余甘子(Phyllanthus emblica L.)为大戟科叶下珠属植物,果实、根、茎和叶都可入药[1]。余甘子含有酚酸类、鞣质类、黄酮类及萜烯类等化合物,具有较好的抗氧化、保肝、抗肿瘤和抗病毒等药理作用[2-4]。目前已知余甘子与35 种民族民间临床治疗功效有关,有17 个国家和民族在使用余甘子,并在中国的中药、傣药、壮药、藏药中广泛应用[5-7]。除药用功能显著外,余甘子还因其富含维生素等营养物质及特殊的回甘风味,成为广受群众喜爱的特优稀水果[8]。此外,余甘子植株耐贫瘠,固土能力强,具有良好的生态功能,成为山区植被修复及水土保持等生态治理的优选物种[9]。
近年来随着余甘子药用价值、营养价值、生物及地理研究的深入,优良品种的选育与改良越来越受到关注。然而,中国余甘子目前无栽培种,基本处于野生状态,各地区种质虽多,但缺乏系统的整理和发掘,种质的遗传背景并不十分清晰。已有学者基于叶形、雄花性状、果形等表型性状对余甘子种质资源的遗传多样性进行研究[10-11],但植物的表型性状是由基因和环境共同调控的,并不能准确地反映余甘子群体的遗传多样性水平。邵雪花等[12]利用ISSR 分子标记技术分析28份余甘子种质的遗传多样性,并构建了指纹图谱。郭林榕等[13]研究表明SRAP分子标记技术可有效运用于南方湿润分布区的22 份余甘子种质资源的遗传多样性分析。蔡英卿等[14]利用RAPD 分子标记技术将34 份福建省余甘子种质分为惠安余甘和莆田余甘两类。
SNP 和InDel 是指基因组上单核酸的插入、替换、缺失引起的核酸序列多态性,与其他种类的分子遗传标记相比,能更好地代表余甘子全基因组的遗传信息,具有分布范围广、密度高、稳定性好、结果准确、容易实现检测自动化和规模化的优点[15-16]。近年来,随着测序技术的快速发展,SNP 和InDel 分子标记已广泛应用于非模式生物的群体遗传结构分析、遗传多样性分析、遗传图谱构建、数量性状相关性分析等领域[17-20]。然而,目前并没有基于SNP 和InDel标记对余甘子群体遗传结构和遗传多样性进行分析的报道。
简化基因组测序技术可为种群遗传学研究提供更为丰富而准确的数据,其中,限制性酶切位点关联DNA 测序技术(restriction-site associated DNA sequencing,RADseq)是简化基因组测序技术中发展较快、应用较广的测序技术,根据不同试验需求,该技术已有ddRAD、mbRAD、ezRAD、GBS 等多种版本[21-22]。ddRADseq 技术具有通量高、成本低、试验时间短及无需参考基因组等优点[23],已用于马铃薯、水稻螟虫等多物种的种质鉴定、系统发育和遗传进化及性状关联分析研究[24-25]。目前余甘子参考基因组并未发布,笔者在本研究中拟采用ddRADseq 技术对112 份余甘子样品进行高通量简化基因组测序,开发SNP和InDel分子标记,分析余甘子种质的遗传多样性及群体遗传结构,探讨不同种质的系统演化关系,为后续余甘子育种亲本选配、优良品系筛选提供材料和理论指导。
1 材料和方法
1.1 材料
试验材料均取自云南省农业科学院热区生态农业研究所余甘子种质资源保护云南创新基地,共有112 份余甘子种质材料,供试名称见表1,其中,1~3号FJ种质自福建引种,4~12号GJ种质自广西引种,13~23号JY种质自广东引种,24号CL(西印度醋栗)种质为近缘种,25~112 号YGZ 种质从云南省内引种。采集各供试材料健康叶片,于-4 ℃保存备用。
表1 用于测序分析的112 份余甘子种质资源
Table 1 112 species of P.emblica germplasm resources for sequencing analysis

