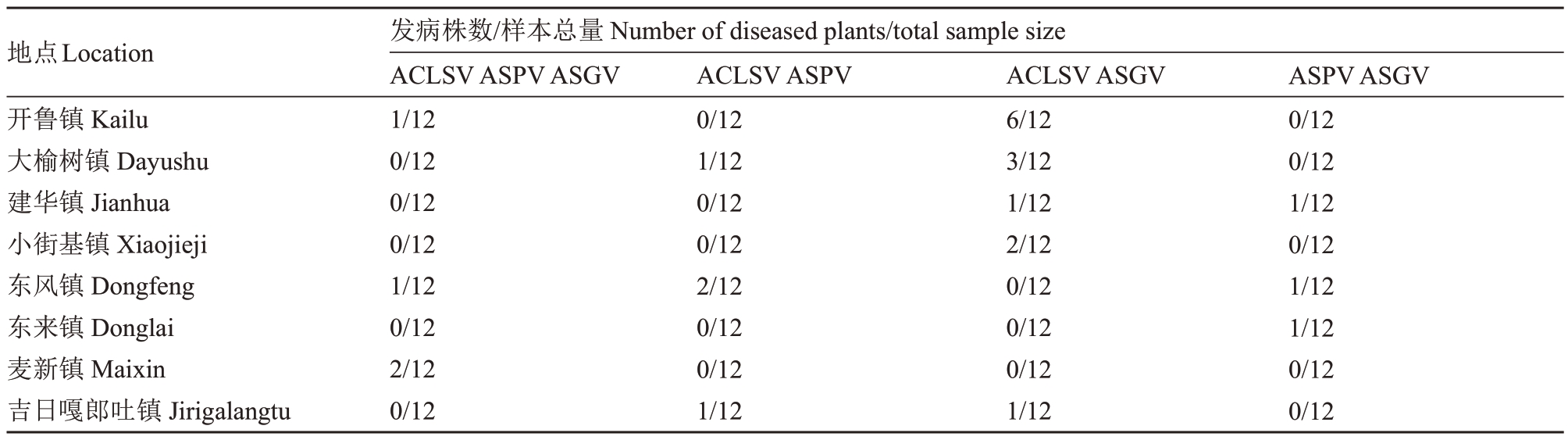
地点Location开鲁镇Kailu大榆树镇Dayushu建华镇Jianhua小街基镇Xiaojieji东风镇Dongfeng东来镇Donglai麦新镇Maixin吉日嘎郎吐镇Jirigalangtu发病株数/样本总量Number of diseased plants/total sample size ACLSV ASPV ASGV 1/12 0/12 0/12 0/12 1/12 0/12 2/12 0/12 ACLSV ASPV 0/12 1/12 0/12 0/12 2/12 0/12 0/12 1/12 ACLSV ASGV 6/12 3/12 1/12 2/12 0/12 0/12 0/12 1/12 ASPV ASGV 0/12 0/12 1/12 0/12 1/12 1/12 0/12 0/12
2.2 序列一致性分析
笔者在本研究中共获得29 个来源于塞外红苹果的ACLSV 分离物(GenBank 登录号:ON001687~ON001715),核苷酸序列一致性为78.1%~99.9%,其中ACLSV Kailu4-8(ON001702)与Kailu5-1(ON001704)的核苷酸序列一致性最高,为99.9%;Kailu3-12(ON001699)与Kailu7-10(ON001713)之间的核苷酸序列一致性最低,为78.1%。该29个分离物与其他15个已知的ACLSV分离物的核苷酸序列一致性为69.1%~93.6%,其中分离物Kailu7-10(ON001713)与来源于中国苹果的分离物XC-HF(MF678819)核苷酸序列一致性最高(93.6%);分离物Kailu1-5(ON001689)与美国桃分离物Ta Tao 5(EU223295)核苷酸序列一致性最低(69.1%)(图1)。根据扩增得到的部分复制蛋白和移动蛋白基因序列构建系统发育树,结果显示,44 个ACLSV 分离物聚集在7 个分支,即JB、MO-5、Balatonl、P863、组1、B6、Ta Tao 5(图2)。来源于塞外红苹果的部分ACLSV分离物(ON001687~689、ON001692、ON001700~704、ON001706~708、ON001713~715)位于JB 组,与中国山楂分离物SY03(KU870525)、苹 果 分 离 物 XC- HF(MF678819)、梨分离物KMS、YH、JB(KC935954、KC935955、KC935956)聚为一支;分离物Kailu3-12(ON001699)与法国樱桃分离物Balaton1(X99752)聚为一支,位于Balatonl组;分离物Kailu6-9、Kailu5-11(ON001709、ON001705)单独形成一个分支,位于1组;其他的分离物(ON001690~691、ON001693~698、ON001710~712)位于B6 组,与日本苹果分离物QD- 13(KJ522693)、中国苹果分离物MS(KC847061)聚为一支。