柚是芸香科柑橘属多年生果树,属于柑橘属3个基本种之一。柚类起源于东南亚或中国南方一带,在中国的栽培历史长达3000多年,古书《吕氏春秋》记载“果之美者,云梦之柚”[1]。中国是柚类资源的起源中心和遗传变异中心之一[2],柚为单胚性,在长期的栽培过程中容易产生遗传变异,长期以来,人类对柚的选择和培育产生了数以百计的柚品种,这些品种具有不同的果实大小和品种风味,形成丰富的种质资源,但至今未发现有真正野生类型[3]。
柚类种质资源开发利用与品种鉴定具有巨大的潜力,柚类品种鉴定、分类同其他果树一样经历了形态分类[3]、孢粉学[4-5]、同工酶[6]等传统的鉴定方法。尽管柑橘可根据枝叶形态等鉴别其属种,但柚类的形态学标记较少,种内品种间差异不明显,随着分子生物学技术的发展,以PCR 技术为代表的DNA 分子标记技术被广泛应用,为柚类种质资源的鉴定、亲缘关系分析和遗传图谱的构建提供了良好的技术手段。其中,SSR(simple sequence repeat)为简单序列重复,由1~6个核苷酸重复多次的基序组成,具有信息含量高、共显性遗传、操作简单、多态性高等特点[7],且不受形态和环境的影响而在柑橘[8]、板栗[9]、蔓越莓[10]上得到应用。柚类种质丰富,遗传多样性低,具有较高的形态变异,所以需要更准确、快速、高效的鉴定方法。
在众多的分子标记中,荧光标记SSR 能精准识别目标基因DNA片段大小,精准识别到只有1 bp差异的碱基,检测结果可靠、稳定且准确[11]。所以,荧光标记SSR被广泛应用于果树遗传结构分析、品种鉴定、分子身份证构建及品种权保护等方面的研究。随着SSR标记的研究,“分子身份证”逐渐被提出和应用,使得在品种鉴别和检索时变得更快捷。目前,通常采用荧光测序技术结合新的带型编码方式构建分子身份证,该技术已成功应用于构建桃[12]、杏[13]、梨[14]、苹果[15]等多种果树的分子身份证。目前,尚未发现利用SSR 荧光毛细管电泳结合新的带型编码方式构建柚类资源分子身份证的相关研究报道,笔者在本研究中以22 份柚类种质为研究材料,采用SSR荧光标记引物,结合荧光毛细管电泳检测技术分析扩增条带分子质量和图谱带型方式构建分子身份证,以期为柚类品种的鉴定和亲缘关系分析提供一定的理论依据。
1 材料和方法
1.1 试验材料
22 份柚类材料分别取自桂林市广西特色作物研究院柑橘种质资源保存圃、桂林市全州县以及南宁市高明农场,每个品种采集叶片后,保存于-80 ℃超低温冰箱中备用。
1.2 试验方法
