自从在玉米中发现knotted1(kn1)基因以来,已在多个植物中发现KNOX(KNOTTED1 like homeobox)基因[1]。KNOX 转录因子属于TALE(three amino-acid loop extension)基因家族中的亚家族,在植物中包含多个家族成员,其显著的特征是含有同源异型盒结构域(homeodomain,HD),此外还含有KNOX1、KNOX2 和ELK 结构域[2]。其中HD 结构域参与蛋白互作的形成和与DNA 的结合[3]。KNOX1 和KNOX2 结构域被称为MEINOX 结构域,对转基因植株表型的变化起到重要作用[3]。ELK结构域编码核定位信号,在与其他蛋白互作和转录抑制过程中也起到一定作用[3]。多项研究表明,KNOX 蛋白能与BLH 蛋白相互作用形成不同组合的异质二聚体,某些异质二聚体还可与MADS-box 或者OVATE family proteins(OFP)转录因子相互作用形成功能复合体,调控植物的生长和发育过程[4]。基于KNOX 基因的序列相似性、结构特征、系统进化关系以及表达模式,可将KNOX 蛋白分为2 个亚家族:Class Ⅰ(KNOX Ⅰ)和Class Ⅱ(KNOX Ⅱ)[2,5-8]。进一步的研究表明,在蒺藜苜蓿(Medicago truncatula L.)、番茄(Solanum lycopersicum L.)和拟南芥(Arabidopsis thaliana L.Heynh.)中鉴定到一个新的KNOX 蛋白亚家族KNOXM,与KNOX Ⅰ和KNOX Ⅱ亚家族不同的是其本身缺少ELK 和HD 结构域[9-11]。KNOX Ⅰ能够进一步分为3 个亚组:STM-like、KNAT2/6-like 和KNAT1/BPlike,而KNOX Ⅱ分为2 个亚组:KNAT7-like 和KANT3/4/5-like[1]。通过拟南芥突变体研究发现,KNOX 基因不仅调控植物细胞增殖和组织分化,还参与调节细胞分裂素和赤霉素信号途径[1,7,12]。拟南芥KNOX Ⅰ亚组包含4 个基因:SHOOTMERISTMELESS(STM)以及KNAT1/2/6。其中,STM 基因在茎顶端分生组织(shoot apical meristem,SAM)早期胚胎发生期间表达,对SAM 形成的起始起到重要作用[12]。KNAT1 和KNAT6 在SAM 功能的形成和花序发育过程中扮演着重要角色[13-14]。KNAT2对花模型的调控起到重要作用[15]。相比KNOX Ⅰ的广泛研究,KNOX Ⅱ已知突变体缺乏表型,研究相对较少,在拟南芥中发现KNAT7 基因在调控次级细胞壁合成转录网络中起到一定作用[16-17]。最近研究表明,KNAT3/4/5 这3 个基因与KNOX Ⅰ成员起到相反的作用,功能冗余地促进结瘤器官的分化[8]。此外,MtKNAT3/4/5-like 基因参与共生根瘤的发育,并可调节豆科植物根瘤细胞分裂素生物合成[18]。在苹果(Malus domestica Borkh.)中发现Md-KNOX15 可通过调节赤霉素水平调控苹果株高和开花[19]。MdKNOX19 能够靶向ABI5 调节ABA 敏感性和种子萌发[20]。
Jia 等[21]通过对苹果基因组数据库序列比对,发现苹果基因组中含有22 个MdKNOX 基因。去除已克隆的MdKNOX15和MdKNOX19基因,笔者获得了7 个苹果MdKNOX 基因,进一步对它们进行了生物信息学、组织器官表达、盐和渗透胁迫表达以及蛋白相互作用分析,为苹果KNOX 转录因子在生长、发育和逆境下生物学功能的解析、调控网络的构建提供了强有力的理论基础和参考。
1 材料和方法
1.1 植物材料与处理方法
盐和渗透胁迫表达分析供试材料为嘎拉组培苗。嘎拉组培苗培养条件和处理方法参照李慧峰等[22]的文献描述进行操作。将嘎拉组培苗放在液氮中速冻以备提取RNA。
1.2 MdKNOX基因克隆
MdKNOX 基因克隆所采用的模板为完全展开的紫弘富士苹果叶片,采用改良热硼酸法进行RNA的提取。嘎拉组培苗RNA 的提取采用QIAGEN 公司的RNA 提取试剂盒RNeasy Plant Mini Kit(货号:74903)。cDNA 的合成采用TaRaKa 公司的反转录试剂盒PrimeScriptTM 1st Strand cDNA Synthesis Kit(货号:6110A)。依据苹果MdKNOX 核苷酸序列使用Primer 3在线软件(https://primer3.ut.ee/)设计Md-KNOX 基因特异性引物(表1)进行RT-PCR 扩增。PCR 反应条件参照Dong 等[23]文献描述进行操作。对PCR反应液进行胶回收后,将目的基因克隆片段导入到pMD18-T 克隆载体上转化Escherichia coli DH5α感受态细胞,涂板后筛选阳性克隆,把摇菌送到公司进行测序。