序号Code 123456789 10 11 12 13 14 15 16 17 18 19 20 21 22 23样品名Species FJ1 FJ2 FJ3 GJ1 GJ2 GJ3 GJ4 GJ5 GJ6 GJ7 GJ8 GJ9 JY10 JY11 JY1 JY2 JY3 JY4 JY5 JY6 JY7 JY8 JY9序号Code 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46样品名Species CL YGZ01 YR02 YGZ02 YGZ03 YGZ04 YGZ05 YGZ06 YGZ07 YGZ104 YGZ105 YGZ106 YGZ10 YGZ111 YGZ115 YGZ11 YGZ121 YGZ12 YGZ133 YGZ137 YGZ13 YGZ144 YGZ145序号Code 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69样品名Species YGZ146 YGZ147 YGZ148 YGZ149 YGZ14 YGZ150 YGZ151 YGZ152 YGZ153 YGZ154 YGZ155 YGZ157 YGZ164 YGZ167 YGZ16 YGZ173 YGZ179 YGZ180-3 YGZ183 YGZ187 YGZ21 YGZ22 YGZ23序号Code 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92样品名Species YGZ24 YGZ25 YGZ26 YGZ28 YGZ2 YGZ32 YGZ33 YGZ35 YGZ37 YGZ3 YGZ42 YGZ50 YGZ51 YGZ53 YGZ58 YGZ67 YGZ72 YGZ73 YGZ78 YGZ7 YGZ81 YGZ82 YGZ8序号Code 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112样品名Species YGZ92 YGZ94 YGZ99 YGZ9 YGZ201B YGZ301B YGZ3104 YGZ109-1 YGZ201-1 YGZ201-2 YGZ201-3 YGZ401-1 YGZ401-2 YGZ4001-3 YGZ514-1 YGZ806-1 YGZ806-2 YGZ62101 YGZ621 YGZDG
1.2 余甘子基因组DNA提取及建库测序
采用改良的CTAB法提取余甘子叶片的基因组DNA,分别用1%琼脂糖凝胶电泳和Nano Drop ND-1000对DNA样品纯度和浓度进行检测,调节样品质量浓度至100 ng·μL-1,-20 ℃保存。采用限制性内切酶SacⅠ和MseⅠ对基因组DNA 进行酶切,再用测序接头与酶切片段进行连接,连接片段纯化后,选择插入长度在300~400 bp 范围内的片段进行扩增,并使用Illumina Hiseq测序,原始测序读长为Pairedend 150 bp。
1.3 SNP和InDel分子标记开发
采用Cutadapt 和Trimmomatic 软件对原始测序数据进行数据质控,统计测序得到的reads数目。使用不依赖参考基因组进行变异发掘的MUNEAK 软件进行SNP 和InDel 多态性标记的发掘以及各样品基因型的分析。
1.4 余甘子群体结构分析
基于筛选的多态性标记,使用STRUCTURE 软件对所有样品进行聚类分析,设定类群数K为1~6,计算ΔK 值,ΔK 值最大对应的K值即为最合理类群数。运用STRUCTURE 软件分析每个余甘子样品归属各类群比率,确定各种质归属的类群。