1.2.1 DNA 提取及PCR 体系 采用磁珠法基因组DNA提取试剂盒提取基因组DNA,并用1%琼脂糖凝胶电泳检测DNA 提取质量。再取2 μL DNA 样本,用NanoDROP8000 超微量分光光度计进行检测核酸浓度和纯度检测,将DNA统一稀释成20 ng·μL-1,用于后续的PCR扩增。
引物选自李益等[16]报道的21 对引物(表1),由武汉天一华煜基因科技有限公司合成。PCR反应体系10 μL,包括:2×Taq PCR Master Mix 5.0 μL,正、反向引物(10 mol·L-1)各0.5 μL,基因组DNA(约20 ng·μL-1),超纯水3.0 μL。PCR反应程序:95 ℃预变性5 min,95 ℃变性30 s,62~52 ℃退火30 s,72 ℃延伸30 s,循环35次;72 ℃延伸20 min,4 ℃保存。
表1 21 对引物信息
Table 1 Information of 21 pairs of SSR primers
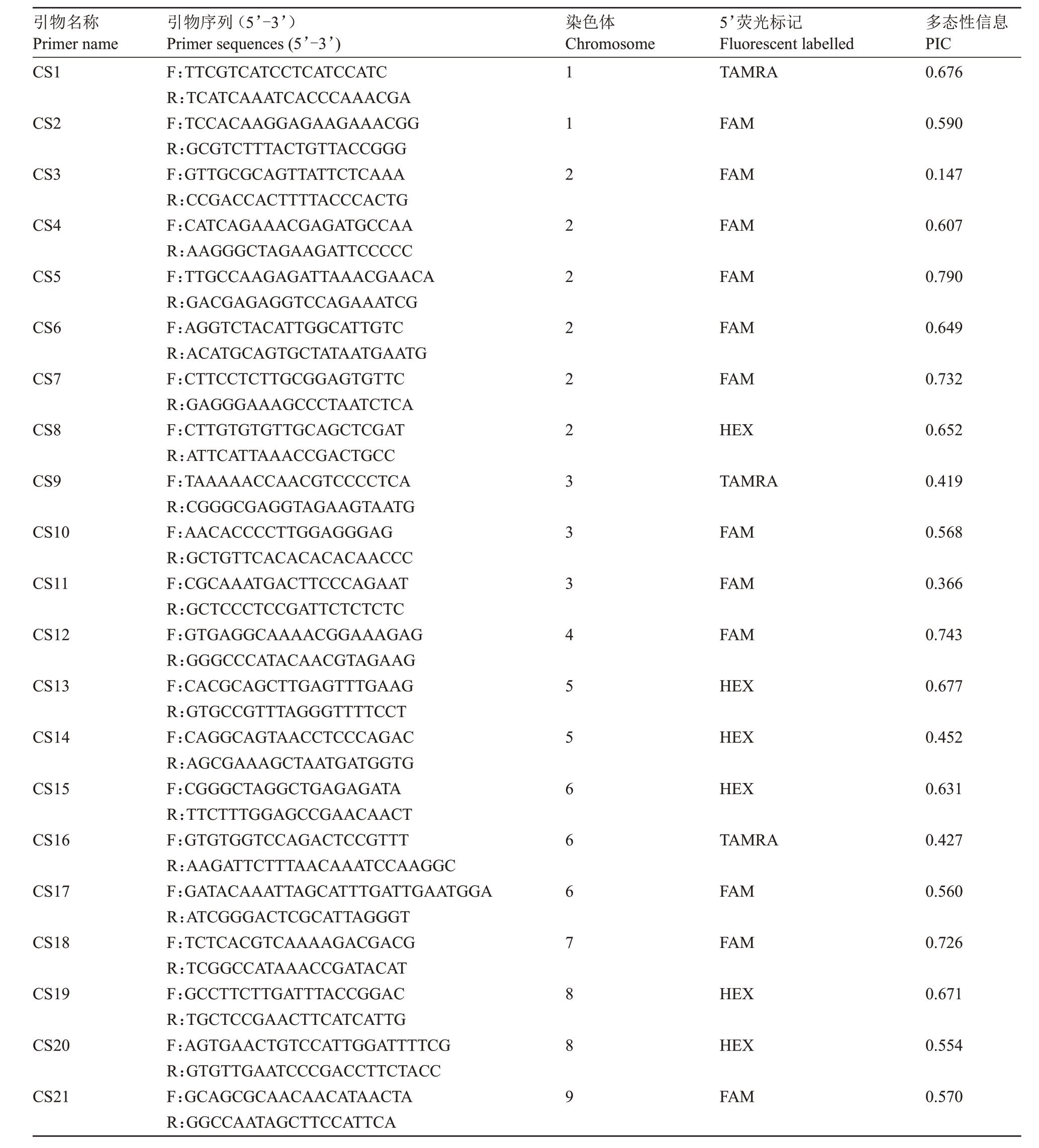
引物名称Primer name CS1染色体Chromosome 5’荧光标记Fluorescent labelled TAMRA多态性信息PIC 0.676 CS2 FAM 0.590 CS3 FAM 0.147 CS4 FAM 0.607 CS5 FAM 0.790 CS6 FAM 0.649 CS7 FAM 0.732 CS8 HEX 0.652 CS9 TAMRA 0.419 CS10 FAM 0.568 CS11 FAM 0.366 CS12 FAM 0.743 CS13 HEX 0.677 CS14 HEX 0.452 CS15 HEX 0.631 CS16 TAMRA 0.427 CS17 FAM 0.560 CS18 FAM 0.726 CS19 HEX 0.671 CS20 HEX 0.554 CS21引物序列(5’-3’)Primer sequences(5’-3’)F:TTCGTCATCCTCATCCATC R:TCATCAAATCACCCAAACGA F:TCCACAAGGAGAAGAAACGG R:GCGTCTTTACTGTTACCGGG F:GTTGCGCAGTTATTCTCAAA R:CCGACCACTTTTACCCACTG F:CATCAGAAACGAGATGCCAA R:AAGGGCTAGAAGATTCCCCC