表1 载体构建和PCR 所用引物
Table 1 Primers for vector construction and PCR
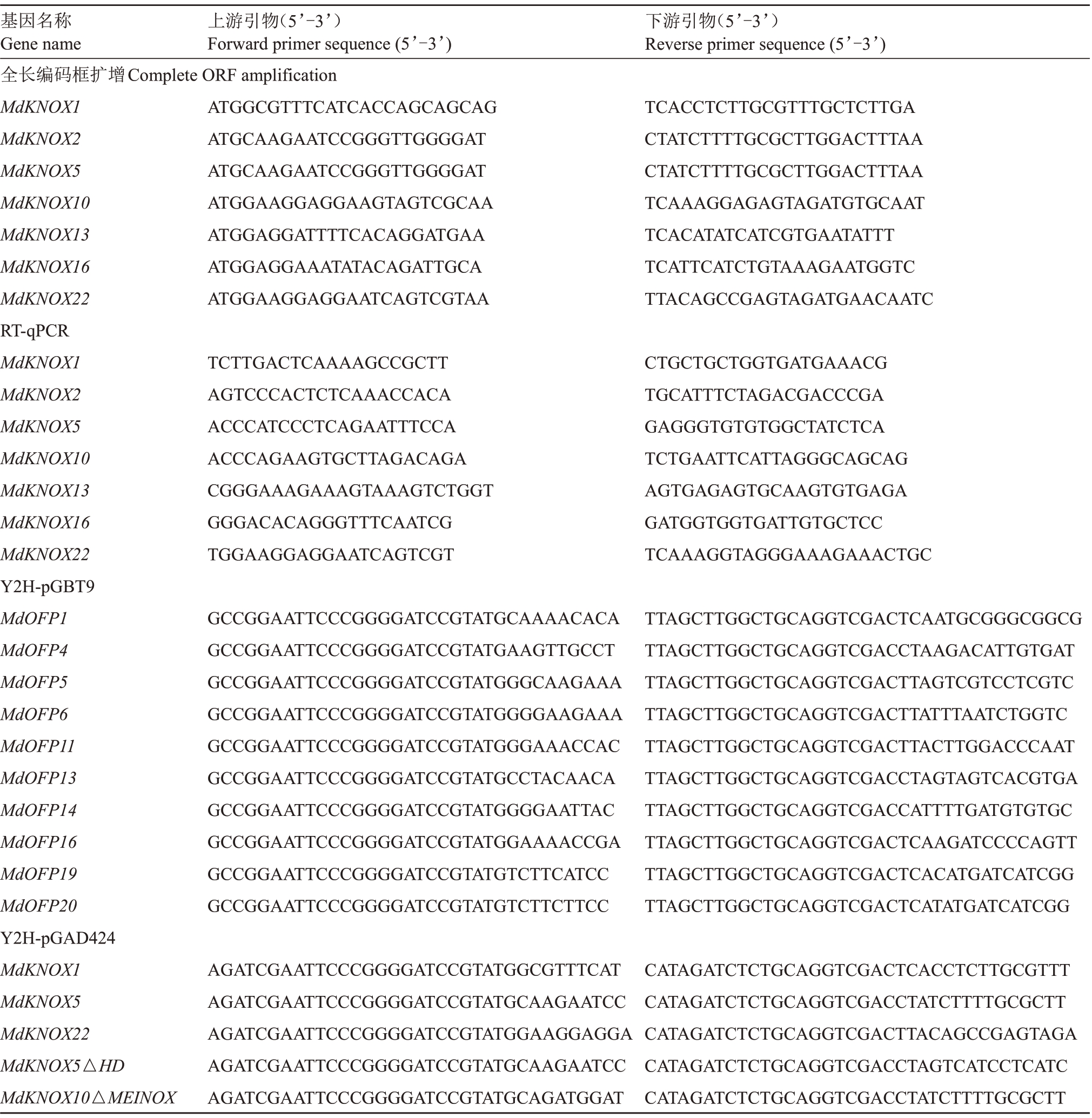
基因名称Gene name全长编码框扩增Complete ORF amplification MdKNOX1 MdKNOX2 MdKNOX5 MdKNOX10 MdKNOX13 MdKNOX16 MdKNOX22 RT-qPCR MdKNOX1 MdKNOX2 MdKNOX5 MdKNOX10 MdKNOX13 MdKNOX16 MdKNOX22 Y2H-pGBT9 MdOFP1 MdOFP4 MdOFP5 MdOFP6 MdOFP11 MdOFP13 MdOFP14 MdOFP16 MdOFP19 MdOFP20 Y2H-pGAD424 MdKNOX1 MdKNOX5 MdKNOX22 MdKNOX5△HD MdKNOX10△MEINOX上游引物(5’-3’)Forward primer sequence(5’-3’)ATGGCGTTTCATCACCAGCAGCAG ATGCAAGAATCCGGGTTGGGGAT ATGCAAGAATCCGGGTTGGGGAT ATGGAAGGAGGAAGTAGTCGCAA ATGGAGGATTTTCACAGGATGAA ATGGAGGAAATATACAGATTGCA ATGGAAGGAGGAATCAGTCGTAA TCTTGACTCAAAAGCCGCTT AGTCCCACTCTCAAACCACA ACCCATCCCTCAGAATTTCCA ACCCAGAAGTGCTTAGACAGA CGGGAAAGAAAGTAAAGTCTGGT GGGACACAGGGTTTCAATCG TGGAAGGAGGAATCAGTCGT GCCGGAATTCCCGGGGATCCGTATGCAAAACACA GCCGGAATTCCCGGGGATCCGTATGAAGTTGCCT GCCGGAATTCCCGGGGATCCGTATGGGCAAGAAA GCCGGAATTCCCGGGGATCCGTATGGGGAAGAAA GCCGGAATTCCCGGGGATCCGTATGGGAAACCAC GCCGGAATTCCCGGGGATCCGTATGCCTACAACA GCCGGAATTCCCGGGGATCCGTATGGGGAATTAC GCCGGAATTCCCGGGGATCCGTATGGAAAACCGA GCCGGAATTCCCGGGGATCCGTATGTCTTCATCC GCCGGAATTCCCGGGGATCCGTATGTCTTCTTCC AGATCGAATTCCCGGGGATCCGTATGGCGTTTCAT AGATCGAATTCCCGGGGATCCGTATGCAAGAATCC AGATCGAATTCCCGGGGATCCGTATGGAAGGAGGA AGATCGAATTCCCGGGGATCCGTATGCAAGAATCC AGATCGAATTCCCGGGGATCCGTATGCAGATGGAT下游引物(5’-3’)Reverse primer sequence(5’-3’)TCACCTCTTGCGTTTGCTCTTGA CTATCTTTTGCGCTTGGACTTTAA CTATCTTTTGCGCTTGGACTTTAA TCAAAGGAGAGTAGATGTGCAAT TCACATATCATCGTGAATATTT TCATTCATCTGTAAAGAATGGTC TTACAGCCGAGTAGATGAACAATC CTGCTGCTGGTGATGAAACG TGCATTTCTAGACGACCCGA GAGGGTGTGTGGCTATCTCA TCTGAATTCATTAGGGCAGCAG AGTGAGAGTGCAAGTGTGAGA GATGGTGGTGATTGTGCTCC TCAAAGGTAGGGAAAGAAACTGC TTAGCTTGGCTGCAGGTCGACTCAATGCGGGCGGCG TTAGCTTGGCTGCAGGTCGACCTAAGACATTGTGAT TTAGCTTGGCTGCAGGTCGACTTAGTCGTCCTCGTC TTAGCTTGGCTGCAGGTCGACTTATTTAATCTGGTC TTAGCTTGGCTGCAGGTCGACTTACTTGGACCCAAT TTAGCTTGGCTGCAGGTCGACCTAGTAGTCACGTGA TTAGCTTGGCTGCAGGTCGACCATTTTGATGTGTGC TTAGCTTGGCTGCAGGTCGACTCAAGATCCCCAGTT TTAGCTTGGCTGCAGGTCGACTCACATGATCATCGG TTAGCTTGGCTGCAGGTCGACTCATATGATCATCGG CATAGATCTCTGCAGGTCGACTCACCTCTTGCGTTT CATAGATCTCTGCAGGTCGACCTATCTTTTGCGCTT CATAGATCTCTGCAGGTCGACTTACAGCCGAGTAGA CATAGATCTCTGCAGGTCGACCTAGTCATCCTCATC CATAGATCTCTGCAGGTCGACCTATCTTTTGCGCTT