利用Plink软件对各样品进行主成分分析,使用R 软件计算各主成分向量,绘制主成分分析散点图。采用MEGA 7 软件计算参试材料的遗传距离,使用iTOL(https://itol.embl.de/)绘制进化树图。
1.5 余甘子群体遗传多样性分析
基于获得的SNP、InDel 标记和群体来源地信息,利用PowerMarker 软件计算余甘子群体的期望杂合度(He)、观测杂合度(Ho)、多态性信息含量(PIC)以及群体分化指数(Fst),以评估余甘子群体的遗传多样性。
2 结果与分析
2.1 建库和测序质量评估
通过对所有余甘子样品进行建库和测序后,总共得到64.70 Gb 数据,共获得233 960 457 条reads,读长数目在288 386~7 202 919 bp之间,平均读长为2 088 933 bp;测序Q30 值在97.46%~98.40%之间,平均Q30 为97.96%,所有样品Q30 值均在95%以上;测序GC 值在36.25%~38.68%之间,平均GC 为37.30%,测序数据质量符合预期,可用于后续分析。
2.2 SNP和InDel分子标记开发
采用MUNEAK 软件,对余甘子样品根据缺失基因型的比例≤0.5、较小等位基因频率≥0.05 进行过滤,共得到4019 个ddRADseq 测序位点和8934 个SNP和InDel标记,用于后续群体结构分析。
2.3 基于SNP和InDel标记的余甘子群体遗传结构分析
基于112 份余甘子样品开发的SNP 和InDel 标记(CL 样品由于基因型数据缺失过多,在后续的分析中去掉),运用STRUCTURE 软件进行群体结构的分析,通过设置K=1~7,得到如图1 的ΔK 随K 值变化的趋势图。从图1中可以看出,ΔK值最大对应的K 值为2,余甘子群体的最佳亚群数为2,表明来自不同地区的余甘子种质可能来自于2个祖先。
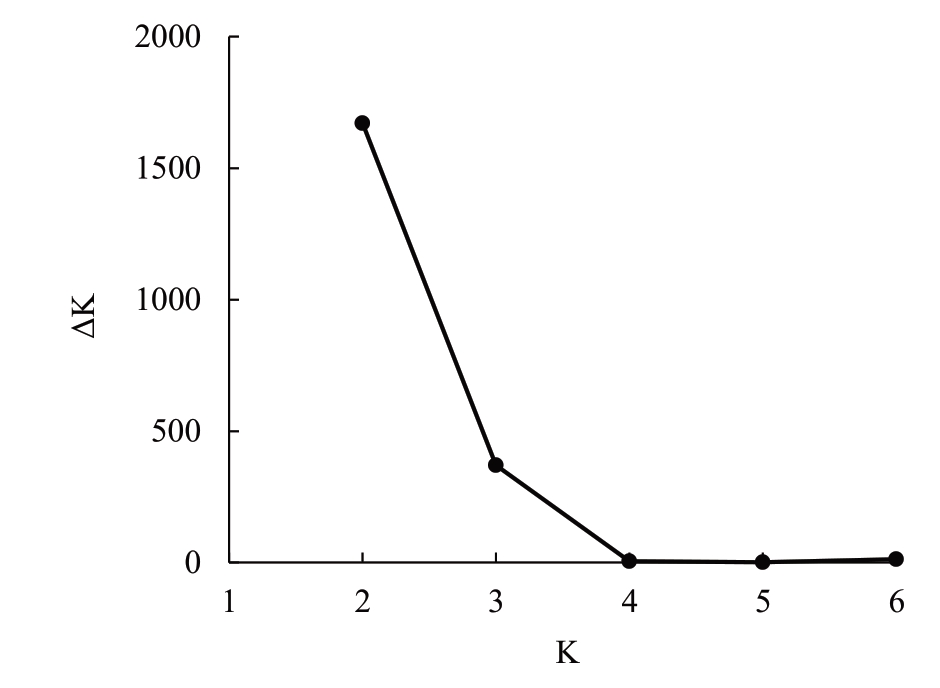
图1 ΔK 随K 值变化的趋势
Fig.1 The trend of ΔK with K value
当K=2 时,STRUCTURE 输出每个样品组分比例数据,基于Q 值,笔者将每个样品分配到各个亚群,如图2 所示。类群1 显示为红色,共有88 份样品,这些种质均来自于云南;其中,61 份样品的Q 值为1,这61份样品代表类群1的基因库。类群2在图中显示为蓝色,有23份样品,分别引种自福建、广西和广东地区;类群2共20份样品的Q值为1,这20份样品代表类群2 的基因库,其中,GJ1 和GJ6 这两个种质属于类群2,对应Q 值分别为0.59 和0.58,说明这两个种质从广西、广东和福建引种后,在种质选育驯化的过程中,与云南的种质产生基因交流,具有混合的遗传背景。
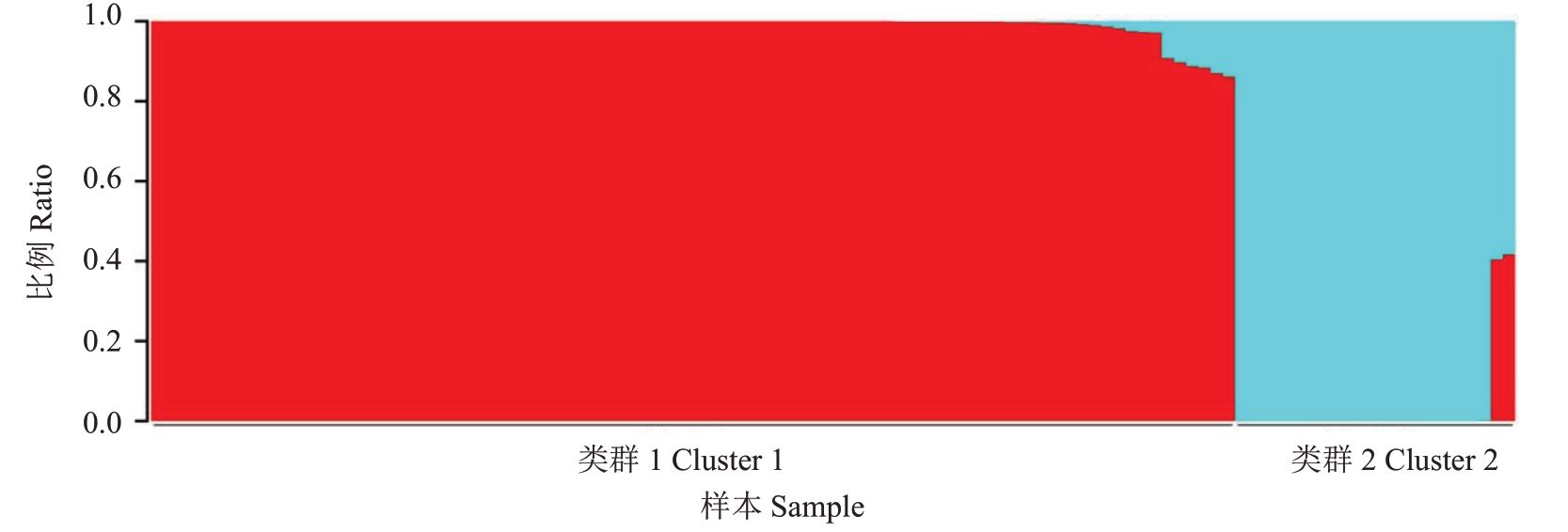
图2 余甘子遗传结构
Fig.2 Population genetic structure of P.emblica
为了验证群体结构分析结果的可靠性,基于所有参试样品的高质量SNP和InDel标记,采用主成分分析法对上述标记进行降维处理,绘制了主成分分析图(图3),图中样品点距离越近,表示样品间遗传背景差距越小。由图3 可知,第一特征向量PC1 和第二特征向量PC2 将余甘子样品分为2 个类群,该结果与群体结构分析结果一致。其中,类群1 在PC1轴上分布较为集中,类群2较为分散,说明类群2样品间遗传背景较类群1差异大。

图3 余甘子主成分分析
Fig.3 Principal component analysis of P.emblica
2.4 基于SNP和InDel标记的余甘子群体系统发育分析