外参为樱桃斑驳叶病毒(cherry mottle leaf virus,CMLV)(NC-002500)。

图1 ACLSV 基因序列两两比对一致性矩阵图
Fig.1 A matrix of pairwise identities among ACLSV gene sequences

图2 来自塞外红苹果的ACLSV 分离物与其他ACLSV 分离物序列构建的系统发育进化树
Fig.2 A phylogenetic tree was constructed based on the sequences of ACLSV isolates from Saiwaihong and the other published ACLSV isolates
笔者在本研究中共获得12 个来源于塞外红苹果的ASPV 分离物(GenBank 登录号:OL953372~OL953383),这12 个分离物间核苷酸序列一致性为83.6%~99.7%,其中来源于塞外红的ASPV 分离物Kailu2-9(OL953373)与Kailu7-12(OL953382)的核苷酸序列一致性最高(99.7%);分离物Kailu2-9(OL953373)、Kailu7- 6(OL953380)与Kailu8- 1(OL953383)之间的核苷酸序列一致性最低(83.6%)。该12个ASPV分离物与其他15个已知的ASPV分离物的核苷酸序列一致性为75.4%~91.5%,其中ASPV 分离物Kailu7-12(OL953382)与加拿大苹果分离物Aurora-1(HE963831)的核苷酸序列一致性最高(91.5% );ASPV 分离物Kailu7- 6(OL953380)与中国梨 ASPV 分离物 PR1(EU095327)的核苷酸序列一致性最低(75.4%)(图3)。根据部分外壳蛋白基因和3’非编码区序列构建系统发育树,27个ASPV分离物聚集为6支,塞外红苹果的ASPV 分离物分别位于组Ⅰ、Ⅳ、Ⅴ(图4)。部分塞外红苹果的ASPV 分离物(OL953372~75、OL953379、OL953381~83)与加拿大苹果分离物Aurora-1(HE963831)聚集在同一个分支上形成组Ⅰ;部分塞外红苹果的ASPV 分离物(OL953379、OL953380)与德国苹果的ASPV 分离物PM8(KF319056)、印度苹果的ASPV 分离物Palampur(FR694186)、日本苹果的ASPV 分离物IF38(AB045371)聚为一支,位于组Ⅳ;其余塞外红苹果的ASPV分离物(OL953378、OL953377)形成单独的一个分支Ⅴ。外参为沙地葡萄茎痘病毒(grapevine rupestris stem pitting- associated virus,GRSPaV)(JX559646)。
