柑橘是世界上最重要和最广泛种植的果树之一。自然界中柑橘及其近缘种基本为二倍体(2n=2x=18)[1],近年也创制、筛选并积累了大量柑橘多倍体材料[2]。染色体细胞遗传研究不仅是柑橘类种属起源进化分析的重要手段[3-4],更是减数分裂同源或部分同源染色体配对遗传利用研究的直接工具[5-6],还能够辅助全基因组测序的验证[7]。而对特定单条染色体特别是减数分裂前期Ⅰ中单条全长染色体的精准识别,是实现以上研究的关键。
由于缺乏染色体全长特异的DNA 探针,在大多数植物中无法跟踪研究单条染色体[8-9]。柑橘类为小染色体(1~4µm)物种[10],已在其有丝分裂中期细胞上开展了广泛的染色体识别和比较鉴定,包括利用染色体色霉素A3(chromomycin A3,CMA)荧光显带[10-13],以及综合CMA 显带、rDNA、基因组中其他重复序列[14-15]和染色体特异细菌人工染色体(bacterial artificial chromosome,BAC)探针标记的荧光原位杂交(fluorescence in situ hybridization,FISH)[16-18]。然而,以上标记仅识别染色体上的特定区段,无法跟踪研究细胞分裂活动,特别是减数分裂各时期的单条全长染色体,例如涉及同源染色体精细配对的粗线期染色体,它相较于中期Ⅰ的配对分析能更准确评估染色体的同源性[6,19]。另外,虽已经完成部分柑橘种的基因组测序,但精准组装仍受基因组中大量重复序列的干扰,可利用粗线期染色体高分辨率的FISH辅助基因组的组装和验证,特别是含有大量重复序列的着丝粒及附近异染色质区[7]。而中期染色体太小[10],FISH 信号的分辨率极低,难以精准观察测定。
最近,基于寡核苷酸的新型DNA文库已迅速成为“新一代”的植物染色体精准研究FISH 探针[9,19]。利用生物信息学手段,实现了从任何植物物种组装的基因组序列中,筛选出针对染色体特定区域或整个染色体鉴定的寡核苷酸序列,经人工合成后标记作为染色体鉴定的特异FISH 探针[19]。利用覆盖整个染色体的寡核苷酸探针能够产生“染色体涂染(chromosome painting)”信号,实现了有丝及减数分裂中特定单条染色体的精准追踪[4-6,19-21]。笔者已在柑橘中研创了所有9条全长染色体特异的寡核苷酸涂染探针,准确鉴定了有丝分裂中期所有染色体[4]。笔者在本研究中旨在利用柑橘1 号染色体的特异涂染FISH探针,首次进行沙田柚减数分裂粗线期染色体的跟踪研究;同时以有丝分裂不同浓缩阶段的染色质做对比,揭示粗线期与有丝分裂各时期染色体结构及FISH信号分辨率的巨大差异,为精准解析粗线期染色体结构和特定区段序列组成,辅助全基因组特别是测序组装困难的着丝粒及附近异染色质区的测序组装验证提供新工具,也为染色体的配对遗传利用,特别是柑橘多倍体减数分裂粗线期染色体的精准研究奠定新的方法基础。
1 材料和方法
1.1 植物材料
选取柑橘类代表种沙田柚[Citrus maxima(Burm.)Merr.‘Shatian pummelo’,2n=2x=18]用作染色体涂染分析。植物材料保存于四川省农业科学院园艺研究所成都试验基地。
1.2 染色体特异涂染探针制备
前期,参照Albert 等[20]的方法开发了柑橘染色体特异涂染探针[4]。利用Chorus2 软件(https://github.com/zhangtaolab/Chorus2)从Citrus maxima的参考基因组中(https://citrus.hzau.edu.cn/orange/index.php)筛选1号染色体上的长45个碱基的单拷贝寡核苷酸。利用Citrus maxima 的基因组序列数据(SRR4294213)[22]过滤重复寡核苷酸序列。寡核苷酸探针文库由Arbor Biosciences(Ann Arbor, 密歇根,美国)合成。按照Han等[19]的方法用地高辛标记染色体涂染探针。
1.3 有丝分裂中期染色体制片