基于余甘子样品的SNP 和InDel 标记构建的系统发育树如图4所示。由图4可知,所有余甘子种质分为2个类群,来源于云南的88份YGZ种质归属于类群1,来源于福建、广西和广东的FJ、JY 和GY 等23份种质归属于类群2,种质聚类结果与群体结构、主成分分析相一致,各个类群能较好地聚在一起。其中,类群1 样品距离进化树圆心较近且紧密的聚类在一起,说明类群1 个体亲缘关系较近;类群2 的23 个样品聚在一起,距离进化树圆心的距离较远,表明该类群个体在进化上积累了更多的遗传差异。余甘子各种质间平均遗传距离为0.248,其中,YGZ179种质和YGZ180-3种质遗传距离值最小,为0.027,表明这两个种质的亲缘关系较近,种质间相似程度比较高;JY1种质和YGZ401-1种质遗传距离值最大,为0.459,表明它们亲缘关系最远,遗传信息差异大。
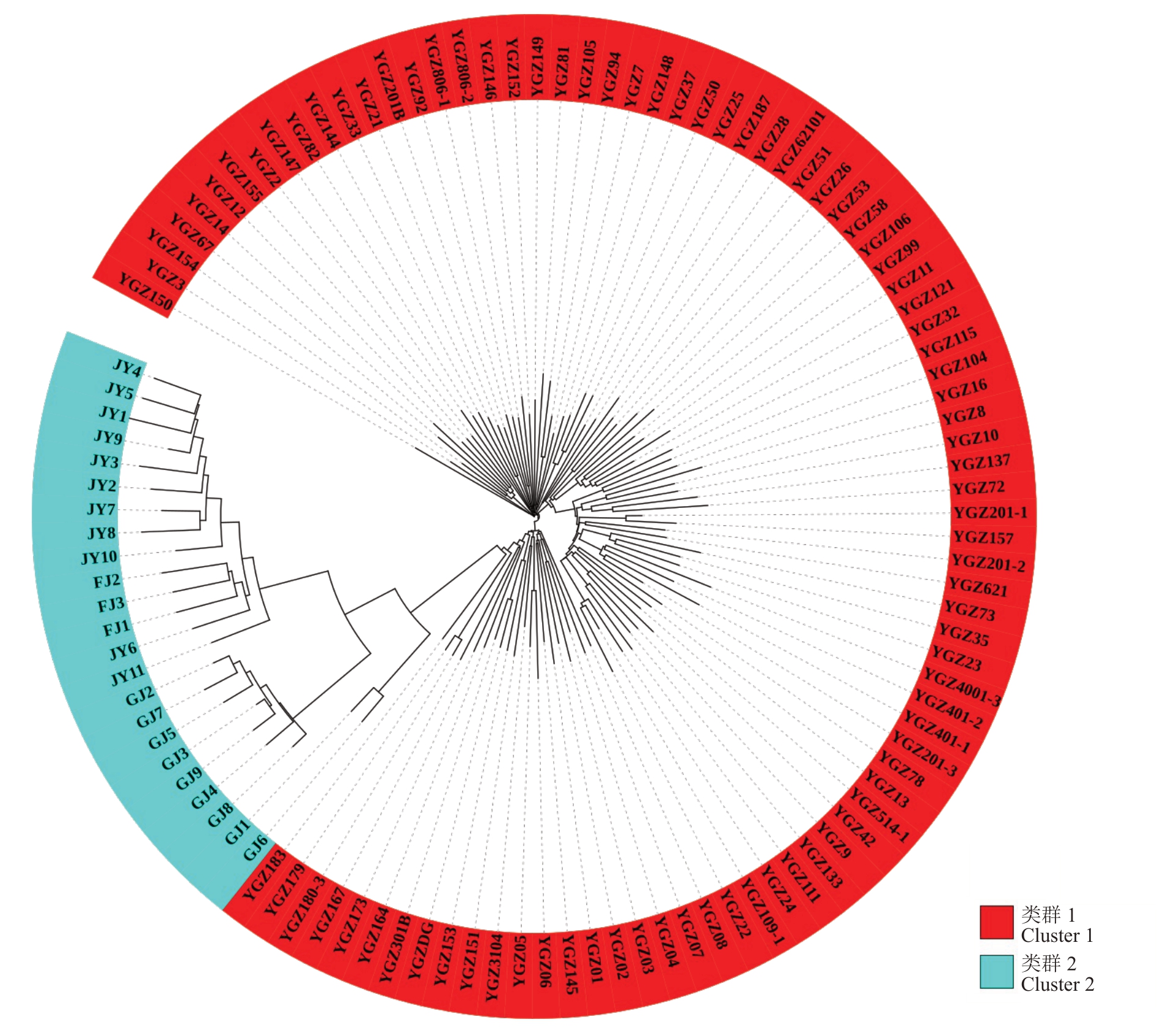
图4 余甘子进化树
Fig.4 Phylogenetic trees of P.emblica
2.5 余甘子群体遗传多样性分析
从上述余甘子群体结构分析可知,余甘子群体结构划分与地理来源地密切相关,为了解余甘子群体的遗传多样性水平,对不同来源地余甘子种质的杂合度、PIC 和Fst 进行分析。杂合度和PIC 值可反映余甘子群体的遗传变异程度,它们的值越大,表示遗传变异越大,则群体的遗传多样性越丰富,其中杂合度包括期望杂合度(He)和观测杂合度(Ho)。当Fst 为0 或1 时,表明群体间没有分化或完全分化;0<Fst<0.05 时,表明群体间遗传分化较弱;0.05≤Fst<0.15 时,群体遗传分化中等;0.15≤Fst<0.25 或0.25≤Fst<1时,表明群体间具有较强或非常强的遗传分化[26]。
余甘子群体的遗传多样性参数和遗传分化系数结果见表2和表3。由表2可知,余甘子群体的He值在0.094~0.267 之间,平均值为0.17;Ho 值为0.071~0.184,平均值为0.113;PIC值在0.095~0.218之间,平均值为0.143。其中,来源于云南的YGZ 种质杂合度和PIC值最大,表现出最多的遗传变异,其次依次为广东引种的JY 种质和广西引种的GJ 种质,福建引种的FJ 种质He、Ho 及PIC 值最小,分别为0.094、0.071及0.095。由表3可知,余甘子群体间的遗传分化指数在0.080~0.266之间,表现为中等偏强的遗传分化。其中,YGZ 和其余种质间均为中等分化,亲缘关系相对近;JY 种质和GJ 种质间Fst 为0.231,遗传分化比较强,亲缘关系远;JY种质和FJ种质Fst间为0.136,属于中等遗传分化;GJ 种质和FJ 种质间Fst最大,为0.266,遗传分化非常强,亲缘关系较远。
表2 余甘子群体遗传多样性参数统计
Table 2 Statistics of genetic diversity parameters of P.emblica populations
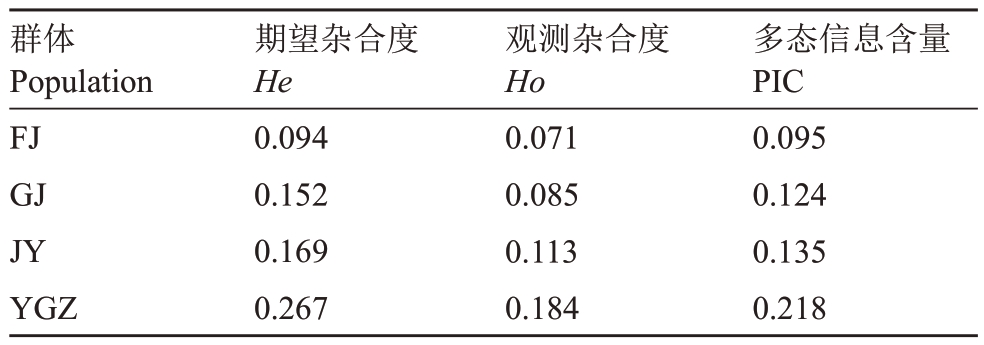
群体Population FJ GJ JY YGZ期望杂合度He 0.094 0.152 0.169 0.267观测杂合度Ho 0.071 0.085 0.113 0.184多态信息含量PIC 0.095 0.124 0.135 0.218
表3 余甘子群体遗传分化指数统计
Table 3 Statistics of population differentiation index of P.emblica populations
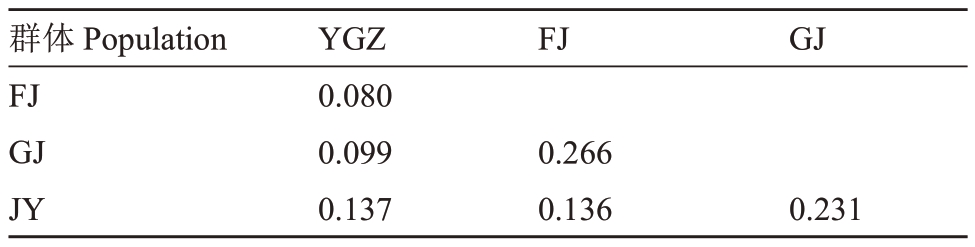
群体Population FJ GJ JY YGZ 0.080 0.099 0.137 FJ 0.266 0.136 GJ 0.231
3 讨 论