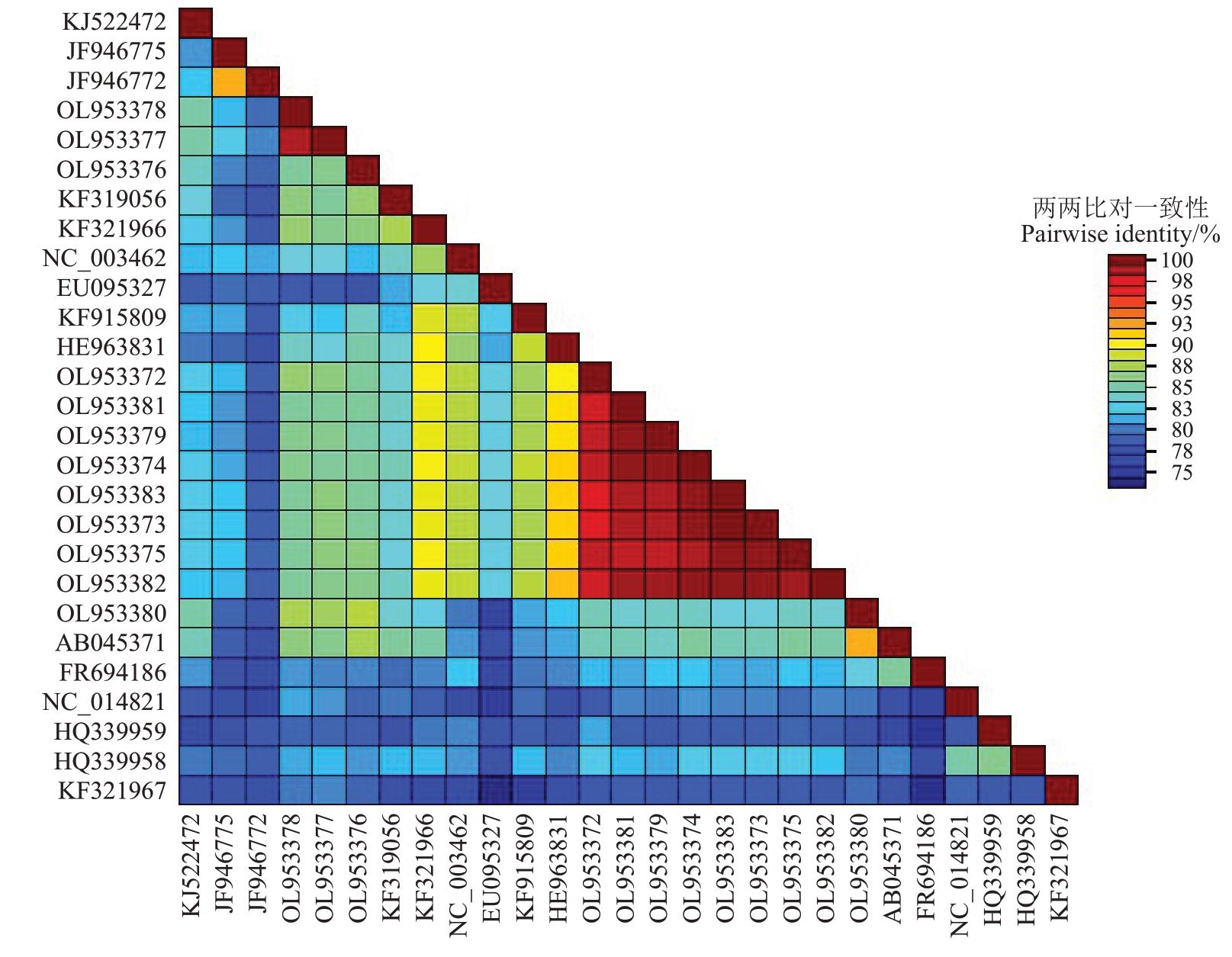
图3 ASPV 基因序列两两比对一致性矩阵图
Fig.3 A matrix of pairwise identities among ASPV gene sequences
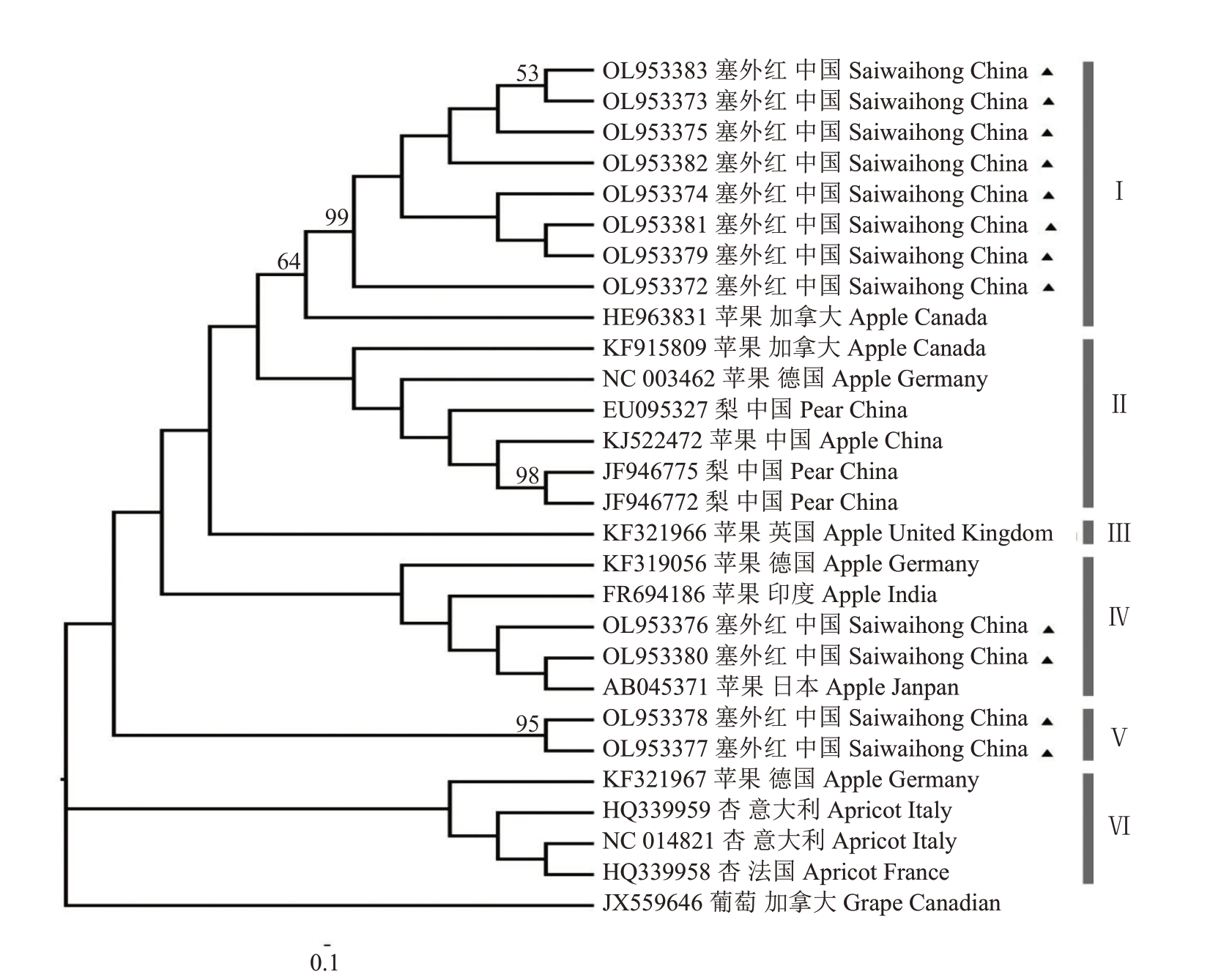
图4 来自塞外红苹果的ASPV 分离物与其他已知ASPV 分离物构建的系统发育进化树
Fig.4 A phylogenetic tree was constructed based on the sequences of ASPV isolates from Saiwaihong and other published ASPV isolates
笔者在本研究中共获得21个来源于塞外红的ASGV分离物(GenBank登录号OL960733~OL960753),核苷酸序列一致性为96.4%~100.0%,来自塞外红苹果的ASGV 分离物Kailu1-1(OL960733)与Kailu1-12(OL960739)核苷酸序列一致性最高(100.0%);ASGV 分离物(OL960734~735、OL960745~747、OL960749~750)与分离物Kailu8-1(OL960753)之间的核苷酸序列一致性最低(96.4%)。来自塞外红苹果的21 个ASGV 分离物与其他已知的38 个ASGV分离物核苷酸序列一致性为89.9%~98.9%,其中塞外红苹果的ASGV 分离物Kailu7-5、Kailu7-10(OL960751、OL960752)与加拿大苹果的ASGV 分离物13TF163B(MZ126539)的核苷酸序列一致性最高(98.9%);分离物Kailu1-7(OL960738)与中国滇黄精ASGV分离物PG(MK427058)核苷酸序列一致性最低(89.9%)(图5)。系统发育分析显示59 个ASGV 分离物聚集成8 个大的分支。塞外红苹果的ASGV 分离物独自聚集形成一支,记为组Ⅰ(图6)。外参为樱桃病毒A(Cherry Virus A,CVA)(AB181355)。

图5 ASGV 基因序列两两比对一致性矩阵图
Fig.5 A matrix of pairwise identities among ASGV gene sequences