F:TTGCCAAGAGATTAAACGAACA R:GACGAGAGGTCCAGAAATCG F:AGGTCTACATTGGCATTGTC R:ACATGCAGTGCTATAATGAATG F:CTTCCTCTTGCGGAGTGTTC R:GAGGGAAAGCCCTAATCTCA F:CTTGTGTGTTGCAGCTCGAT R:ATTCATTAAACCGACTGCC F:TAAAAACCAACGTCCCCTCA R:CGGGCGAGGTAGAAGTAATG F:AACACCCCTTGGAGGGAG R:GCTGTTCACACACACAACCC F:CGCAAATGACTTCCCAGAAT R:GCTCCCTCCGATTCTCTCTC F:GTGAGGCAAAACGGAAAGAG R:GGGCCCATACAACGTAGAAG F:CACGCAGCTTGAGTTTGAAG R:GTGCCGTTTAGGGTTTTCCT F:CAGGCAGTAACCTCCCAGAC R:AGCGAAAGCTAATGATGGTG F:CGGGCTAGGCTGAGAGATA R:TTCTTTGGAGCCGAACAACT F:GTGTGGTCCAGACTCCGTTT R:AAGATTCTTTAACAAATCCAAGGC F:GATACAAATTAGCATTTGATTGAATGGA R:ATCGGGACTCGCATTAGGGT F:TCTCACGTCAAAAGACGACG R:TCGGCCATAAACCGATACAT F:GCCTTCTTGATTTACCGGAC R:TGCTCCGAACTTCATCATTG F:AGTGAACTGTCCATTGGATTTTCG R:GTGTTGAATCCCGACCTTCTACC F:GCAGCGCAACAACATAACTA R:GGCCAATAGCTTCCATTCA 112222223334556667889 FAM 0.570
1.2.2 毛细管电泳检测 将甲酰胺与分子质量内标混合,取9 μL 混合物于上机板中,再加入1 μL 稀释的荧光PCR 产物,上样于96 通道全自动ABI3730XL 遗传分析仪进行毛细管电泳。利用GeneMarker分析软件对原始数据进行分析,将各泳道内分子质量内标的位置与各样品峰值的位置进行比较,确定片段大小和基因型类别。