ddRADseq 测序技术是基于全基因组酶切位点的高通量简化基因组测序技术,该技术无需参考基因组,采用的SacⅠ和MseⅠ两种限制性内切酶目的性较强,减少了早期RAD文库在物理打断及片段选择等制备过程中造成的DNA 损失,简化RAD 文库制备过程,降低待测基因组复杂度[27-28]。笔者在本试验中采用该测序技术对112份余甘子样品进行建库测序,共得到64.70 Gb 数据,开发了8934 个SNP 和InDel标记,可达到余甘子种质资源群体结构和遗传多样性分析的目的。其中,余甘子近缘种CL 种质由于标记缺失过多,在群体和系统发育分析中被去掉,这应该是由本试验中采用的标记是根据序列相似性进行聚类,然而该种质和其余种质基因型差异大,没与其余种质的序列聚到一起所导致的。
全面了解种质资源的遗传结构是有效保护和高效利用的前提,其中群体结构是遗传结构的核心。不同地理来源的余甘子种质由于地理和环境等因素的影响,会出现遗传信息差异,不同地理来源地种质的遗传信息分析是遗传学分析的重要内容。王建超等[29]研究表明余甘子存在明显的地域分布规律,生境相似、来源相近的种质资源亲缘关系较近;熊仪俊[30]也认为余甘子遗传距离与空间距离有一定相关性。笔者在本研究中利用3种群体结构分析方法将所有参试余甘子样品分为两个类群,其中,类群1有88 份样品,为云南收集的种质,类群2 为有23 份样品,为从福建、广东、广西收集的种质,表明余甘子分子水平的群体聚类结果与地理来源地相关,地理区域近的种质遗传距离较为接近。类群2部分种质具有混合的遗传背景,且福建、广东、广西3 个引种地的种质并未划分开,该结果可能与3 个引种地的地理距离较近,种质间基因交流较为频繁有关。此外,类群1 的YGZ179 和YGZ180-3 种质遗传距离值最小,加之两者收集时间和地点相近,推测可能为同一材料。同时,类群1的云南种质遗传多样性较高,可作为种质创新利用的备选材料。近年来,笔者在云南种质的基础上已成功选育出果型大的鲜食余甘子品种盈玉,并在生产中大面积推广应用。
笔者计算并统计了余甘子群体的He、Ho、PIC以及Fst 值,以评估余甘子群体的遗传多样性,参试样品中,来源于云南的YGZ种质杂合度和PIC值最大,表现出最多的遗传变异。李巧明等[31]采用ISSR分子标记技术检测云南干热河谷区的4个余甘子居群的遗传多样性,也认为云南余甘子居群具有较高的遗传多样水平。前人研究表明,云南余甘子居群间遗传分化系数为0.122[31],熊仪俊[30]基于RAPD 技术的研究结果表明64份余甘子种群地理分化明显,种群间遗传分化较强。笔者在本文中发现归属于类群2 的FJ、GJ、JY 群体较类群1 的YGZ 群体遗传分化强,表现为整体中等偏强,这与主成分分析和系统发育分析一致,即类群2 的个体分布更为离散,其中,来源于广西的种质与来源于福建的种质间Fst为0.266,遗传分化非常强,后期可加大福建、广西、广东等地种质收集力度,丰富余甘子群体的遗传多样性。笔者在本文中解析了112份余甘子种质的群体结构和遗传多样性,为后续余甘子遗传图谱构建、分子标记开发、系统发育研究、品种选育及产业开发利用提供理论依据。
4 结 论
笔者在本文中基于高通量测序技术开发的8934 个SNP 和InDel 标记解析112 份余甘子种质的群体结构和遗传多样性。群体结构分析、主成分分析及系统发育分析将余甘子群体分为2 个类群,其中,类群1 有88 份样品,主要为从云南收集的种质,类群2 为有23 份样品,为从福建、广东、广西收集的种质,3种分析方法相互补充印证,表明余甘子群体结构划分可靠。遗传多样性分析结果表明,余甘子群体He 平均值为0.17,Ho 平均值为0.113,PIC 平均值为0.143,群体间的Fst 在0.080~0.266 之间,群体遗传分化整体中等偏强。遗传距离分析显示,余甘子群体遗传距离在0.027~0.459之间,平均遗传距离为0.248。本研究可为后续余甘子优良基因挖掘、优良种质引进及创新利用提供参考依据。