以根尖为材料制备有丝分裂中期染色体制片。截取新鲜的根尖在1.10 mPa压力下用N2O处理2 h,然后在固定液(3 倍乙醇/1 倍冰醋酸)中固定备用。根尖用蒸馏水洗涤10 min 后,用2%(w,后同)纤维素酶(Yakult Pharmaceutical,日本)和1%果胶酶(Sigma,美国)在37 ℃水浴锅中酶解140 min。采用滴片法制取有丝分裂中期染色体[4]。
1.4 减数分裂粗线期染色体制片
以花蕾为材料制取减数分裂粗线期染色体。选新鲜并未变白的花蕾,用固定液(3倍乙醇/1倍冰醋酸)固定备用。用花蕾中的1 个花药压片镜检花粉母细胞是否处于粗线期,其余花药用作酶解制片。花药用蒸馏水洗涤20 min,后用3%纤维素酶(Yakult Pharmaceutical,日本)和2%果胶酶(Sigma,美国)的酶解液在37 ℃水浴锅中酶解5 h。采用滴片法制取粗线期染色体[6]。
1.5 寡核苷酸荧光原位杂交(Oligo-FISH)
参照He等[4]的方法开展Oligo-FISH和检测。杂交液含寡核苷酸探针200 ng,去离子甲酰胺10µL,50%硫酸葡聚糖4 µL,20×SSC 2 µL,加蒸馏水至20µL。地高辛标记的探针用抗地高辛异硫氰酸荧光 素(fluorescein isothiocyanate isomer,FITC)(Roche Diagnostics,美国)检测。用含4’,6-二脒基-2-苯基吲哚(4’, 6-diamidino-2-phenylindole,DAPI)的VectaShield 抗淬灭剂(Vector Laboratories,美国)对染色体进行复染。用蔡司Axio Scope A1 荧光显微镜上耦合的电荷耦合器件(charge coupled device,CCD)相机(ORCA Flash4.0,滨松,日本)拍摄FISH图像。用Adobe Photoshop 5.0调整图像的亮度和对比度。
用Image J 软件测定中期和粗线期染色体的长度,并用其中的“Straighten”插件拉直弯曲的粗线期染色体。
2 结果与分析
2.1 柚有丝分裂中期染色体涂染鉴定
笔者前期利用Chorus2 软件(https://github.com/zhangtaolab/Chorus2),以柚基因组DNA 序列开发了柑橘类物种所有9 条染色体的特异性涂染探针[4]。为了筛选能够均匀覆盖1 号染色体的寡核苷酸,从1号染色体的DNA序列中筛选了27 392个长为45 bp的单拷贝寡核苷酸序列(寡核苷酸分布密度为0.85个·kb-1)作为混合池,用作该染色体鉴定的特异涂染探针。
经绿色荧光检测,在柚有丝分裂中期细胞中,能清楚鉴定出1 号染色体的2 条同源染色体(图1)。然而,FISH信号在1号染色体上的分布并不均匀,部分区域没有观察到信号或信号非常微弱(图1-B)。这可能是1号染色体对应区域单拷贝寡核苷酸的密度相对较低(小于0.1个·kb-1)引起的。
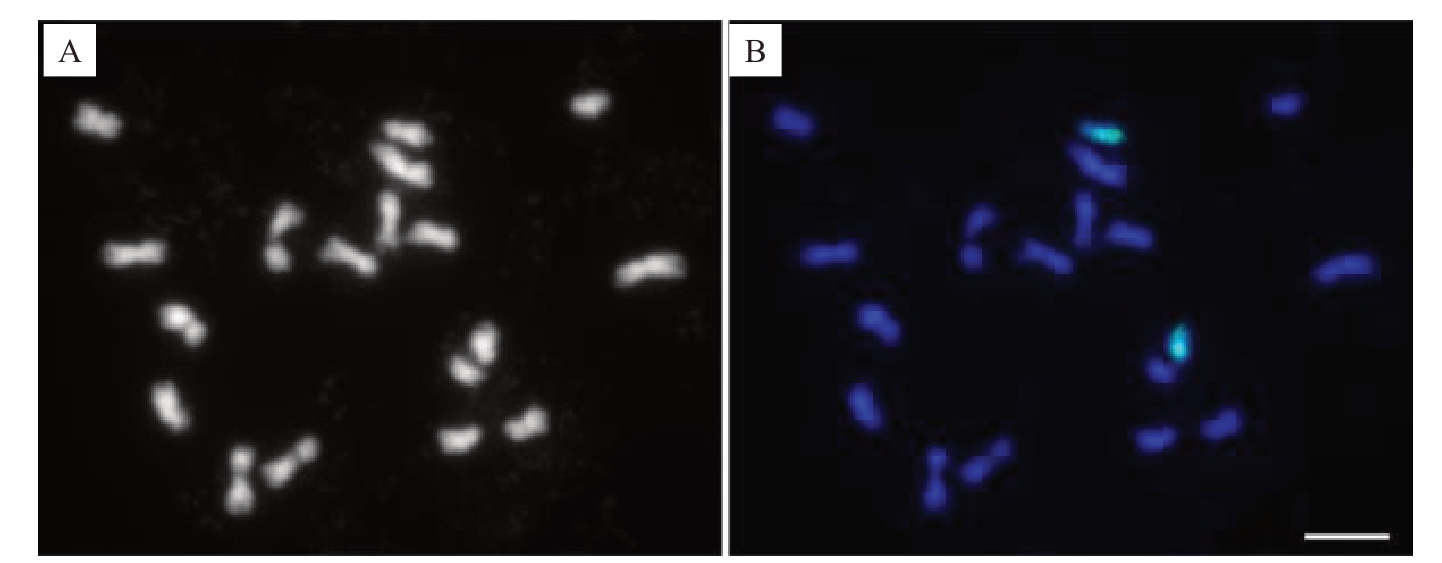
图1 柚有丝分裂中期细胞1 号染色体的Oligo-FISH 涂染鉴定
Fig.1 Identification of chromosome 1 in a mitotic metaphase cell of Citrus maxima by Oligo-FISH painting
A.单色显示DAPI 染色的柚中期细胞染色体;B.柑橘1 号染色体特异涂染探针(绿色)在蓝色显示来自A 中柚染色体的FISH 涂染。标尺为5µm。
A. DAPI stained metaphase chromosomes of C. maxima that were shown in monochrome; B. FISH painting signals (green) from Citrus chromosome 1-specific probe shown on blue stained chromosomes in A.Bar=5µm.