1.3 苹果KNOX转录因子蛋白序列、进化关系和顺式作用元件分析
使用苹果、拟南芥和水稻全长KNOX氨基酸序列,采用MEGA 6 软件(http://www.megasoftware.net)进行进化树分析。软件具体参数:NJ方法、pairwise deletion、Poission correction 和bootstrap(1000 repeat)[24]。利用序列分析软件DNAMAN 6.0.3.99分析苹果KNOX全长蛋白序列。MdKNOX基因启动子上含有的顺式作用元件利用PlantCARE网站(http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)进行分析。
1.4 MdKNOX的表达分析
根据NCBI 网站中GEO 数据库GSE42873 生成苹果MdKNOX 组织器官表达数据。该转录谱(Array)包含10 个不同苹果基因型的16 个器官组织数据。通过TIGR MeV v4.8.1软件进行表达热图的绘制。MdKNOX 基因在盐和渗透胁迫下的表达分析采用实时荧光定量PCR(RT-qPCR)进行检测,使用MdMDH 基因作为苹果内参基因,相对表达水平结果采用2–ΔΔCT法进行计算[25]。RT-qPCR 反应体系和程序参照李慧峰等[22]的文献描述进行操作。Md-KNOX基因的荧光定量引物见表1。
1.5 酵母双杂交(Y2H)实验
Y2H方法参照Clontech公司公布的方法进行操作。将MdKNOX1、MdKNOX5和MdKNOX22全长编码框、MdKNOX5 删除MEINOX 结构域片段和Md-KNOX5 删除HD 结构域片段插入到pGAD424(GAL4 activation domain,AD)捕获载体中;将MdOFP1/4/5/6/11/13/14/16/19/20全长编码框克隆到pGBT9(GAL4 DNA-binding domain,BD)诱饵载体中[26]。将不同融合载体以AD和BT配对形式分别转化酵母菌株Y2HGold,然后涂布在二缺培养基SD/-Trp/-Leu和四缺培养基SD/-Trp/-Leu/-His/-Ade+X-α-Gal 中进行培养,通过观察菌体是否变蓝判断蛋白是否存在相互作用。采用pGBT9-MdOFP 和pGBT9-MdWKRY52融合载体与pGAD424空载体质粒共转化的酵母菌作为阴性对照;采用pGBT9-Md-WRKY52 和pGAD424-MdVQ10 融合载体共转化的酵母菌作为阳性对照[23]。
1.6 数据分析
MdKNOX 基因在盐和渗透胁迫处理下的表达水平采用IBM SPSS Statistics v.20 软件中One-way ANOVA 方法及Duncan 检验(p<0.05)进行差异显著性分析。
2 结果与分析
2.1 苹果MdKNOX基因的克隆
使用紫弘富士叶片的cDNA 作为PCR 反应模板,进行RT-PCR扩增。测序结果显示,7个苹果Md-KNOX基因克隆成功。根据前人对MdKNOX基因家族鉴定的研究结果,对7 个苹果MdKNOX 基因进行命名,各个基因的GenBank登录号、基因组ID号、染色体定位、开放阅读框、分子质量以及等电点信息详见表2。DNAMAN 软件分析结果显示,7 个Md-KNOX转录因子含有典型的KNOX1、KNOX2、ELK和HD结构域(图1)。
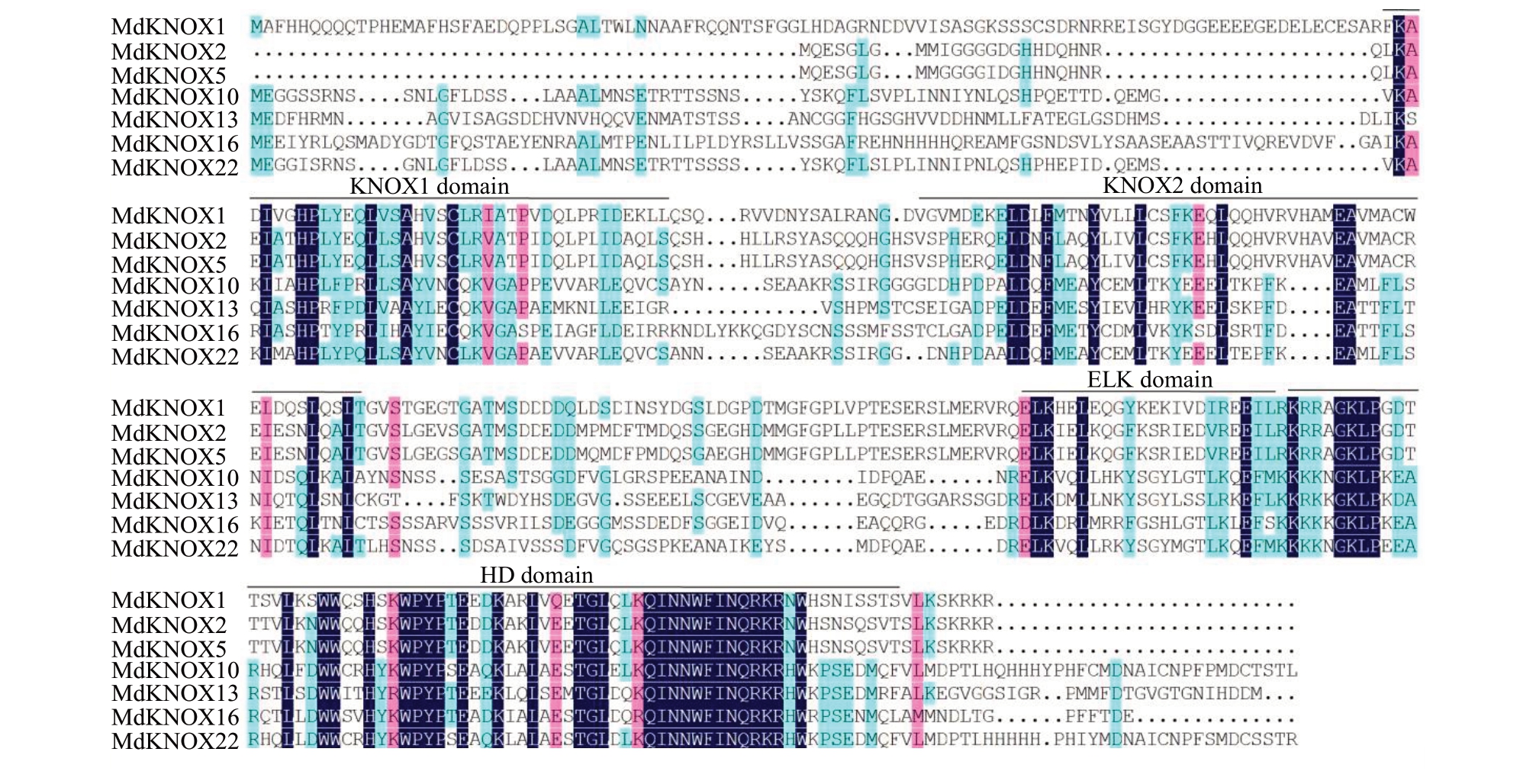
图1 苹果MdKNOX 氨基酸序列同源性比对
Fig.1 Homology comparison of the deduced amino acid sequence alignment of cloned MdKNOX proteins
不同颜色字体代表相同或相似氨基酸残基。KNOX1、KNOX2、ELK 和HD 保守结构域使用上划线标出。
Different color-boxed letters represent identical or similar residues.The conserved KNOX1,KNOX2,ELK and HD domains have been underlined.