1.2.3 品种分子身份证构建 对25 个样品进行扩增,得到各样品扩增的指纹数据,将指纹数据转换为数字编码,即分子身份证。编码方式为:将每对引物在25 份样品中扩增时所产生的等位基因按从小到大的顺序排序,然后用个位阿拉伯数字1,2,3,……,9 作为不同等位基因的代码,等位基因数大于9时,用a,b,c代表第10、11、12个等位基因,依次类推,无扩增产物时用0表示;然后,以2位代码对某个标记的图谱带型进行编码,杂合带型以2 位不同代码编码,纯合带型以2 位相同代码编码。根据各个标记带型数的多少,确定用于构建分子身份证的标记;再按照带型数由大到小的顺序,将某一供试种质各标记的编码串联,即形成8 位数字或字母表示的分子身份证[13]。
1.2.4 聚类分析 使用Cervus 软件计算引物的多态性信息含量(polymorphism information content,PIC)和基因多样性(gene diversity,D),将各样品各位点的等位基因编码成0,1 矩阵形式的指纹图谱。利用GenAIex 软件?计算各个样品间的遗传距离(Nei’s genetic distance,GD),根据遗传距离构建进化树。
2 结果与分析
2.1 引物筛选
经荧光毛细管电泳后,每对引物均获得精确的DNA 片段大小,如引物CS5 的SSR 检测结果(图1)。21 对引物中,有4 对引物多态性(PIC)大于0.7,属于高多态性位点;PIC 介于0.3~0.7 之间的引物有16 对,属于中高多态性位点;仅有1 对引物CS3 PIC 为0.147,低于0.2,属于低多态性位点,表明该基因较保守(表2)。经筛选确定4 对引物CS5、CS7、CS12、CS18 用于后续柚类种质分子身份证的构建。
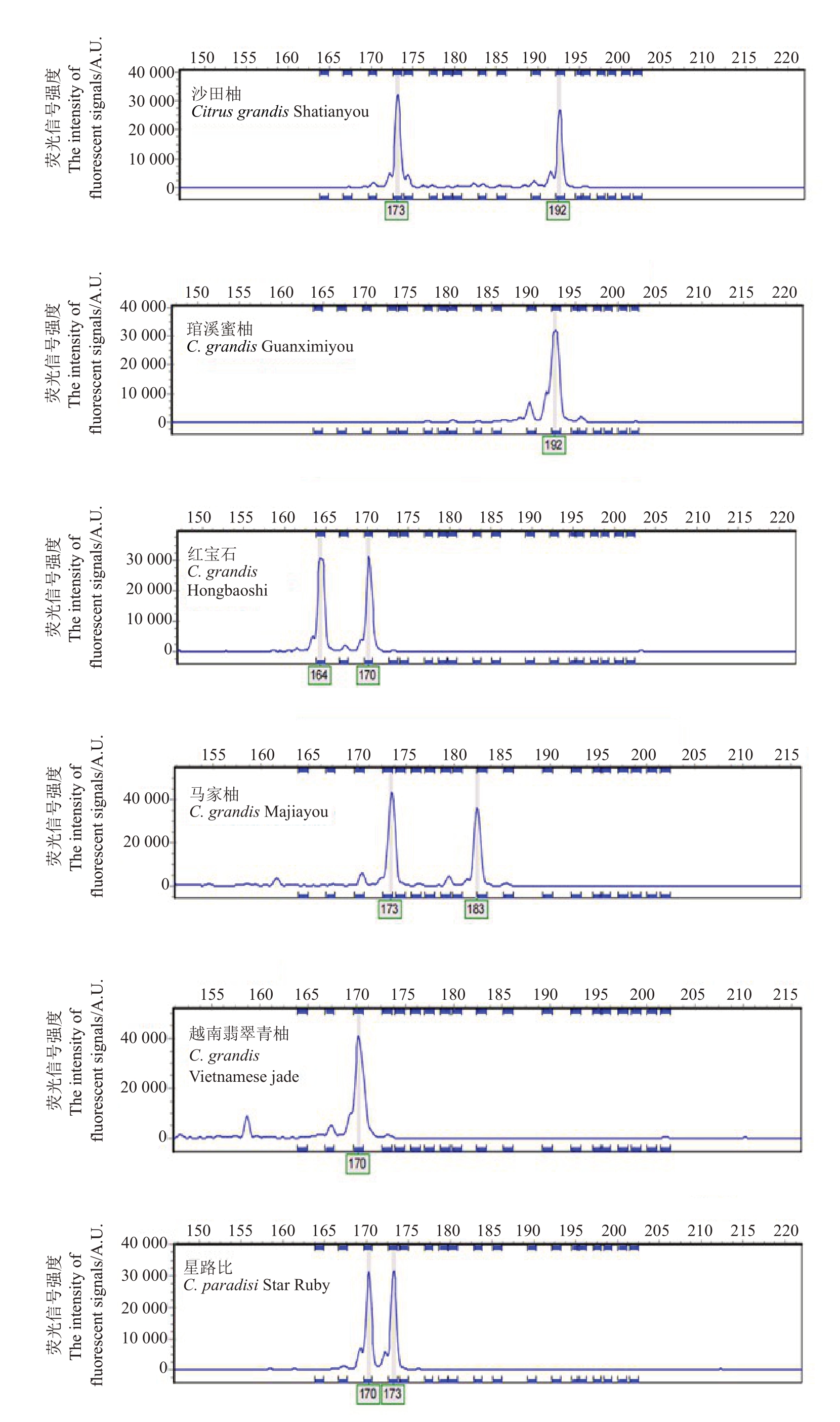
图1 引物CS5 在6 个柚类品种中检测到的等位变异
Fig.1 Polymorphism detected among six pomelo cultivars with SSR primer CS5
表2 4 对荧光标记引物扩增结果
Table 2 Amplified results of 4 pairs of fluorescent-labelled SSR primers
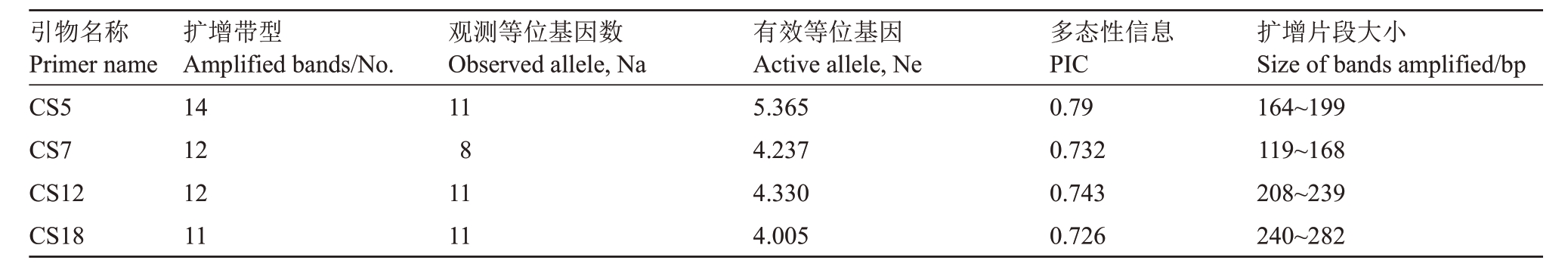