2.2 间期细胞核及前中期染色体可视化
为了识别中期染色体外其他不同分裂阶段浓缩的染色质,并展现可视化单个染色体占据的区域,利用1号染色体特异涂染探针开展了FISH(图2)。对柚根尖细胞进行FISH涂染,结果清晰显示细胞核中的2个同源染色体分别位于独立的区域(图2-B),且染色质呈离散状态,浓缩程度极低。前中期细胞上的探针信号显示,每个同源染色体相较间期时更浓缩,但相对中期时浓缩程度更低。中部无信号或信号微弱的染色体区段清晰可见(图2-D,箭头)。然而,此时染色体各部位的特征,特别是着丝粒不易辨认。试验表明,特异探针涂染能有效示踪有丝分裂各周期的1号染色体。
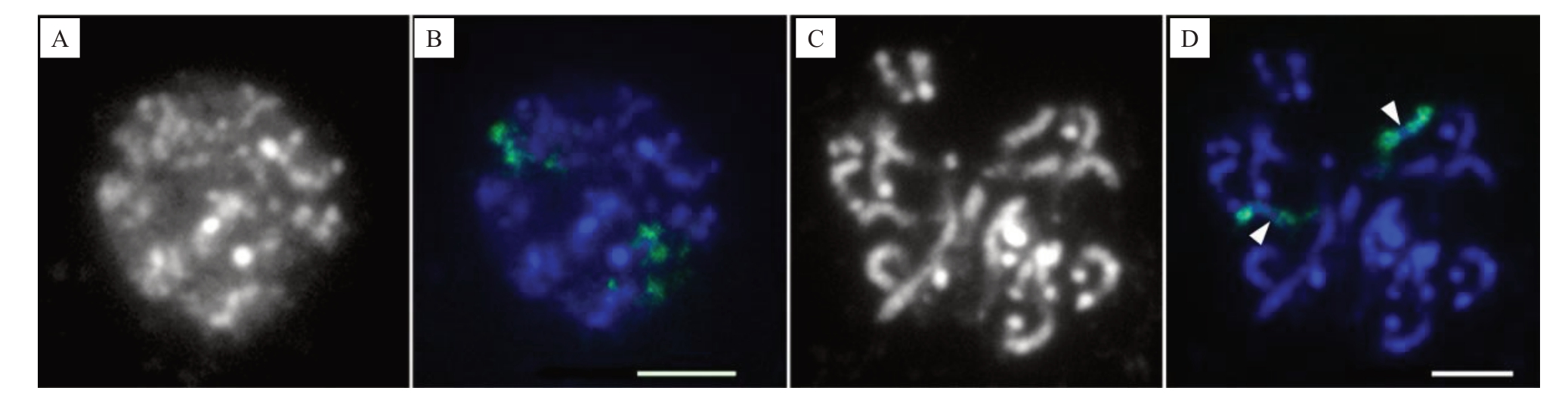
图2 柑橘1 号染色体特异探针在柚有丝分裂间期细胞核和前中期染色体的FISH 涂染
Fig.2 FISH painting on mitotic interphase nuclei and prometaphase chromosomes of C.maxima
with Citrus chromosome 1-specific probe
A.单色显示DAPI 染色的柚间期细胞核;B.柑橘1 号染色体涂染探针(绿色)在用蓝色显示来自A 中柚细胞核上的FISH 涂染;C.单色显示DAPI 染色的柚前中期染色体;D.柑橘1 号染色体涂染探针(绿色)在蓝色显示来自C 中柚前中期染色体上的FISH 涂染。箭头指示染色体上信号微弱的区段。标尺为5µm。
A.A DAPI stained interphase nuclei of C. maxima that was shown in monochrome; B. FISH painting signals (green) from Citrus chromosome 1-specific probe displayed in the interphase nuclei of C. maxima that were pseudo colored in blue from A; C.A DAPI stained prometaphase cell of C.maxima that was shown in monochrome; D. FISH painting signals (green) from Citrus chromosome 1-specific probe shown in the blue stained prometaphase cell(blue)of C.maxima in C.Arrow heads denote regions showing weak signals.Bar=5µm.
2.3 减数分裂粗线期染色体Oligo-FISH涂染
为了精准示踪减数分裂粗线期1 号染色体,利用特异涂染探针(图3)进行了FISH 标记。除着丝粒及紧邻的亚着丝粒小区段外,足够密度的探针信号将该染色体从末端到末端标记了出来,清晰地展示了粗线期2条1号同源染色体从末端到末端充分配对的情况。利用探针信号(图3-B),清晰展示了减数分裂粗线期的状态,即使在众多染色体相互缠绕以致几乎无法辨认其形态的情况下(图3-A),也能够准确反映特定粗线期染色体在细胞中的分布及配对情况,并进行精准跟踪研究。在15个粗线期细胞中观察到,1号染色体的2条同源染色体均表现从末端到末端的完整配对,展示了它们之间高度的同源性。结果表明,采用特异探针涂染能精准有效示踪粗线期染色体及同源染色体配对。
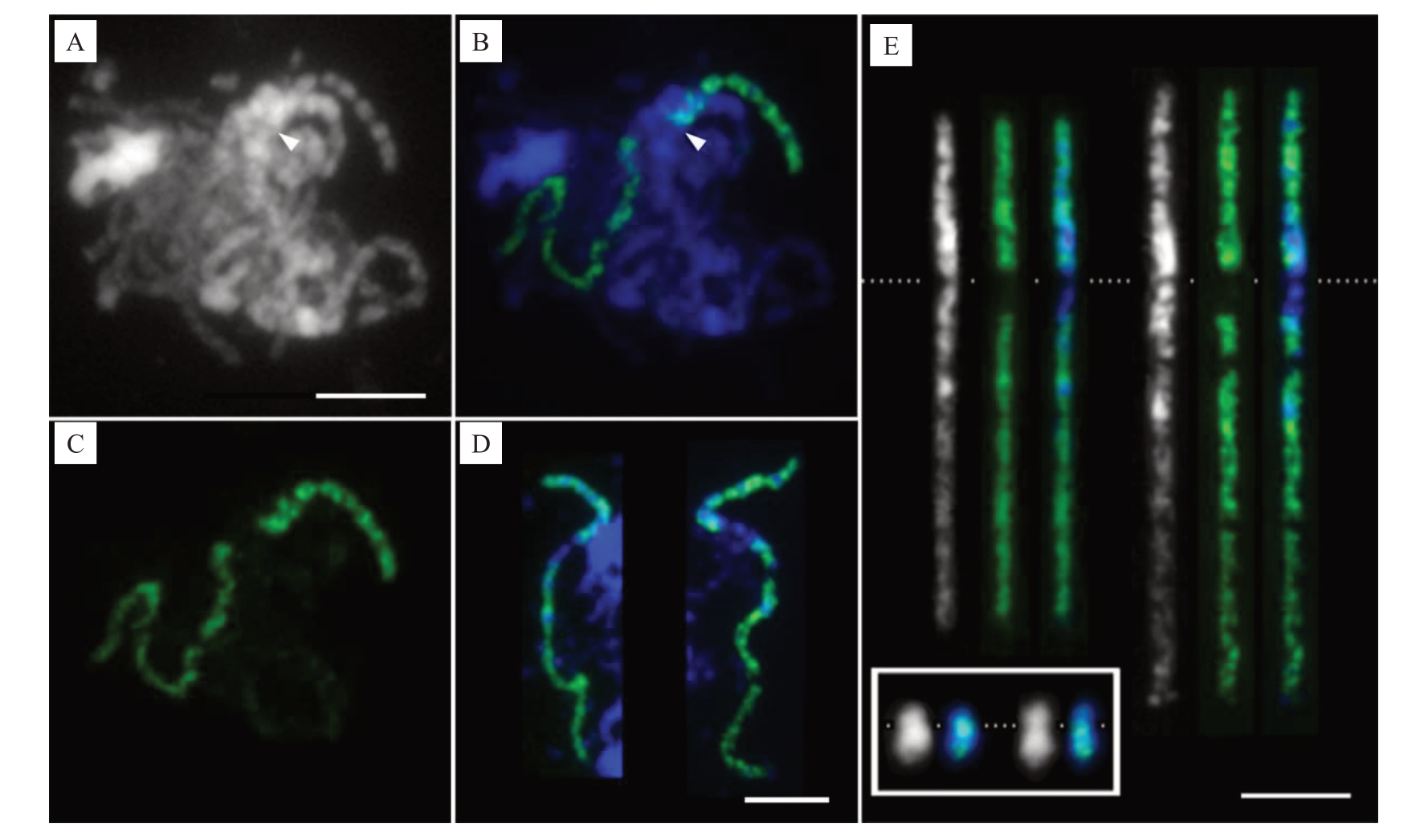
图3 柑橘1 号染色体特异探针FISH 涂染柚减数分裂粗线期染色体