表2 MdKNOX 基因的基本信息
Table 2 Basic information of MdKNOX genes in apple
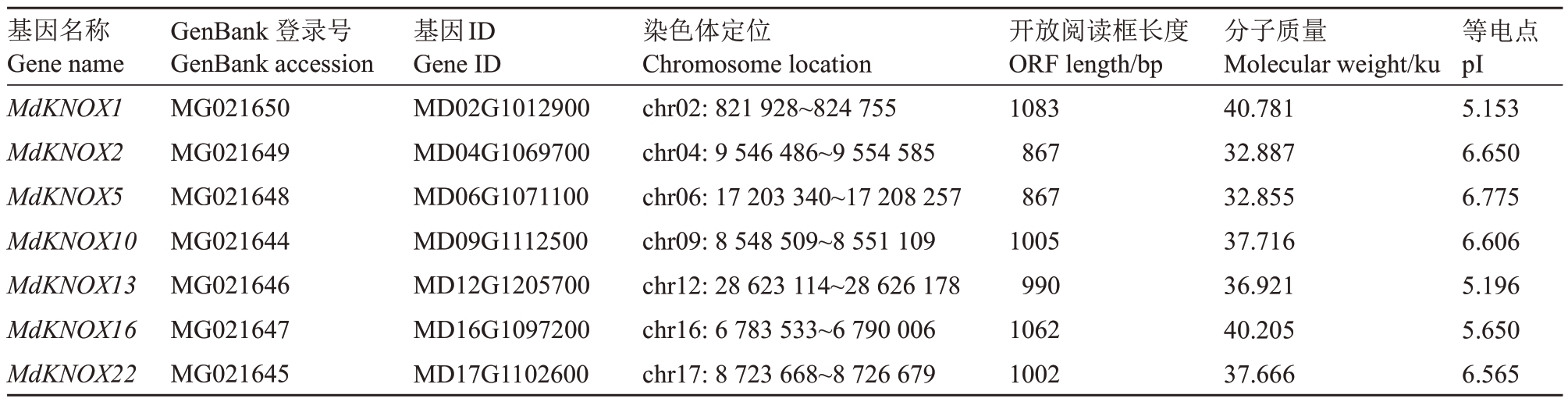
基因名称Gene name MdKNOX1 MdKNOX2 MdKNOX5 MdKNOX10 MdKNOX13 MdKNOX16 MdKNOX22 GenBank 登录号GenBank accession MG021650 MG021649 MG021648 MG021644 MG021646 MG021647 MG021645基因ID Gene ID MD02G1012900 MD04G1069700 MD06G1071100 MD09G1112500 MD12G1205700 MD16G1097200 MD17G1102600染色体定位Chromosome location chr02:821 928~824 755 chr04:9 546 486~9 554 585 chr06:17 203 340~17 208 257 chr09:8 548 509~8 551 109 chr12:28 623 114~28 626 178 chr16:6 783 533~6 790 006 chr17:8 723 668~8 726 679开放阅读框长度ORF length/bp 1083 867 867 1005 990 1062 1002分子质量Molecular weight/ku 40.781 32.887 32.855 37.716 36.921 40.205 37.666等电点pI 5.153 6.650 6.775 6.606 5.196 5.650 6.565
2.2 苹果KNOX转录因子的进化分析
利用进化树分析软件MEGA6 对9 个苹果Md-KNOX(7 个本文克隆的MdKNOX、MdKNOX15 和MdKNOX19)、9 个拟南芥KNOX 和11 个水稻(Oryza sativa L.)KNOX 全长蛋白序列进行进化分析。图2结果显示,KNOX成员被清楚地分为3个亚家族(KNOX Ⅰ、KNOX Ⅱ和KNOX M),其中KNOX Ⅰ中可进一步分为3 个亚组(STM-like、KNAT2/6-like和KNAT1/BP- like),KNOX Ⅱ分为2 个亚组(KNAT7-like和KANT3/4/5-like)。STM-like亚组中包含MdKNOX10 和MdKNOX22;KNAT2/6-like 亚组中包含 MdKNOX13、MdKNOX15 和 Md-KNOX16;KNAT7-like 亚组中包含MdKNOX2 和MdKNOX5;KNAT3/4/5-like亚组中包含MdKNOX1和MdKNOX19。
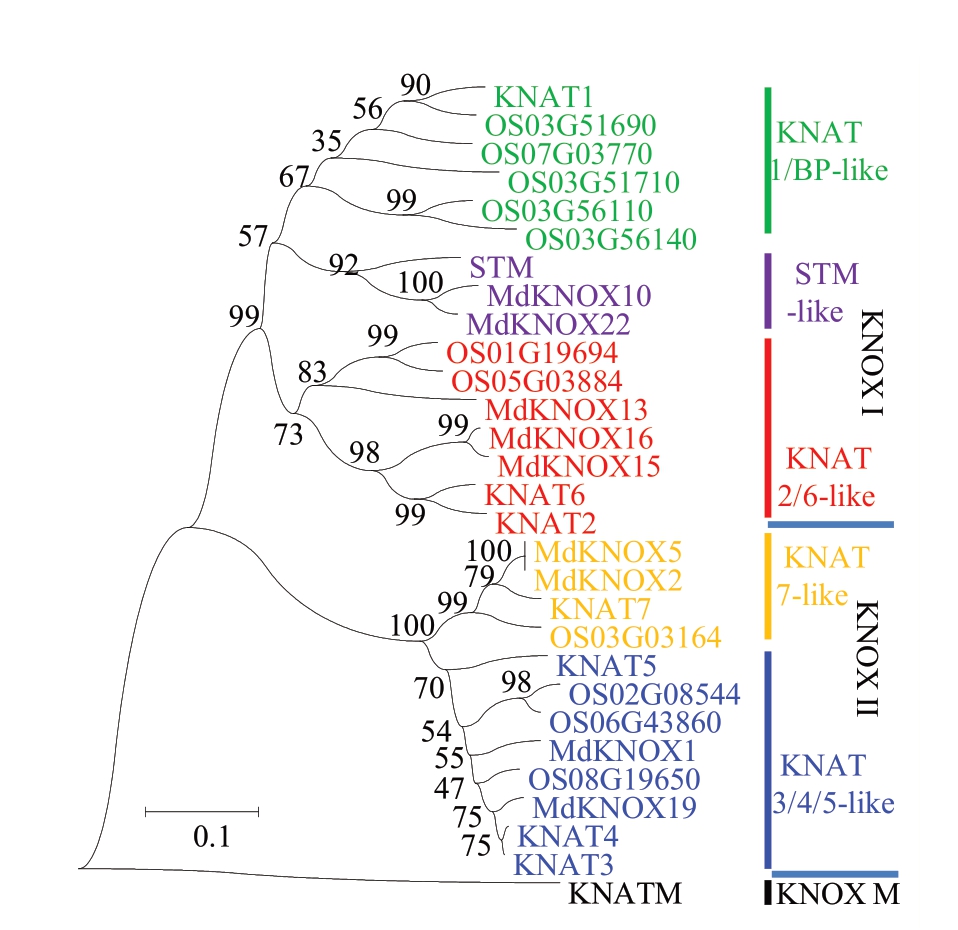
图2 苹果、拟南芥和水稻KNOX 蛋白的进化分析
Fig.2 Phylogenetic relationship of KNOX proteins in apple,Arabidopsis and rice
利用29 个苹果、拟南芥和水稻KNOX 蛋白全长序列,使用MEGA 6 软件构建进化树。
The phylogenetic tree was constructed with MEGA 6 software using full-length amino acid sequences from the 29 KNOX proteins of apple,
Arabidopsis and rice.