图6 来自塞外红苹果的ASGV 分离物与其他已知ASGV 分离物构建的系统发育树
Fig.6 A phylogenetic tree was constructed based on the ASGV isolates from Saiwaihong and other published ASGV isolates
3 讨 论
对病毒的种群多样性和遗传特性进一步深入分析,能够更好地掌握病毒的流行特点,笔者在本研究中对在开鲁县塞外红苹果上发生的3种苹果潜隐病毒病进行了报道。通过RT-PCR获得了29个塞外红苹果ACLSV 分离物,GenBank 登录号:ON001687~ON001715;12个塞外红苹果ASPV分离物,GenBank登录号:OL953372~OL953383;21个塞外红苹果ASGV分离物,GenBank登录号:OL960733~OL960753。根据第九次国际病毒分类会议的建议标准可知,凹陷病毒属不同种的RdRp 基因或者CP 基因的核苷酸序列一致性应高于72%或者氨基酸序列一致性高于80%。塞外红苹果的ACLSV 分离物之间核苷酸序列一致性为78.1%~99.9%,与其他15 个ACLSV分离物之间核苷酸序列一致性为69.1%~93.6%,其中塞外红苹果的ACLSV分离物(ON001689)与美国桃分离物Ta Tao 5(EU223295)核苷酸序列一致性最低(69.1%),基因组序列一致性低于国际病毒分类委员会第九次会议中给出的范围。但通过与其他已经报道过的ACLSV 核苷酸序列一致性比对分析均大于分类标准,本研究认为来自塞外红苹果的ACLSV分离物Kailu1-5(ON001689)仍属于ACLSV。通过对ACLSV 进化树分析,ACLSV 未表现地理位置相关性和寄主相关性。通过核苷酸序列一致性分析可以发现塞外红苹果的ASPV分离物之间核苷酸序列一致性为83.6%~99.7%,与其他15 个ASPV 分离物之间的核苷酸序列一致性为75.4%~91.5%,塞外红苹果的ASPV 分离物与已经报道的ASPV 分离物具有较高的核苷酸一致性,这些数据可以说明该分离物属于ASPV。根据Ma 等[17]的研究结果,ASPV 分离物与地理位置和寄主有相关性,但笔者在本研究中构建的系统发育进化树不支持ASPV分离物间呈现地理位置相关性和寄主相关性,这可能与样本量较少有关。本研究结果表明,塞外红苹果的ASGV分离物之间核苷酸序列一致性为96.4%~100.0%,与其他38 个ASGV 分离物之间核苷酸序列一致性为89.9%~98.9%。塞外红苹果的ASGV分离物与已经报道的ASGV 分离物具有较高的核苷酸序列一致性,这些数据表明该分离物属于ASGV。基于基因序列的系统发育分析表明,这些ASGV 分离物在地理来源特异性或寄主专一性方面没有明确的归类。