Fig.3 FISH painting of meiotic pachytene chromosomes in C.maxima with Citrus chromosome 1-specific probe
A.单色显示DAPI 染色的柚粗线期细胞,其中的染色体相互缠绕。箭头指示DAPI 染色浅的着丝粒;B.在蓝色显示来自A 中柚粗线期染色体上柑橘1 号染色体特异探针的FISH 信号(绿色)。箭头指示着丝粒位置;C.从B 中分离单独显示的绿色FISH 探针信号;D.一左一右分别为从2 个不同粗线期细胞中得到的与其他染色体无交叉重叠经FISH 涂染识别的粗线期1 号染色体;E.用Image J 软件拉直显示D 中的2 条粗线期染色体。染色体左右排列的顺序与D 中相同,从左到右依次为单色显示的粗线期染色体,其上的绿色涂染探针信号,以及绿色信号与蓝色显示染色体的叠加图。白色虚线指示着丝粒位置。E 中插入的白框内为从图1-A~B 中分离的柚中期单色显示及探针信号叠加后的2 条1 号同源染色体,放大倍数与E 中的粗线期染色体相同。标尺为5µm。
A. DAPI stained cell with intertwined chromosomes at pachytene stage that was shown in monochrome.Arrow head indicates centromere lightly stained with DAPI.B.FISH painting signal(green)from Citrus chromosome 1-specific probe in the pachytene chromosome(blue)in A.Arrow head indicates the position of the centromere;C.FISH painting signal in green that separated from B;D.Two unoverlapped pachytene chromosome 1 identified by painting of specific probe were isolated from two different cells and arranged from left to right.E.Both of the pachytene chromosomes in D were straightened with Image J software.The pachytene chromosomes were arranged from left to right as shown in D,showing monochromatic chromosomes,painting signals(green),and overlayed images with blue pseudo colored chromosomes in order.The position of centromeres were indicated with a dashed white line.The two metaphase homologous chromosomes isolated from Fig.1-A and B, which were displayed in monochrome and overlayed images with painting signals (green) on chromosomes (blue), respectively, were inserted within the white box.The metaphase and pachytene chromosomes share the same magnification in E.Bar=5µm.