2.3 苹果MdKNOX启动子序列分析
下载已克隆MdKNOX 基因翻译起始位点上游1500 bp序列,获得了7个MdKNOX基因的启动子序列。通过PlantCARE数据库分析启动子序列上的顺式作用元件,除了光响应元件,还有多个逆境和激素响应元件(图3)。在7个MdKNOX基因的启动子上总共存在13种不同类型的顺式作用元件,分别是对响应逆境的低氧、低温、热、干旱、机械创伤、病原菌和激素响应的茉莉酸甲酯、水杨酸、生长素、赤霉素、乙烯和ABA 等顺式作用元件(图3)。这些结果表明,MdKNOX启动子上的顺式作用元件可能在苹果生长和发育以及逆境胁迫响应中起到重要的作用。
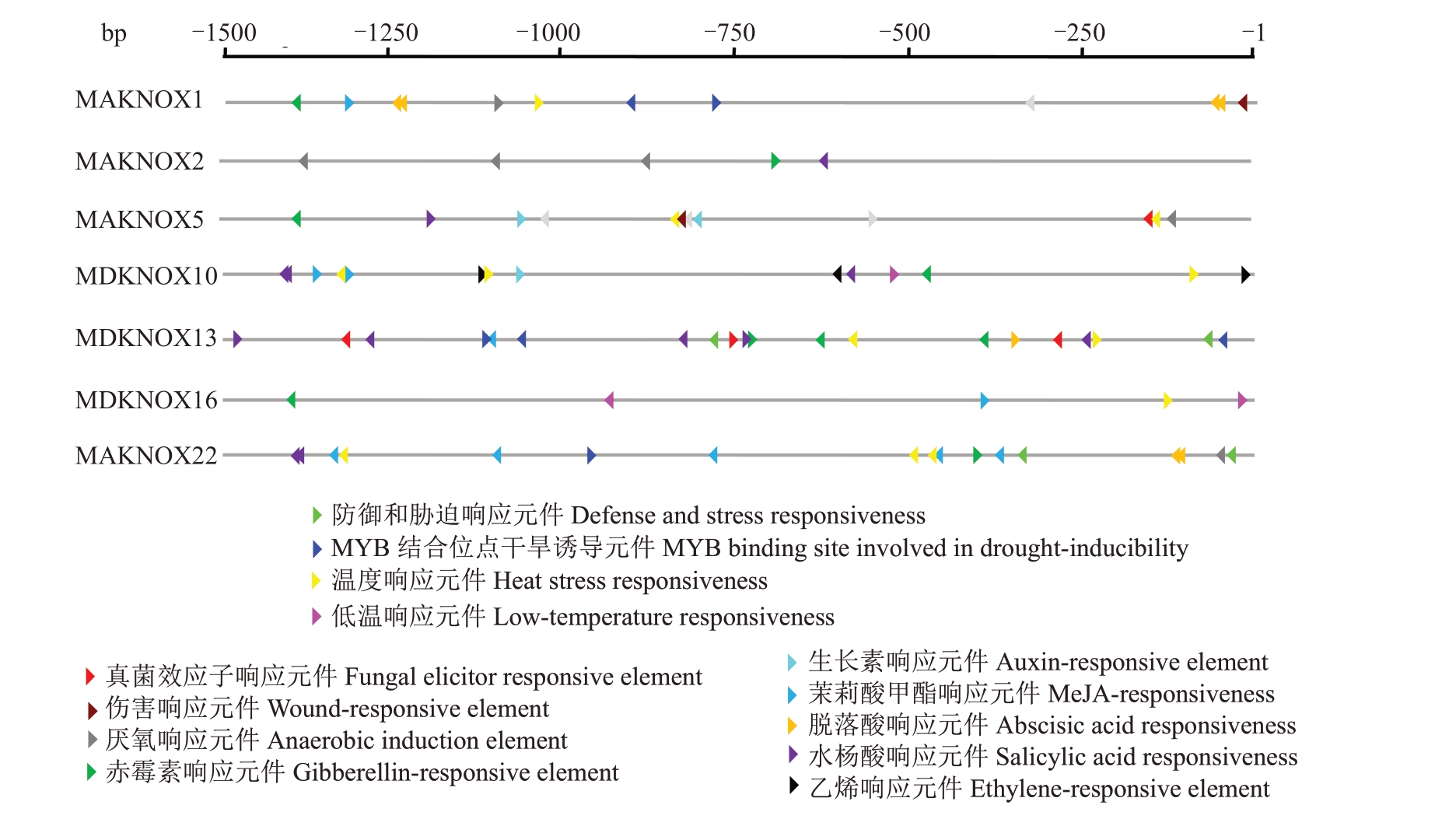
图3 7 个MdKNOX 基因启动子顺式作用元件分析
Fig.3 The cis-acting elements of 7 promoters in MdKNOX genes
2.4 苹果MdKNOX基因的表达分析
利用NCBI 网站的GEO 数据库下载苹果16 个不同组织的转录谱(GSE42873),检测MdKNOX 在不同组织中的表达情况(图4)。结果显示,Md-KNOX基因在被检测的组织中均有表达。其中,Md-KNOX1、MdKNOX2 和MdKNOX5 在被检测的组织中有相对较高的表达水平;在叶_M49(Leaf M49)、花_M74(Flower M74)、果实_M20 花后100 d [Fruit M20(100DAM)]和果实_M20 收获期[Fruit M20(harvest)]组织中,MdKNOX10 、MdKNOX13、Md-KNOX16 和MdKNOX22 基因有相对较高的表达水平(图4)。
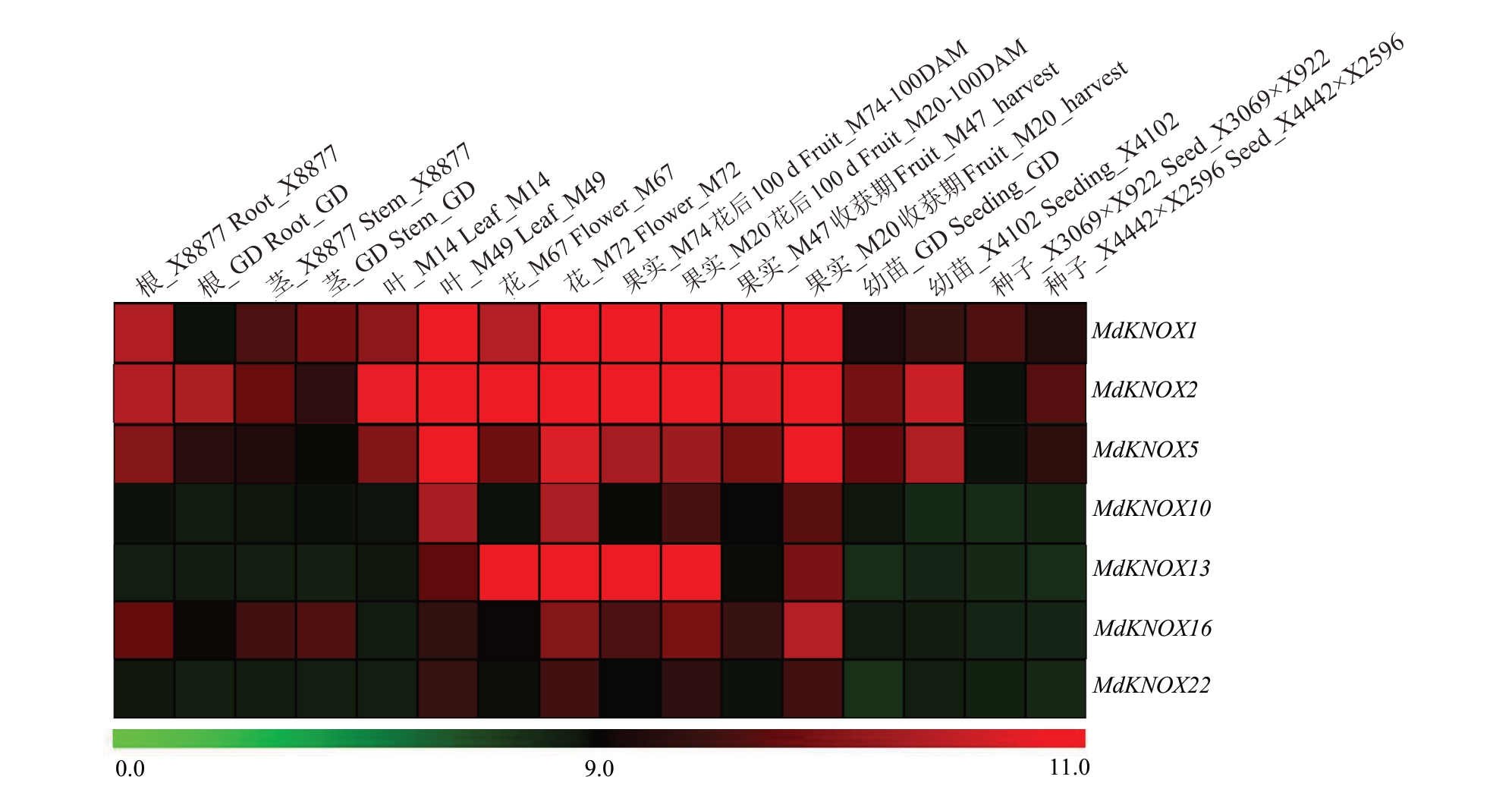
图4 MdKNOX 基因在苹果不同组织中的表达模式
Fig.4 Expression profiles of MdKNOX genes in various tissues
进一步利用荧光实时定量PCR分析盐和甘露醇处理下嘎拉组培苗中MdKNOX基因的表达情况,结果显示,在正常条件下,MdKNOX的相对表达水平无明显变化(图5-A);在NaCl处理条件下,MdKNOX13与对照相比,相对表达水平升高,在处理48 h后相对表达水平达到对照的1.96倍,MdKNOX1、MdKNOX2和MdKNOX5 的相对表达量与对照相比下降,Md-KNOX1 在处理24 h 后相对表达水平为对照的0.28倍;MdKNOX2 和MdKNOX5 在处理48 h 后,相对表达水平分别是对照的0.32和0.29倍(图5-B);在甘露醇处理条件下,MdKNOX2与对照相比,表达水平降低,在48 h时为对照的0.47倍(图5-C)。

图5 MdKNOX 基因在正常生长(A)、盐处理(B)和甘露醇处理下(C)的表达模式
Fig.5 Expression analysis of MdKNOX genes under normal growth(A),salt(B)and mannitol(C)treatments
数据以(平均值±SD)表示,*表示差异显著(p<0.05)。
The data were expressed as(mean±SD),*indicates the difference was significant(p<0.05).