笔者在本研究中对开鲁县塞外红上3种苹果潜隐性病毒的分子变异情况进行分析,进一步明确了各个病毒分子变异规律及相应株系的分类情况,以及开鲁不同塞外红苹果产区的病毒分离物系统发育的亲缘关系,本研究对更好地了解塞外红苹果潜隐性病毒的发生规律及制定有效的防治措施具有重要的理论指导意义。
4 结 论
ACLSV、ASPV和ASGV均能侵染塞外红苹果,双重复合侵染和三重复合侵染也有发生。与ASPV和ASGV相比,ACLSV的发生率较高,达30%。对比于其他已经报道的病毒分离物之间的一致性,来自于塞外红苹果的3种病毒种内分离物的核苷酸序列一致性均较高,并且进化关系较近。研究结果为塞外红苹果产业绿色、健康、持续发展提供基本理论依据。
[1] 弟豆豆,曹晓敏,李裕旗,胡慧,姜中武,赵玲玲.苹果病毒病致病机制研究进展[J].烟台果树,2019(3):7-9.DI Doudou,CAO Xiaomin,LI Yuqi,HU Hui,JIANG Zhongwu,ZHAO Lingling. Research progress on pathogenic mechaism of apple virus disease[J].Yantai Fruits,2019(3):7-9.
[2] MINK G I,SHAY J R.Preliminary evolution of some russian apple varieties as indicators for apple viruses[J]. Supplement Plant Disease Reporter,1959,254:13-17.
[3] 郭巍.山楂属植物中苹果褪绿叶斑病毒小RNA 解析及其靶基因功能的研究[D].沈阳:沈阳农业大学,2019.GUO Wei.Analysis of apple chlorotic leaf spot virus small RNA in hawthorn plants and its function of target gene[D].Shenyang:Shenyang Agricultural University,2019.
[4] 王海荣,刘伟,安淼,董晓民,余贤美.桃上苹果褪绿叶斑病毒的发生与检测[J].山东农业科学,2016,48(4):109-110.WANG Hairong,LIU Wei,AN Miao,DONG Xiaomin,YU Xianmei. Occurrence and detection of apple chlorotic leaf spot virus on peach[J].Shandong Agricultural Sciences,2016,48(4):109-110.