经DAPI 染色后,着丝粒区染色浅且两侧分布DAPI染色明亮的亚着丝粒异染色质区(图3-E),染色体臂上的常染色质区染色较浅。短臂上绿色探针信号覆盖至非常接近DAPI 染色浅的着丝粒区(图3-E)。结合着丝粒位置及短臂挨着丝粒区信号消失的特征,甚至能在众多相互缠绕的粗线期染色体上定位着丝粒(图3-A~B 箭头指示),并能据此准确测定染色体臂长及总长。测得15个粗线期细胞1号染色体的平均总长为(27.48±1.89)µm,是有丝分裂中期高度浓缩染色体长度的13.93倍(表1);粗线期长臂平均长度为中期长臂的16.54倍,高于染色体全长的倍率,表明粗线期长臂的伸展程度高于整条染色体的平均值;粗线期短臂长为中期的10.44 倍,低于染色体全长的倍率,更低于长臂的倍率,表明粗线期短臂的伸展程度除低于整条染色体的平均值外,更远低于长臂的伸展程度。
表1 柚减数分裂粗线期和有丝分裂中期1 号染色体之间长度的差异
Table 1 The differences of lengths between meiotic pachytene and mitotic metaphase chromosome 1 of C.maxima
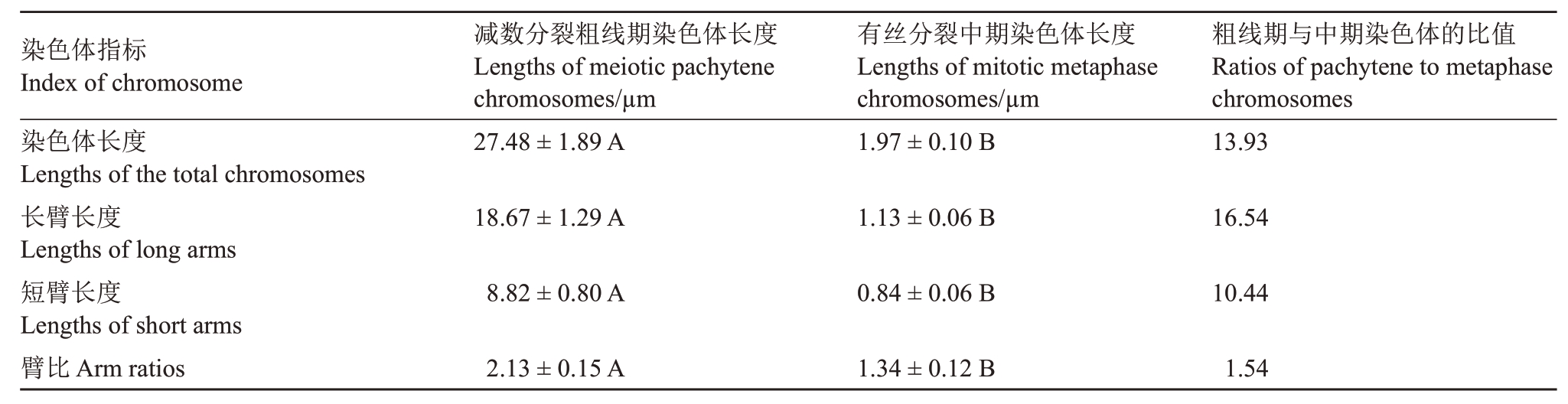
注:染色体的臂比等于长臂的长度除以其短臂的长度。不同大写字母表示差异极显著(p<0.01)。
Note:The arm ratio of a chromosome is calculated by the length of the long arm divided by the length of its short arm.Different capital letters indicate significant differences(p<0.01).
染色体指标Index of chromosome染色体长度Lengths of the total chromosomes长臂长度Lengths of long arms短臂长度Lengths of short arms臂比Arm ratios减数分裂粗线期染色体长度Lengths of meiotic pachytene chromosomes/µm 27.48±1.89 A有丝分裂中期染色体长度Lengths of mitotic metaphase chromosomes/µm 1.97±0.10 B粗线期与中期染色体的比值Ratios of pachytene to metaphase chromosomes 13.93 18.67±1.29 A 1.13±0.06 B 16.54 8.82±0.80 A 0.84±0.06 B 10.44 1.54 2.13±0.15 A 1.34±0.12 B
虽在有丝分裂中期和前中期1号染色体上能够观察到探针信号较弱或缺失的区域,但因染色体浓缩程度较大,分辨率低,导致难以准确鉴定其所处的区段。借助涂染探针信号,在粗线期染色体上清晰观察到1 段信号缺失区,但在众多染色体相互缠绕的减数分裂相中,较难厘清该区段所处区域及染色质特征(图3-A~B)。因此,利用涂染探针信号作指示,选取与其他染色体无交叉重叠的粗线期1 号染色体(图3-D~E),以准确鉴定整条染色体上常染色质和异染色质区的分布特征,以及涂染探针信号在不同染色质区的精准分布。用Image J 软件分离并拉直了图3-D 来自2 个细胞中无交叉重叠的2 条粗线期1 号染色体。染色体灰度图非常清晰地显示,即使2 条粗线期染色体的浓缩程度存在差异,它们的常染色质(DAPI 染色较浅)和异染色质(DAPI 染色明亮)分布模式,以及探针信号在对应位置的覆盖情况仍极为相似(图3-E)。短臂绝大多数区域为DAPI染色明亮的异染色质区。占长臂约1/3长度且靠近着丝粒区域为较为集中的异染色质区,余下近端粒段2/3 的长臂均为DAPI 染色较浅的常染色质区。然而,从细胞学的角度认为富含重复序列的异染色质区几乎均被单拷贝寡核苷酸涂染探针信号覆盖,说明相应区域仍含有一定比例的单拷贝DNA序列,能够进行基因组组装。在DAPI 染色最强烈的区域,探针信号相对其他区域明显更微弱,如短臂和长臂上各有1 个明亮的异染色质区域;在包括着丝粒及长臂紧邻着丝粒DAPI染色明亮的整个亚着丝粒异染色质区几乎没有任何信号。
经Image J软件测定,粗线期1号染色体上连续无信号的着丝粒及紧邻亚着丝粒区长度为(2.36 ±0.34)µm,占染色体总长的8.58%。由于涂染探针筛选自基因组测序组装大小为32.08 Mb的柚1号染色体[22],很可能完全对应探针信号覆盖区域的大小。假定DNA 含量在全长染色体上呈均衡分布,1号染色体的大小可计算为32.08 Mb÷(100%-8.58%)=35.09 Mb,而着丝粒附近信号缺失区段的大小为35.09 Mb×8.58%=3.01 Mb。
3 讨 论