2.5 MdKNOX蛋白与MdOFP蛋白相互作用
笔者前期研究发现苹果基因组中存在26 个MdOFP 基因家族成员,并且成功克隆了10 个MdOFP 基因(MdOFP1/4/5/6/11/13/14/16/19/20)[26]。本研究利用酵母双杂交试验,检测了MdKNOX 蛋白与这10 个MdOFP 蛋白的相互作用情况。3 个MdKNOX 全长cDNA 序列也融合到AD 捕获载体上,同时10个MdOFP全长cDNA序列融合到BD诱饵载体中[26]。将AD-MdKNOX 和BD-MdOFP 融合载体共转化到酵母细胞,通过观察β-galactosidase活性来测试LacZ报告基因的表达。如图6-A所示,作为阴性对照,10 个BD-MdOFP 融合诱饵载体与空AD 捕获载体在四缺培养基上不变蓝,说明这10 个MdOFP 蛋白没有转录自激活活性。MdKNOX1 和MdKNOX22 蛋白能与MdOFP6 蛋白相互作用,MdKNOX5 蛋白能与MdOFP1、MdOFP4、MdOFP14和MdOFP16 蛋白相互作用(图6-A)。为了检测MdKNOX 蛋白哪一段区域对于MdOFP 蛋白相互作用是必需的,构建了MdKNOX10 删除MEINOX结构域捕获载体(AD-MdKNOX10△MEINOX)和MdKNOX10 删除HD 结构域捕获载体(AD-Md-KNOX10△HD)(图6-B)。如图6-C 所示,作为阳性对照,MdVQ10 蛋白和MdWRKY52 蛋白相互作用变蓝[23]。MdKNOX5 △MEINOX 与MdOFP1、MdOFP4、MdOFP14 和MdOFP16 蛋白相互作用,而MdKNOX5△HD与这些蛋白失去相互作用能力,说明MdKNOX5 与MdOFP1、MdOFP4、MdOFP14 和MdOFP16相互作用,HD区域是互作必需的。
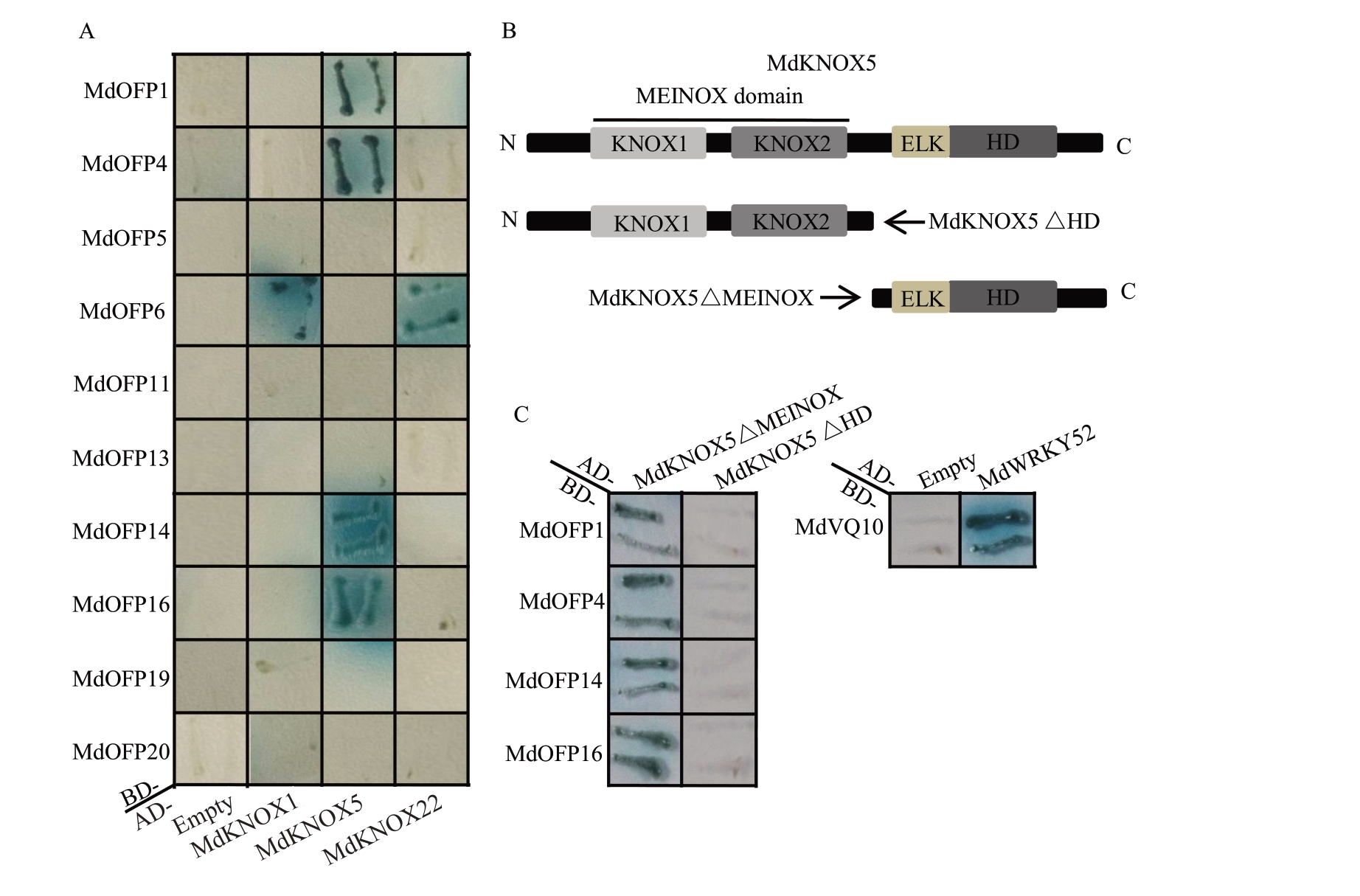
图6 MdKNOX 蛋白和MdOFP 蛋白在酵母细胞中的互作分析
Fig.6 Interactions of MdKNOX proteins with MdOFP proteins in yeast cells
A.AD-MdKNOX 融合捕获载体与BD-MdOFP 融合诱饵载体共转化酵母细胞。酵母细胞在缺乏Leu-Trp-His-Ade 四缺培养基外加x-α-gal上正常生长变蓝代表存在相互作用。空AD 捕获载体加BD-MdOFP 融合诱饵载体作为阴性对照;B.分离MdKNOX5 不同片段用于酵母双杂交实验;C.AD-MdKNOX 片段融合捕获载体与BD-MdOFP 融合诱饵载体互作分析。空AD 捕获载体加BD-MdWRKY52 融合诱饵载体作为阴性对照。AD-MdVQ10 捕获载体加BD-MdWRKY52 融合诱饵载体作为阳性对照。
A.The AD-MdKNOX fusion prey vectors were co-transformed with the BD-MdOFP fusion bait vectors into yeast cells.Postive interactions were indicated by the ability of cells to grow on synthetic dropout medium additive x-α-gal and lacking Leu,Trp,His and Ade.The empty AD prey vector plus BD-MdOFP fusion bait vectors were used as negative controls;B.Names and Locations of MdKNOX5 fragments cloned separately and used for Y2H;C.The fragments of AD-MdKNOX5 interacted with BD-MdOFP1/4/14/16.The empty AD prey vector plus BD-MdWRKY52 fusion bait vectors were used as negative controls.The AD-MdVQ10 plus BD-MdWRKY52 was used as positive control.