[1] 杨崇仁,张颖君,王海涛,王建刚,丁艳芬,邵艳红.余甘子应用源流考[J].亚太传统医药,2021,17(2):197-200.YANG Chongren,ZHANG Yingjun,WANG Haitao,WANG Jiangang,DING Yanfen,SHAO Yanhong. Study on the source of fruit emblic[J].Asia-Pacific Traditional Medicine,2021,17(2):197-200.
[2] 林艺青.余甘子多酚的化学组成及生物活性研究进展[J].食品工业,2020,41(10):241-244.LIN Yiqing.Research progress on chemical compounds and biological activities of polyphenols of Phyllanthus emblica[J]. The Food Industry,2020,41(10):241-244.
[3] 尹可欢,罗晓敏,丁翼,阙涵韵,谭睿,李大鹏,龚普阳,顾健.余甘子及其活性成分肝保护作用及机制的研究进展[J].中草药,2022,53(1):295-307.YIN Kehuan,LUO Xiaomin,DING Yi,QUE Hanyun,TAN Rui,LI Dapeng,GONG Puyang,GU Jian.Research progress on hepatoprotective effect and mechanism of Phyllanthus emblica and its active components[J]. Chinese Traditional and Herbal Drugs,2022,53(1):295-307.
[4] 陈静梅,郝二伟,杜正彩,李思维,王星圆,侯小涛,邓家刚.基于化学成分、药理作用和网络药理学的余甘子质量标志物(QMarker)预测分析[J].中草药,2022,53(5):1570-1586.CHEN Jingmei,HAO Erwei,DU Zhengcai,LI Siwei,WANG Xingyuan,HOU Xiaotao,DENG Jiagang.Predictive analysis on quality marker of Phyllanthus emblica based on chemical composition, pharmacological effects and network pharmacology[J].Chinese Traditional and Herbal Drugs,2022,53(5):1570-1586.
[5] 陈文静,李师,梁文仪,吴玲芳,崔雅萍,亓旗,张兰珍.余甘子在藏医药中的应用[J].世界科学技术-中医药现代化,2016,18(7):1154-1158.CHEN Wenjing,LI Shi,LIANG Wenyi,WU Lingfang,CUI Yaping,QI Qi,ZHANG Lanzhen. Application of Phyllanthus emblica L. in Tibetan medicine[J]. Modernization of Traditional Chinese Medicine and Materia Medica-World Science and Technology,2016,18(7):1154-1158.