标准的单条染色体鉴定方法可实现中期染色体及其他不同细胞分裂阶段浓缩染色质的识别。最近的研究利用生物信息学手段,研发基于单拷贝寡核苷酸序列的FISH涂染探针,能够在任何已获得基因组序列的植物种中应用[9,21],并已在许多植物种中取得成功,包括黄瓜[19]、马铃薯[6,23]、水稻[24]和杨树[25]等。即使在大基因组的小麦[26]和玉米[20]中也获得了成功。这相对于传统在小基因组模式植物拟南芥和二穗短柄草中采用大量BAC混合池作FISH涂染探针[27-28],取得了革命性的进步,将极大地提升染色体FISH 涂染在众多植物种中的应用。已有柑橘及多个相关种属完成了全基因组测序组装[22,29-30],可用作该新型染色体精准鉴定单拷贝FISH 寡核苷酸探针的发掘。笔者新近研发了柑橘所有9条染色体特异的涂染探针[4],为基于FISH 技术开展丰富的柑橘及相关种属间系统精准染色体结构变异鉴定和进化分析提供了有力工具[9,21]。笔者在本研究中利用柑橘1号染色体特异FISH涂染探针,实现了在有丝分裂间期细胞核、前中期和中期细胞染色体及减数分裂粗线期染色体和其所在区域的可视化,可为开展柑橘染色体研究及育种利用提供强大工具,开拓柑橘染色体结构及特定区段序列组成精准研究,特别是柑橘多倍体材料减数分裂粗线期染色体精准配对和遗传鉴定利用的新领域。
减数分裂是真核生物生命周期中染色体数目减少并产生配子的重要且特殊过程,有效地实现从末端到末端鉴定单条粗线期染色体,将大力推动减数分裂染色体配对机制的精准研究。前人研创了染色体全长涂染FISH探针,实现了玉米1号染色体[20]、马铃薯7号和11号染色体[6]、黄瓜3号染色体的全长示踪[19],以及不同倍性茄属物种的粗线期染色体精细配对研究[6]。在柑橘中,前人采用的染色体鉴定方法包括CMA 显带[10-13],以及结合CMA 显带、rDNA、基因组中筛选的其他重复序列[14-15]和染色体特异BAC 开展FISH[16-18],均无法对减数分裂粗线期特定的全长染色体进行跟踪研究。笔者在本研究中首次在柑橘粗线期染色体上开展FISH实验,实现了单条全长粗线期染色体的精准鉴定,可解决对柑橘粗线期特定全长染色体进行跟踪研究的难题。
染色体涂染探针能否有效示踪鉴定粗线期染色体,与探针设计时染色体上筛选到的寡核苷酸分布密度有关[9]。在几个不同的植物中,染色体上寡核苷酸的密度为0.1~0.5个·kb-1时,足以在浓缩的中期染色体上产生高质量的全染色体涂染信号[9]。但粗线染色体的长度为体细胞中期染色体的10~40倍[31],因此,用涂染探针鉴定减数分裂粗线染色体,需要采用更高的寡核苷酸密度。1个密度为2个·kb-1的水稻9号染色体探针在粗线染色体上显示出良好的信号[24]。1 个密度为0.6 个·kb-1的马铃薯11 号染色体探针足以显示整个粗线染色体[6]。玉米1 号染色体上密度为0.3 个·kb-1,可基本覆盖粗线期染色体[20],但信号覆盖的密度明显更低。本研究中柑橘1号染色体上寡核苷酸的密度为0.85 个·kb-1,在粗线期染色体上产生了很好的信号覆盖,起到了很好的示踪效果。涂染探针信号显示,粗线期15个细胞中的1 号染色体均表现从末端到末端的完整配对,证明2 条同源染色体的同源性高,与测序报道柚类基因组杂合度低的特征相吻合[29]。
前人研究表明,利用粗线期染色体开展FISH定位,可大大提高信号分辨率。在不同基因组大小的代表性物种中,将粗线期与有丝分裂中期的染色体长度进行比较,发现黑麦中的长度差异大于7倍、玉米为10 倍、番茄为15 倍、拟南芥大于20 倍、水稻为40 倍[31]。因此,以高度伸展的粗线期染色体相较于高度收缩的中期染色体作靶标,具有高出数倍的FISH信号分辨率[31-32],是将DNA序列信息与染色体结构特征相结合极有效的方法,是高分辨率细胞遗传图谱构建的强大工具。例如在水稻早粗线期10号染色体上,可区分相距40 kb 的DNA 探针信号[32]。在番茄粗线期染色体常染色质区FISH 定位的分辨率为120 kb,在异染色质区为1.20 Mb[31]。本研究揭示,柑橘粗线期1 号染色体的平均长度为中期的13.93 倍,实现了柑橘粗线期染色体高分辨率的FISH 鉴定,为深入开展其结构精准研究提供了新方法。
本研究中测算出着丝粒及紧邻的信号缺失区约为3.01 Mb,这在高度收缩的柑橘中期染色体上是极难测定的。由于着丝粒及附近异染色质区DNA的浓缩程度高于全染色体上的平均值[31],因此,该区段实际应大于3.01 Mb。利用着丝粒功能蛋白CENH3免疫共沉淀测序,估算1号染色体着丝粒区大小为7.6 Mb,含有大量长末端重复序列(long terminal repeat,LTR)[33]。在基因组测序组装时,很可能由于该区域富含高度重复DNA 序列而未被当前的参考基因组覆盖,因此未筛选到单拷贝寡核苷酸,表现为探针信号的缺失。研究结果显示着丝粒及其紧邻异染色质的特定区段无信号覆盖,提示该区域可能未完全实现基因组的组装。
本研究表明高分辨率粗线期染色体FISH 涂染可应用于植物的基因组研究,检查基因组序列组装的完整性[34],未来可用作测序组装困难着丝粒和附近异染色质区的辅助验证,以及其他植物细胞遗传学新研究。
4 结 论
首次在柑橘上利用染色体全长特异探针涂染,揭示了粗线期与有丝分裂各时期染色体结构及FISH信号分辨率的巨大差异,为进一步探究柑橘减数分裂其余染色体的结构组成及不同倍性材料中的配对遗传利用奠定了新的试验基础。
[1] NAITHANI S P,RAGHUVANSHI S S.Cytogenetical studies in the genus Citrus[J].Nature,1958,181(4620):1406-1407.