3 讨 论
根据植物KNOX 蛋白的高度保守结构域和苹果基因组数据库公布的数据,Jia 等[22]对苹果KNOX基因进行了基因组范围内的家族鉴定,筛选出了22个MdKNOX 基因,随后对MdKNOX15 和Md-KNOX19转录因子进行了功能鉴定[19-20]。本研究根据前人研究结果,克隆了7个MdKNOX基因,对结构域、进化分类和启动子顺式作用元件进行了分析,检测了组织器官表达、非生物胁迫应答以及与MdOFP蛋白互作模式,表明这些MdKNOX基因在苹果调控生长、发育以及应对非生物胁迫过程中起到不同的作用。
研究表明,KNOX蛋白能够与BLH形成异质二聚体或者与OFP转录因子相互作用或者它们3者形成功能复合体参与调控植物生长和发育[12,16,27]。例如,KANT3 与BLH1 形成异质二聚体,AtOFP5 介导它们活性的抑制参与到胚囊的正常发育以及细胞命运的决定[28];KNAT7与OFP4相互作用,增强KNAT7的转录抑制活性调控次生细胞壁的形成[29]。此外,KNAT7 蛋白还可与BLH6 蛋白相互作用,通过抑制homeodomain-leucine zipper transcription factor REVOLUTA/INTERFASCICULAR FIBERLESS1 (REV/IFL1)的表达,进而调控次生细胞壁的形成[16]。进一步研究表明,KNAT7-BLH6 异质二聚体还可与AtOFP1 和AtOFP4 相互作用形成功能复合体,参与到次生细胞壁的形成[30];水稻OsKNAT7 可与Os-OFP2、BLH6-like和BLH-like2相互作用调控维管发育[31]。水稻KNOX 蛋白OSH15 与BEL-like homeodomain 蛋白SH5 相互作用形成二聚体,通过抑制木质素生物合成基因改善落粒性[32];玉米(Zea mays L.)KNOTTED1 与BLH12和BLH14相互作用,在茎中的脉管和节间结构中起到重要作用[27]。此外,KNAT7可与MYB75相互作用在茎和种皮中对细胞壁的形成起到重要的调控作用[33-34]。在本文中,通过酵母双杂交实验,发现MdKNOX1 和MdKNOX22蛋白能与MdOFP6 蛋白相互作用,MdKNOX5 蛋白能与多个MdOFP蛋白相互作用。MdKNOX蛋白与MdOFP 之间的这种不同的相互作用可能影响到苹果可能的BLH-KNOX异质二聚体的活性,从而改变它们所调控靶基因的表达水平,进而调控苹果的生长和发育,但还需进一步的试验来分析这种潜在的相关分子机制。
尽管KNOX 蛋白能够调控植物生长和发育等多个方面,但其对非生物和生物胁迫的响应机制还研究甚少。最近研究表明,在干旱、盐和冷处理条件下,鹰嘴豆(Cicer arietinum Linn.)根和茎中部分KNOX 基因响应胁迫处理应答[35]。在干旱处理下,大豆少数KNOX基因受胁迫响应;在病原菌侵染下,大豆KNOX 基因Glyma17g14180、Glyma04g06810、Glyma09g01000 和Glyma14gg37550 受到不同的响应[35]。在本文中,MdKNOX13 受盐胁迫诱导;Md-KNOX1、MdKNOX2 和MdKNOX5 受盐胁迫下调;MdKNOX2 受甘露醇胁迫下调,并且这些MdKNOX基因启动子上含有多个逆境胁迫顺式作用元件,表明MdKNOX 蛋白可能在应对苹果非生物胁迫中起到一定的作用。相信随着关于KNOX基因研究的深入,其在非生物胁迫中作用和机制也会日益清晰。
4 结 论
本研究克隆获得了苹果中7 个MdKNOX 基因,发现均含有保守的KNOX1、KNOX2、ELK 和HD 结构域。通过进化分析表明7 个MdKNOX 转录因子分别属于KNOX Ⅰ亚组和KNOX Ⅱ亚组。Array结果发现,MdKNOX基因在16个组织中均有不同的表达水平。RT-qPCR 结果表明,MdKNOX13 的相对表达水平受到盐胁迫诱导,而MdKNOX1、MdKNOX2和MdKNOX5的相对表达水平下调;MdKNOX2的转录水平在渗透胁迫处理下下调表达。酵母双杂交分析表明,MdKNOX 蛋白能够与多个MdOFP 蛋白相互作用,且HD 区域是互作必需的。这些结果为苹果KNOX转录因子在生长、发育和逆境下生物学功能的解析、调控网络的构建提供了强有力的理论基础和参考。
[1] HAMANT O,PAUTOT V. Plant development:A tale story[J].Comptes Rendus Biologies,2010,333(4):371-381.
[2] MUKHERJEE K,BROCCHIERI L,BURGLIN T R.A comprehensive classification and evolutionary analysis of plant homeobox genes[J]. Molecular Biology and Evolution,2009,26(12):2775-2794.
[3] SAKAMOTO T,NISHIMURA A,TAMAOKI M,KUBA M,TANAKA H,IWAHORI S,MATSUOKA M. The conserved KNOX domain mediates specificity of tobacco KNOTTED1-type homeodomain proteins[J]. The Plant Cell,1999,11(8):1419-1431.
[4] ARNAUD N,PAUTOT V. Ring the BELL and tie the KNOX:Roles for TALEs in gynoecium development[J]. Frontiers in Plant Science,2014,5:93.