[6] 李雪冬,潘烨华,田雨闪,杨烨,龚普阳.余甘子的本草考证及其现代研究中若干问题的探讨[J].中草药,2022,53(18):5873-5883.LI Xuedong,PAN Yehua,TIAN Yushan,YANG Ye,GONG Puyang. Herbal textual and key problems in modern research of Phyllanthus emblica[J]. Chinese Traditional and Herbal Drugs,2022,53(18):5873-5883
[7] 毛晓健,江菊,赵杰.傣药麻夯板(余甘子)的研究概况[J].中国民族医药杂志,2008,14(2):33-34.MAO Xiaojian,JIANG Ju,ZHAO Jie.Review on the DAI medicine of Phyllanthus emblica[J]. Journal of Medicine & Pharmacy of Chinese Minorities,2008,14(2):33-34.
[8] 张沐棠,陈少虹,程子贤,李湘銮,陈椰娜,姜浩,白卫东.余甘子的回甘风味研究进展[J].食品与发酵工业,2023,49(6):324-331.ZHANG Mutang,CHEN Shaohong,CHENG Zixian,LI Xiangluan,CHEN Yena,JIANG Hao,BAI Weidong.Research development of the back sweet flavour of Phyllanthus emblica L.[J].Food and Fermentation Industries,2023,49(6):324-331.
[9] 段曰汤,瞿文林,宋子波,赵琼玲,马开华,金杰,方海东,雷虓,沙毓沧. 云南野生余甘子保护及开发利用[J]. 农学学报,2019,9(9):49-54.DUAN Yuetang,QU Wenlin,SONG Zibo,ZHAO Qiongling,MA Kaihua,JIN Jie,FANG Haidong,LEI Xiao,SHA Yucang.Wild Phyllanthus emblica L.in Yunnan:Protection and exploitation[J].Journal of Agriculture,2019,9(9):49-54.
[10] 赵琼玲,李丽,沙毓沧,段曰汤,杨子祥,马开华.云南不同种源余甘子植物形态变异研究[J].热带作物学报,2012,33(1):178-181.ZHAO Qiongling,LI Li,SHA Yucang,DUAN Yuetang,YANG Zixiang,MA Kaihua.Morphological variations of different Phyllanthus emblica L. provenances in Yunnan[J]. Chinese Journal of Tropical Crops,2012,33(1):178-181.
[11] 瞿文林,段曰汤,马开华,杨子祥,谭红,沙毓沧.余甘子天然居群果实形态变异研究[J]. 西北植物学报,2012,32(12):2444-2449.QU Wenlin,DUAN Yuetang,MA Kaihua,YANG Zixiang,TAN Hong,SHA Yucang. Fruit morphological variation of natural populations in Phyllanthus emblica[J]. Acta Botanica Boreali-Occidentalia Sinica,2012,32(12):2444-2449.
[12] 邵雪花,刘牛,赖多,肖维强,匡石滋.28 份余甘子品种遗传多样性的ISSR 分析及指纹图谱构建[J].西北农林科技大学学报(自然科学版),2020,48(8):129-136.SHAO Xuehua,LIU Niu,LAI Duo,XIAO Weiqiang,KUANG Shizi. Genetic diversity analysis and DNA fingerprint mapping of 28 varieties of Phyllanthus emblica L.based on ISSR molecular marker[J]. Journal of Northwest A & F University (Natural Science Edition),2020,48(8):129-136.
[13] 郭林榕,周平,陈志峰.22 份余甘子核心种质资源遗传多样性的SRAP 分析[J].热带作物学报,2014,35(7):1382-1387.GUO Linrong,ZHOU Ping,CHEN Zhifeng.Analysis of genetic diversity of 22 Phyllanthus emblica germlasms with SRAP markers[J]. Chinese Journal of Tropical Crops,2014,35(7):1382-1387.
[14] 蔡英卿,赖钟雄,陈义挺,郭玉琼,潘东明.福建余甘子遗传资源的RAPD 分析[J].热带作物学报,2007,28(2):74-79.CAI Yingqing,LAI Zhongxiong,CHEN Yiting,GUO Yuqiong,PAN Dongming.RAPD analysis of emblic(Phyllanthus emblica L.) genetic resources in Fujian province[J]. Chinese Journal of Tropical Crops,2007,28(2):74-79.
[15] YOU Q,YANG X P,PENG Z,XU L P,WANG J P.Development and applications of a high throughput genotyping tool for polyploid crops:Single nucleotide polymorphism (SNP) array[J].Frontiers in Plant Science,2018,9:104.
[16] 谢中艺,党江波,温国,王海燕,郭启高,梁国鲁.植物外源基因组成分鉴定方法的研究进展[J]. 生物工程学报,2021,37(8):2703-2718.XIE Zhongyi,DANG Jiangbo,WEN Guo,WANG Haiyan,GUO Qigao,LIANG Guolu.Advances in identification methods of alien genomic components in plants[J]. Chinese Journal of Biotechnology,2021,37(8):2703-2718.