[5] 朱慧.来源梨的苹果褪绿叶斑病毒和两种类病毒的分子特性研究[D].武汉:华中农业大学,2014.ZHU Hui. Study on the molecular characteristics of apple chlorotic leaf spot virus and two viriods from pear[D].Wuhan:Huazhong Agricultural University,2014.
[6] SMITH W W. Occurrence of stem pitting and necrosis in some body stocks for apple trees[J]. Journal of American Society for Horticultural Science,1954,63:101-113.
[7] 高德航,何成勇,邢飞,侯婉莹,李世访,战斌慧,王红清.苹果茎痘病毒在山楂中的首次鉴定与序列分析[J].植物病理学报,2021,51(4):515-524.GAO Dehang,HE Chengyong,XING Fei,HOU Wanying,LI Shifang,ZHAN Binhui,WANG Hongqing. Identification and sequence analysis of apple stem pitting virus from hawthorn tree[J].Acta Phytopathologica Sinica,2021,51(4):515-524.
[8] 刘华珍,杨作坤,周友斌,王国平,洪霓.侵染梨的苹果茎痘病毒基因组序列及小RNA 分析[J].植物病理学报,2017,47(5):584-590.LIU Huazhen,YANG Zuokun,ZHOU Youbin,WANG Guoping,HONG Ni. Study on genome sequence and small RNA of apple stem pitting virus infecting a pear plant[J].Acta Phytopathologica Sinica,2017,47(5):584-590.
[9] MORÁN F,CANALES C,OLMOS A,RUIZ-GARCÍA A B.Loquat (Eriobotrya japonica) is a new natural host of apple stem pitting virus[J].Plants,2020,9(11):1560.
[10] LISTER R M,BANCROFT J B,NADAKAVUKAREN M J.Some saptransmissible viruses from apple[J]. Phytopathology,1965,55:859-870.
[11] 孙晓霞,牛建新,王博慧,裴茂松,李陈静,曹福军.库尔勒香梨上苹果茎沟病毒全序列的鉴定与分析[J]. 分子植物育种,2015,13(8):1771-1778.SUN Xiaoxia,NIU Jianxin,WANG Bohui,PEI Maosong,LI Chenjing,CAO Fujun. Identification and analysis of an apple stem groove virus complete genomic sequence from Korla fragrant pear[J].Molecular Plant Breeding,2015,13(8):1771-1778.
[12] 周洲.印度猕猴桃感染苹果茎沟病毒的特征[J].中国果业信息,2014,31(9):56-57.ZHOU Zhou.Characteristics of Actindia indian infected with apple stem groov virus[J].China Fruit Industry Information,2014,31(9):56-57.
[13] 张秀琪,刘松誉,杨一舟,李梦林,胡安安,刘唱,张宗英,韩成贵,王颖.北京月季病原病毒的高通量测序鉴定和RT-PCR 检测[J].植物病理学报,2021,51(4):525-535.ZHANG Xiuqi,LIU Songyu,YANG Yizhou,LI Menglin,HU An’an,LIU Chang,ZHANG Zongying,HAN Chenggui,WANG Ying.Virus identification and detection by high-throughput sequencing and RT-PCR from rose plants in Beijing[J].Acta Phytopathologica Sinica,2021,51(4):525-535.
[14] BHARDWAJ P,AWASTHI P,PRAKASH O,SOOD A,ZAIDI A A,HALLAN V. Molecular evidence of natural occurrence of apple stem grooving virus on bamboos[J]. Trees,2017,31(1):367-375.
[15] 贾栋,贾小云,马瑞燕.生物信息学数据库及查询[J].山西农业大学学报(自然科学版),2013,33(6):520-525.JIA Dong,JIA Xiaoyun,MA Ruiyan. Overview in bioinformatics database and the introduction of inquiry[J].Journal of Shanxi Agricultural University(Natural Science Edition),2013,33(6):520-525.
[16] 邵西群,巴恒星,李志鹏,章秀婷,杨福合.生物软件在序列分析过程中的运用[J].生物信息学,2010,8(4):321-324.SHAO Xiqun,BA Hengxing,LI Zhipeng,ZHANG Xiuting,YANG Fuhe. Some tips about applications for biological sequence retrieval and analyses[J]. China Journal of Bioinformatics,2010,8(4):321-324.
[17] MA X F,HONG N,MOFFETT P,WANG G P.Genetic diversity and evolution of apple stem pitting virus isolates from pear in china[J].Canadian Journal of Plant Pathology,2016,38(2):218-230.