[2] 郭文武,叶俊丽,邓秀新.新中国果树科学研究70 年:柑橘[J].果树学报,2019,36(10):1264-1272.
GUO Wenwu,YE Junli,DENG Xiuxin. Fruit scientific research in New China in the past 70 years:Citrus[J].Journal of Fruit Science,2019,36(10):1264-1272.
[3] GUERRA M,DOS SANTOS K G,BARROS E S A E,EHRENDORFER F J. Heterochromatin banding patterns in Rutaceae-Aurantioideae:A case of parallel chromosomal evolution[J].American Journal of Botany,2000,87(5):735-747.
[4] HE L,ZHAO H N,HE J,YANG Z J,GUAN B,CHEN K L,HONG Q B,WANG J H,LIU J J,JIANG J M. Extraordinarily conserved chromosomal synteny of Citrus species revealed by chromosome-specific painting[J]. The Plant Journal,2020,103(6):2225-2235.
[5] BRAZ G T,YU F,ZHAO H N,DENG Z,BIRCHLER J A,JIANG J M. Preferential meiotic chromosome pairing among homologous chromosomes with cryptic sequence variation in tetraploid maize[J].New Phytologist,2021,229(6):3294-3302.
[6] HE L,BRAZ G T,TORRES GA,JIANG J M.Chromosome painting in meiosis reveals pairing of specific chromosomes in polyploid Solanum species[J].Chromosoma,2018,127(4):505-513.
[7] WANG K,XIANG D,XIA K,SUN B,KHURSHID H,ESH A M H,ZHANG H.Characterization of repetitive DNA in Saccharum officinarum and Saccharum spontaneum by genome sequencing and cytological assays[J]. Frontiers in Plant Science,2022,13:814620.
[8] SCHUBERT I,FRANSZ P F,FUCHS J,DE JONG J H. Chromosome painting in plants[J].Methods in Cell Science,2021,23(1):57-69.
[9] JIANG J M. Fluorescence in situ hybridization in plants:Recent developments and future applications[J]. Chromosome Research,2019,27(3):153-165.
[10] GUERRA M. Cytogenetics of Rutaceae. V. High chromosomal variability in Citrus species revealed by CMA/DAPI staining[J].Heredity,1993,71(3):234-241.
[11] GUERRA M D S. Cytogenetics of Rutaceae. III. heterochromatin patterns[J].Caryologia,1985,38(3/4):335-346.
[12] 梁国鲁.部分柑桔属及其近缘属Giemsa C-带带型研究[J].遗传学报,1988,15(6):409-415.
LIANG Guolu. Studies on the Giemsa C-banding patterns of some Citrus and its related genera[J]. Acta Genetica Sinica,1988,15(6):409-415.
[13] 魏文娜,程尧楚,李润唐,段映池.从染色体核型及Giemsa 显带探讨柑桔类的演化[J].园艺学报,1988,15(4):223-228.
WEI Wenna,CHENG Yaochu,LI Runtang,DUAN Yingchi.Studies on the evolution of Citrus based on karyotype and Giemsa C-banding patterns[J].Acta Horticulturae Sinica,1988,15(4):223-228.
[14] DENG H H,XIANG S Q,GUO Q G,JIN W,CAI Z,LIANG G.Molecular cytogenetic analysis of genome-specific repetitive elements in Citrus clementina Hort.Ex Tan.and its taxonomic implications[J].BMC Plant Biology,2019,19(1):77.
[15] YU C X,DENG X X,CHEN C L. Chromosomal characterization of a potential model mini-Citrus (Fortunella hindsii)[J].Tree Genetics&Genomes,2019,15(5):1-10.
[16] MENDES S,MORAES A P,MIRKOV T E,PEDROSAHARAND A. Chromosome homeologies and high variation in heterochromatin distribution between Citrus L. and Poncirus Raf.as evidenced by comparative cytogenetic mapping[J].Chromosome Research,2011,19(4):521-530.
[17] DA COSTA SILVA S,MENDES S,RÉGIS T,PASSOS O S,FILHO W D S S,PEDROSA-HARAND A.Cytogenetic map of pummelo and chromosome evolution of true Citrus species and the hybrid sweet orange[J]. Journal of Agricultural Science,2019,11(14):148.