[17] ALI KHAN S,CHEN H,DENG Y,CHEN Y H,ZHANG C,CAI T C,ALI N,MAMADOU G,XIE D Y,GUO B Z,VARSHNEY R K,ZHUANG W J. High-density SNP map facilitates fine mapping of QTLs and candidate genes discovery for Aspergillus flavus resistance in peanut (Arachis hypogaea)[J]. Theoretical and Applied Genetics,2020,133(7):2239-2257.
[18] ZHU S P,WANG F S,SHEN W X,JIANG D,HONG Q B,ZHAO X C. Genetic diversity of Poncirus and phylogenetic relationships with its relatives revealed by SSR and SNP/InDel markers[J].Acta Physiologiae Plantarum,2015,37(7):141.
[19] SEO E,KIM K,JUN T H,CHOI J,KIM S H,MUÑOZ-AMATRIAÍN M,SUN H,HA B K.Population structure and genetic diversity in Korean cowpea germplasm based on SNP markers[J].Plants,2020,9(9):1190.
[20] 易丽聪,王运强,焦春海,姚明华,龚钰,王舒景,戴照义.基于SNP 标记的西瓜种质资源遗传多样性分析[J]. 中国瓜菜,2020,33(12):8-13.YI Licong,WANG Yunqiang,JIAO Chunhai,YAO Minghua,GONG Yu,WANG Shujing,DAI Zhaoyi.Genetic diversity analysis of 64 watermelon germplasms by SNP markers[J]. China Cucurbits and Vegetables,2020,33(12):8-13.
[21] ANDREWS K R,GOOD J M,MILLER M R,LUIKART G,HOHENLOHE P A. Harnessing the power of RADseq for ecological and evolutionary genomics[J].Nature Reviews Genetics,2016,17(2):81-92.
[22] KONAR A,CHOUDHURY O,BULLIS R,FIEDLER L,KRUSER J M,STEPHENS M T,GAILING O,SCHLARBAUM S,COGGESHALL M V,STATON M E,CARLSON J E,EMRICH S,ROMERO-SEVERSON J. High-quality genetic mapping with ddRADseq in the non-model tree Quercus rubra[J].BMC Genomics,2017,18(1):417.
[23] PETERSON B K,WEBER J N,KAY E H,FISHER H S,HOEKSTRA H E. Double digest RADseq:An inexpensive method for de novo SNP discovery and genotyping in model and non-model species[J].PLoS One,2012,7(5):e37135.
[24] 杨琼,孙娜,郑敏,罗除成,罗光华,方继朝.水稻二化螟和三化螟基因组ddRADseq 文库的构建[J].环境昆虫学报,2016,38(6):1114-1120.YANG Qiong,SUN Na,ZHENG Min,LUO Chucheng,LUO Guanghua,FANG Jichao. Construction of genomic ddRADseq libraries of Chilo suppressalis and Scirpophaga incertulas(Lepidoptera: Pyralidae)[J]. Journal of Environmental Entomology,2016,38(6):1114-1120.
[25] 单建伟,索海翠,王丽,安康,刘计涛,李成晨,白建明,李小波.基于ddRADseq 的马铃薯品种遗传多样性分析[J].广东农业科学,2021,48(12):120-128.SHAN Jianwei,SUO Haicui,WANG Li,AN Kang,LIU Jitao,LI Chengchen,BAI Jianming,LI Xiaobo.Analysis on genetic diversity of potato varieties based on ddRADseq[J]. Guangdong Agricultural Sciences,2021,48(12):120-128.
[26] WRIGHT S. Evolution and the genetics of population,variability within and among natural populations:4 Vol.[M]. Chicago:The University of Chicago Press,1978.
[27] BUS A,HECHT J,HUETTEL B,REINHARDT R,STICH B.High- throughput polymorphism detection and genotyping in Brassica napus using next- generation RAD sequencing[J].BMC Genomics,2012,13:281.
[28] PUKK L,AHMAD F,HASAN S,KISAND V,GROSS R,VASEMÄGI A.Less is more:extreme genome complexity reduction with ddRAD using ion torrent semiconductor technology[J].Molecular Ecology Resources,2015,15(5):1145-1152.
[29] 王建超,何银莺,黄旭萍,陈发兴,郭林榕.余甘子种质资源的遗传多样性分析[J].森林与环境学报,2021,41(4):396-401.WANG Jianchao,HE Yinying,HUANG Xuping,CHEN Faxing,GUO Linrong. Genetic diversity analysis of Phyllanthus emblica germplasm resources[J]. Journal of Forest and Environment,2021,41(4):396-401.
[30] 熊仪俊.余甘子不同生态型特征与分化初步研究[D].北京:中国林业科学研究院,2003.XIONG Yijun. Preliminary study on the characteristics and differentiations of ecological types of Phyllanthus emblica[D].Beijing:Chinese Academy of Forestry,2003.
[31] 李巧明,赵建立.云南干热河谷地区余甘子居群的遗传多样性研究[J].生物多样性,2007,15(1):84-91.LI Qiaoming,ZHAO Jianli.Genetic diversity of Phyllanthus emblica populations in dry-hot valleys in Yunnan [J]. Biodiversity Science,2007,15(1):84-91.