引物名称Primer name CS5 CS7 CS12 CS18扩增带型Amplified bands/No.14 12 12 11观测等位基因数Observed allele,Na 11 8 11 11有效等位基因Active allele,Ne 5.365 4.237 4.330 4.005多态性信息PIC 0.79 0.732 0.743 0.726扩增片段大小Size of bands amplified/bp 164~199 119~168 208~239 240~282
2.2 毛细管电泳结果
利用4 对引物对25 份品种资源进行扩增,共检测到等位基因数为41个,每个引物检测到的等位基因数为8~11个,平均每个引物扩增10.25个,其中扩增等位基因数CS5、CS12、CS18 相同,为11 个,最少的是CS7,为8 个;扩增片段长度在119~282 bp 之间,其中引物CS18扩增的基因片段较长。每个引物扩增有效等位基因(Ne)介于4.005~5.365之间,平均为4.484;引物多态性信息(PIC)值变化范围为0.726~0.79,平均为0.748,其中CS5 最大,CS18 最小。4 对引物在22 份柚类种质中共检测基因型(带型数)49 个,每个引物扩增的基因型数为11~14 个,平均为12.25 个,其中CS5 最大,CS18 最小,CS7 和CS12相同(表2,表3)。
表3 25 份柑橘资源在4 对SSR 引物上的等位基因片段大小
Table 3 Allelic fragment size of 4 pairs of SSR primers from 25 accession of citrus germplasm
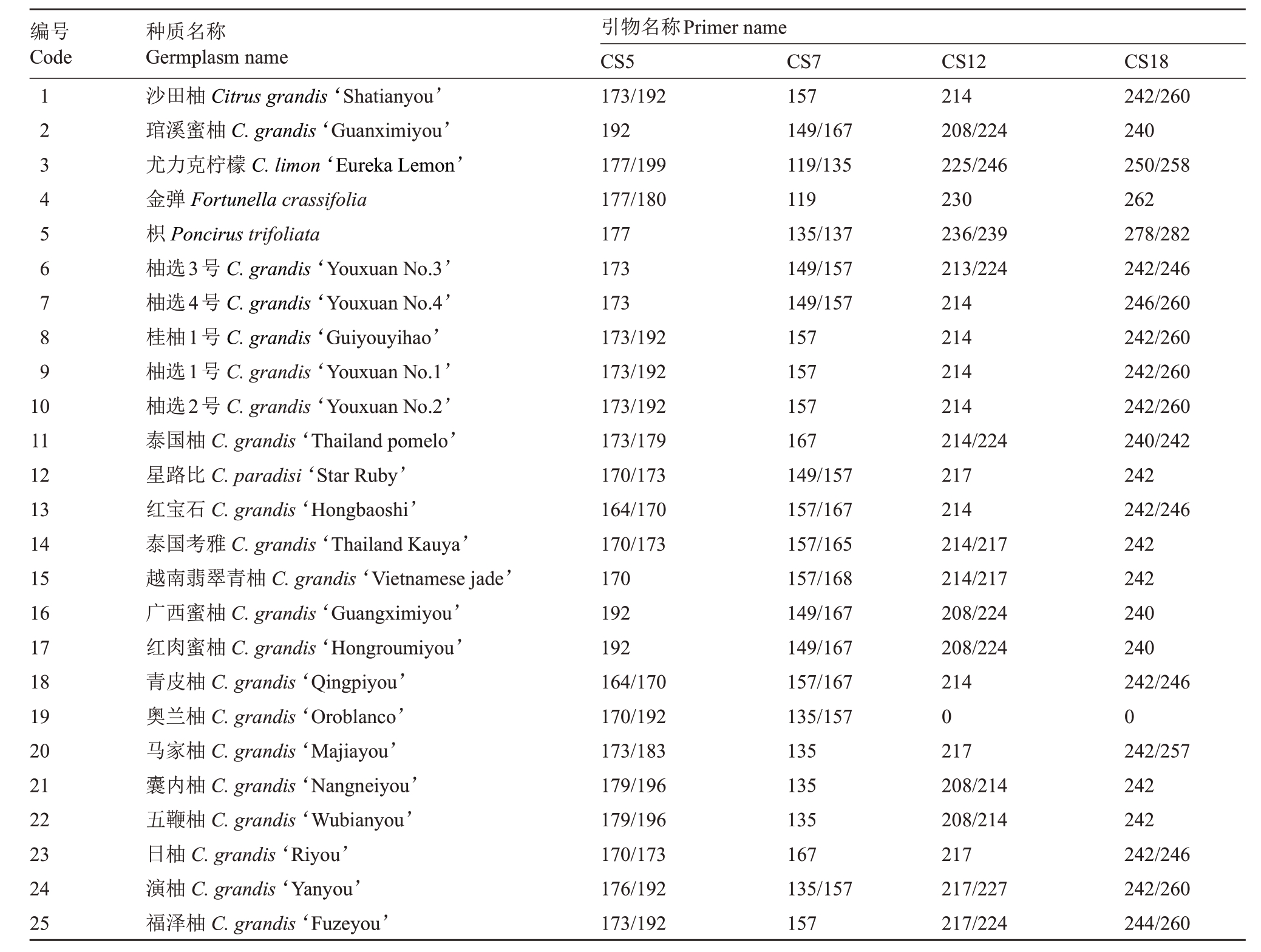
编号Code 123456789 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25种质名称Germplasm name沙田柚Citrus grandis‘Shatianyou’琯溪蜜柚C.grandis‘Guanximiyou’尤力克柠檬C.limon‘Eureka Lemon’金弹Fortunella crassifolia枳Poncirus trifoliata柚选3号C.grandis‘Youxuan No.3’柚选4号C.grandis‘Youxuan No.4’桂柚1号C.grandis‘Guiyouyihao’柚选1号C.grandis‘Youxuan No.1’柚选2号C.grandis‘Youxuan No.2’泰国柚C.grandis‘Thailand