[18] COSTA S S,MARQUES A,SANTOS S F W,MIRKOV T E,PEDROSA-HARAND A,GUERRA M.The cytogenetic map of the Poncirus trifoliata (L.) Raf.:A nomenclature system for chromosomes of all citric species[J]. Tropical Plant Biology,2011,4(2):99-105.
[19] HAN Y H,ZHANG T,THAMMAPICHAI P,WENG Y,JIANG J.Chromosome-specific painting in Cucumis species using bulked oligonucleotides[J].Genetics,2015,200(3):771-779.
[20] ALBERT P S,ZHANG T,SEMRAU K,ROUILLARD J M,KAO Y H,WANG C R,DANILOVA T V,JIANG J,BIRCHLER J A.Whole-chromosome paints in maize reveal rearrangements,nuclear domains,and chromosomal relationships[J]. Proceedings of the National Academy of Sciences of the United States of America,2019,116(5):1679-1685.
[21] ZHANG T,LIU G Q,ZHAO H N,BRAZ G T,JIANG J. Chorus2:Design of genome-scale oligonucleotide-based probes for fluorescence in situ hybridization[J]. Plant Biotechnology Journal,2021,19(10):1967-1978.
[22] WANG X,XU Y,ZHANG S,CAO L,HUANG Y,CHENG J,WU G Z,TIAN S L,CHEN C L,LIU Y,YU H W,YANG X M,LAN H,WANG N,WANG L,XU J D,JIANG X L,XIE Z Z,TAN M L,LARKIN R M,CHEN L L,MA B G,RUAN Y J,DENG X X,XU Q. Genomic analyses of primitive,wild and cultivated Citrus provide insights into asexual reproduction[J].Nature Genetics,2017,49(5):765-772.
[23] BRAZ G T,HE L,ZHAO H N,ZHANG T,SEMRAU K,ROUILLARD J M,TORRES G A,JIANG J M. Comparative oligo-FISH mapping:An efficient and powerful methodology to reveal karyotypic and chromosomal evolution[J]. Genetics,2018,208(2):513-523.
[24] HOU L L,XU M,ZHANG T,XU Z H,WANG W Y,ZHANG J X,YU M M,JI W,ZHU C W,GONG Z Y,GU M H,JIANG J M,YU H X. Chromosome painting and its applications in cultivated and wild rice[J].BMC Plant Biology,2018,18(1):110.
[25] XIN H Y,ZHANG T,WU Y F,ZHANG W,ZHANG P,XI M,JIANG J M. An extraordinarily stable karyotype of the woody Populus species revealed by chromosome painting[J]. The Plant Journal,2020,101(2):253-264.
[26] LI G R,ZHANG T,YU Z H,WANG H,YANG E,YANG Z.An efficient Oligo-FISH painting system for revealing chromosome rearrangements and polyploidization in Triticeae[J]. The Plant Journal,2021,105(4):978-993.
[27] IDZIAK D,BETEKHTIN A,WOLNY E,LESNIEWSKA K,WRIGHT J,FEBRER M,BEVAN M W,JENKINS G,HASTEROK R. Painting the chromosomes of Brachypodium:Current status and future prospects[J]. Chromosoma,2011,120(5):469-479.
[28] LYSAK M A,FRANSZ P F,ALI H B,SCHUBERT I. Chromosome painting in Arabidopsis thaliana[J]. The Plant Journal 2001,28(6):689-697.
[29] WU G A,TEROL J,IBANEZ V,LOPEZ-GARCIA A,PEREZROMAN E,BORREDA C,DOMINGO C,TADEO F R,CARBONELL-CABALLERO J,ALONSO R,CURK F,DU D L,OLLITRAULT P,ROOSE M,DOPAZO J,GMITTER F G,ROKHSAR D S,TALON M.Genomics of the origin and evolution of Citrus[J].Nature,2018,554(7692):311-316.
[30] XU Q,CHEN L L,RUAN X,CHEN D,ZHU A,CHEN C L,BERTRAND D,JIAO W B,HAO B H,LYON M P,CHEN J J,GAO S,XING F,LAN H,CHANG J W,GE X H,LEI Y,HU Q,MIAO Y,WANG L,XIAO S X,BISWAS M K,ZENG W F,GUO F,CAO H B,YANG X M,XU X W,CHENG Y J,XU J,LIU J H,LUO O J H,TANG Z H,GUO W W,KUANG H H,ZHANG H Y,ROOSE M L,NAGARAJAN N R J,DENG X X,RUAN Y J.The draft genome of sweet orange(Citrus sinensis)[J].Nature Genetics,2013,45(1):59-66.
[31] DE JONG J H,FRANSZ P,ZABEL P. High resolution FISH in plants- techniques and applications[J].Trends in Plant Science,1999,4(7):258-263.
[32] CHENG Z K,PRESTING G G,BUELL C R,WING R A,JIANG J M. High-resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice[J]. Genetics,2001,157(4):1749-1757.
[33] XIA Q M,MIAO L K,XIE K D,YIN Z P,WU X M,CHEN C L,GROSSER J W,GUO W W. Localization and characterization of Citrus centromeres by combining half-tetrad analysis and CenH3-associated sequence profiling[J].Plant Cell Reports,2020,39(12):1609-1622.
[34] XIN H Y,ZHANG T,HAN Y H,WU Y F,SHI J S,XI M L,JIANG J M.Chromosome painting and comparative physical mapping of the sex chromosomes in Populus tomentosa and Populus deltoides[J].Chromosoma,2018,127(3):313-321.