[5] BÜRGLIN T R. Analysis of TALE superclass homeobox genes(MEIS,PBC,KNOX,Iroquois,TGIF) reveals a novel domain conserved between plants and animals[J]. Nucleic Acids Research,1997,25(21):4173-4180.
[6] BELLAOUI M,PIDKOWICH M S,SAMACH A,KUSHALAPPA K,KOHALMI S E,MODRUSAN Z,CROSBY W L,HAUGHN G W.The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals[J].The Plant Cell,2001,13(11):2455-2470.
[7] HAKE S,SMITH H M S,HOLTAN H,MAGNANI E,MELE G,RAMIREZ J. The role of KNOX genes in plant development[J].Annual Review of Cell and Developmental Biology,2004,20:125-151.
[8] FURUMIZU C,ALVAREZ J P,SAKAKIBARA K,BOWMAN J L. Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication[J].PLoS Genetics,2015,11(2):e1004980.
[9] KIMURA S,KOENIG D,KANG J L,YOONG F Y,SINHA N.Natural variation in leaf morphology results from mutation of a novel KNOX gene[J].Current Biology,2008,18(9):672-677.
[10] MAGNANI E,HAKE S. KNOX lost the OX:The Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain[J]. The Plant Cell,2008,20(4):875-887.
[11] PENG J L,YU J B,WANG H L,GUO Y Q,LI G M,BAI G H,CHEN R J.Regulation of compound leaf development in Medicago truncatula by fused compound leaf1,a class M KNOX gene[J].The Plant Cell,2011,23(11):3929-3943.
[12] HAY A,TSIANTIS M. KNOX genes:Versatile regulators of plant development and diversity[J]. Development,2010,137(19):3153-3165.
[13] DOUGLAS S J,CHUCK G,DENGLER R E,PELECANDA L,RIGGS C D. KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis[J].The Plant Cell,2002,14(3):547-558.
[14] RAGNI L,BELLES-BOIX E,GÜNL M,PAUTOT V. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences[J].The Plant Cell,2008,20(4):888-900.
[15] LI Y,PI L M,HUANG H,XU L. ATH1 and KNAT2 proteins act together in regulation of plant inflorescence architecture[J].Journal of Experimental Botany,2012,63(3):1423-1433.
[16] LIU Y Y,YOU S J,TAYLOR-TEEPLES M,LI W L,SCHUETZ M,BRADY S M,DOUGLAS C J. BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA[J].The Plant Cell,2015,26(12):4843-4861.
[17] HE J B,ZHAO X H,DU P Z,ZENG W,BEAHAN C T,WANG Y Q,LI H L,BACIC A,WU A M. KNAT7 positively regulates xylan biosynthesis by directly activating IRX9 expression in Arabidopsis[J]. Journal of Integrative Plant Biology,2018,60(6):514-528.
[18] DI GIACOMO E,LAFFONT C,SCIARRA F,IANNELLI M A,FRUGIER F,FRUGIS G. KNAT3/4/5-like class 2 KNOX transcription factors are involved in Medicago truncatula symbiotic nodule organ development[J]. New Phytologist,2017,213(2):822-837.
[19] JIA P,XING L B,ZHANG C G,CHEN H,LI Y M,ZHANG D,MA J J,ZHAO C P,HAN M Y,REN X L,AN N.MdKNOX15,a class I knotted-like transcription factor of apple,controls flowering and plant height by regulating GA levels through promoting the MdGA2ox7 transcription[J]. Environmental and Experimental Botany,2021,185:104411.
[20] JIA P,XING L B,ZHANG C G,ZHANG D,MA J J,ZHAO C P,HAN M Y,REN X L,AN N. MdKNOX19,a class Ⅱknotted-like transcription factor of apple,plays roles in ABA signalling/sensitivity by targeting ABI5 during organ development[J].Plant Science,2021,302:110701.
[21] JIA P,ZHANG C G,XING L B,LI Y M,SHAH K,ZUO X Y,ZHANG D,AN N,HAN M Y,REN X L.Genome-wide identification of the MdKNOX gene family and characterization of its transcriptional regulation in Malus domestica[J]. Frontiers in Plant Science,2020,11:128.
[22] 李慧峰,张文芹,董庆龙,王小非,冉昆.苹果生长素响应因子(MdARF)基因克隆与表达分析[J]. 果树学报,2018,35(10):1170-1181.LI Huifeng,ZHANG Wenqin,DONG Qinglong,WANG Xiaofei,RAN Kun. Cloning,sequencing and expression analysis of auxin response factors (MdARF) in apple[J]. Journal of Fruit Science,2018,35(10):1170-1181.
[23] DONG Q L,ZHAO S,DUAN D Y,TIAN Y,WANG Y P,MAO K,ZHOU Z S,MA F W. Structural and functional analyses of genes encoding VQ proteins in apple[J]. Plant Science,2018,272:208-219.
[24] TAMURA K,STECHER G,PETERSON D,FILIPSKI A,KUMAR S. MEGA6:Molecular evolutionary genetics analysis version 6.0[J]. Molecular Biology and Evolution,2013,30(12):2725-2729.
[25] LIVAK K J,SCHMITTGEN T D.Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method[J].Methods,2001,25(4):402-408.
[26] LI H F,DONG Q L,ZHAO Q,RAN K. Genome-wide identification,expression profiling,and protein-protein interaction properties of ovate family proteins in apple[J]. Tree Genetics & Genomes,2019,15(3):45.
[27] TSUDA K,ABRAHAM-JUAREZ M J,MAENO A,DONG Z B,AROMDEE D,MEELEY R,SHIROISHI T,NONOMURA K I,HAKE S.KNOTTED1 cofactors,BLH12 and BLH14,regulate internode patterning and vein anastomosis in maize[J]. The Plant Cell,2017,29(5):1105-1118.
[28] PAGNUSSAT G C,YU H J,SUNDARESAN V.Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1[J].The Plant Cell,2007,19(11):3578-3592.
[29] LI E Y,WANG S C,LIU Y Y,CHEN J G,DOUGLAS C J.OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana[J]. The Plant Journal,2011,67(2):328-341.
[30] LIU Y Y,DOUGLAS C J. A role for OVATE FAMILY PROTEIN1 (OFP1) and OFP4 in a BLH6-KNAT7 multi-protein complex regulating secondary cell wall formation in Arabidopsis thaliana[J]. Plant Signaling & Behavior,2015,10(7):e1033126.
[31] SCHMITZ A J,BEGCY K,SARATH G,WALIA H. Rice Ovate Family Protein 2 (OFP2) alters hormonal homeostasis and vasculature development[J].Plant Science,2015,241:177-188.
[32] YOON J,CHO L H,ANTT H W,KOH H J,AN G.KNOX protein OSH15 induces grain shattering by repressing lignin biosynthesis genes[J].Plant Physiology,2017,174(1):312-325.
[33] BHARGAVA A,MANSFIELD S D,HALL H C,DOUGLAS C J,ELLIS B E. MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem[J]. Plant Physiology,2010,154(3):1428-1438.
[34] BHARGAVA A,AHAD A,WANG S C,MANSFIELD S D,HAUGHN G W,DOUGLAS C J,ELLIS B E. The interacting MYB75 and KNAT7 transcription factors modulate secondary cell wall deposition both in stems and seed coat in Arabidopsis[J].Planta,2013,237(5):1199-1211.
[35] BHATTACHARJEE A,GHANGAL R,GARG R,JAIN M. Genome-wide analysis of homeobox gene family in legumes:Identification,gene duplication and expression profiling[J]. PLoS One,2015,10(3):e0119198.