pomelo’星路比C.paradisi‘Star Ruby’红宝石C.grandis‘Hongbaoshi’泰国考雅C.grandis‘Thailand Kauya’越南翡翠青柚C.grandis‘Vietnamese jade’广西蜜柚C.grandis‘Guangximiyou’红肉蜜柚C.grandis‘Hongroumiyou’青皮柚C.grandis‘Qingpiyou’奥兰柚C.grandis‘Oroblanco’马家柚C.grandis‘Majiayou’囊内柚C.grandis‘Nangneiyou’五鞭柚C.grandis‘Wubianyou’日柚C.grandis‘Riyou’演柚C.grandis‘Yanyou’福泽柚C.grandis‘Fuzeyou’引物名称Primer name CS5 173/192 192 177/199 177/180 177 173 173 173/192 173/192 173/192 173/179 170/173 164/170 170/173 170 192 192 164/170 170/192 173/183 179/196 179/196 170/173 176/192 173/192 CS7 157 149/167 119/135 119 135/137 149/157 149/157 157 157 157 167 149/157 157/167 157/165 157/168 149/167 149/167 157/167 135/157 135 135 135 167 135/157 157 CS12 214 208/224 225/246 230 236/239 213/224 214 214 214 214 214/224 217 214 214/217 214/217 208/224 208/224 214 0 217 208/214 208/214 217 217/227 217/224 CS18 242/260 240 250/258 262 278/282 242/246 246/260 242/260 242/260 242/260 240/242 242 242/246 242 242 240 240 242/246 0 242/257 242 242 242/246 242/260 244/260
2.3 种质分子身份证编码
根据荧光毛细管电泳及引物多态性信息,选择4对多态性(PIC>0.7)的引物,去除多态性较低的引物,得到各样品扩增的指纹数据,将指纹数据转换为数字编码,构建22份柚类种质分子身份证。按照表2 的引物顺序,将每个荧光标记引物扩增带型数按从多到少的顺序排序编码,将每份种质用CS5、CS7、CS12、CS18 这4 个标记得到的等位基因编码串联,得到各种质的8位分子身份证(表4)。每个字符串的数字从左到右每2个数字分布在同一对染色体上,对应位置编码相同表明该引物在不同样品上得到了相同的扩增带型和等位基因。结果表明,在供试的22份柚类资源中,得到了15份不同类型的分子身份证代码,其中,桂柚1 号、柚选1 号、柚选2 号与沙田柚共享一套分子身份证代码;青皮柚与红宝石共享一套分子身份证代码;广西蜜柚与红肉蜜柚分子身份证代码相同;囊内柚与五鞭柚分子身份证代码相同。共享一套分子身份证代码的样品需进一步从已知柑橘基因组水平上分析差异碱基,进而验证是否存在亲缘关系或者是芽变品种。
表4 25 份柑橘种质的分子身份证代码
Table 4 Molecular identification code of 25 citrus germplasm
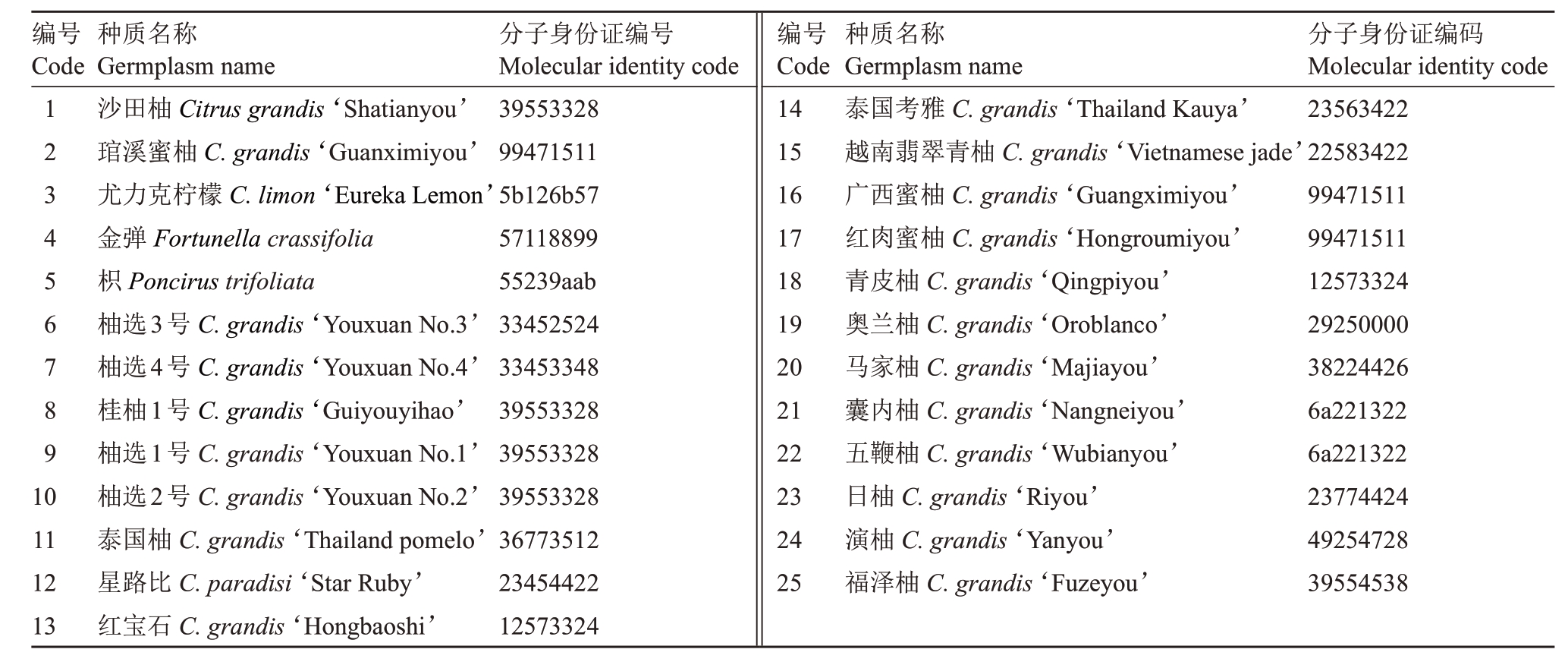
编号Code 123456789 10 11 12 13种质名称Germplasm name沙田柚Citrus grandis‘Shatianyou’琯溪蜜柚C.grandis‘Guanximiyou’尤力克柠檬C.limon‘Eureka Lemon’金弹Fortunella crassifolia枳Poncirus trifoliata柚选3号C.grandis‘Youxuan No.3’柚选4号C.grandis‘Youxuan No.4’桂柚1号C.grandis‘Guiyouyihao’柚选1号C.grandis‘Youxuan No.1’柚选2号C.grandis‘Youxuan No.2’泰国柚C.grandis‘Thailand pomelo’星路比C.paradisi‘Star Ruby’红宝石C.grandis‘Hongbaoshi’分子身份证编号Molecular identity code 39553328 99471511 5b126b57 57118899 55239aab 33452524 33453348 39553328 39553328 39553328 36773512 23454422 12573324编号Code 14 15 16 17 18 19 20 21 22 23 24 25种质名称Germplasm name泰国考雅C.grandis‘Thailand Kauya’越南翡翠青柚C.grandis‘Vietnamese jade’广西蜜柚C.grandis‘Guangximiyou’红肉蜜柚C.grandis‘Hongroumiyou’青皮柚C.grandis‘Qingpiyou’奥兰柚C.grandis‘Oroblanco’马家柚C.grandis‘Majiayou’囊内柚C.grandis‘Nangneiyou’五鞭柚C.grandis‘Wubianyou’日柚C.grandis‘Riyou’演柚C.grandis‘Yanyou’福泽柚C.grandis‘Fuzeyou’分子身份证编码Molecular identity code 23563422 22583422 99471511 99471511 12573324 29250000 38224426 6a221322 6a221322 23774424 49254728 39554538
2.4 聚类分析
根据遗传距离(genetic distance,GD)和遗传相似系数(genetic similarity,GS),使用UPGMA 的方法对22 份柚类开展聚类分析,得到聚类关系树(图2)。可以发现,在遗传距离为0.84 时分为3 组,第1组为枳;第2 组为金弹和尤力克柠檬;第3 组包括22 份柚类种质;在遗传距离为0.36 处,22 份柚类可分为2 个亚组,第1 个亚组为红肉蜜柚、广西蜜柚和琯溪蜜柚;第2 个亚组包含19 份资源,包括沙田柚及变异单株桂柚1 号、青柚等栽培品种和地方柚类品种。从中可以看出,在遗传距离为0.18 时,6 个柚品种被区分开,日柚和考雅聚为一支,与越南翡翠青柚分离;而青皮柚和红宝石聚在一起,本研究构建的两者的分子身份证相同,说明其亲缘关系很近。在遗传距离为0.12 处,桂柚1 号、沙田柚、柚选1 号、柚选2 号聚为一支,而与柚选3 号和柚选4 号分离,前四者共享编码构建的分子身份特征代码,说明四者亲缘关系很近。从聚类关系树中可以看出,五鞭柚和囊内柚、红肉蜜柚和广西蜜柚聚在一起,又与琯溪蜜柚分离,说明五鞭柚和囊内柚、红肉蜜柚和广西蜜柚的基因组相似性序列相似,亲缘关系很近,红肉蜜柚、广西蜜柚与琯溪蜜柚可能是属于同一个柚类群。关系树上显示枳、柠檬、金弹单独分支,说明SSR荧光标记引物具有可重复性和准确性。
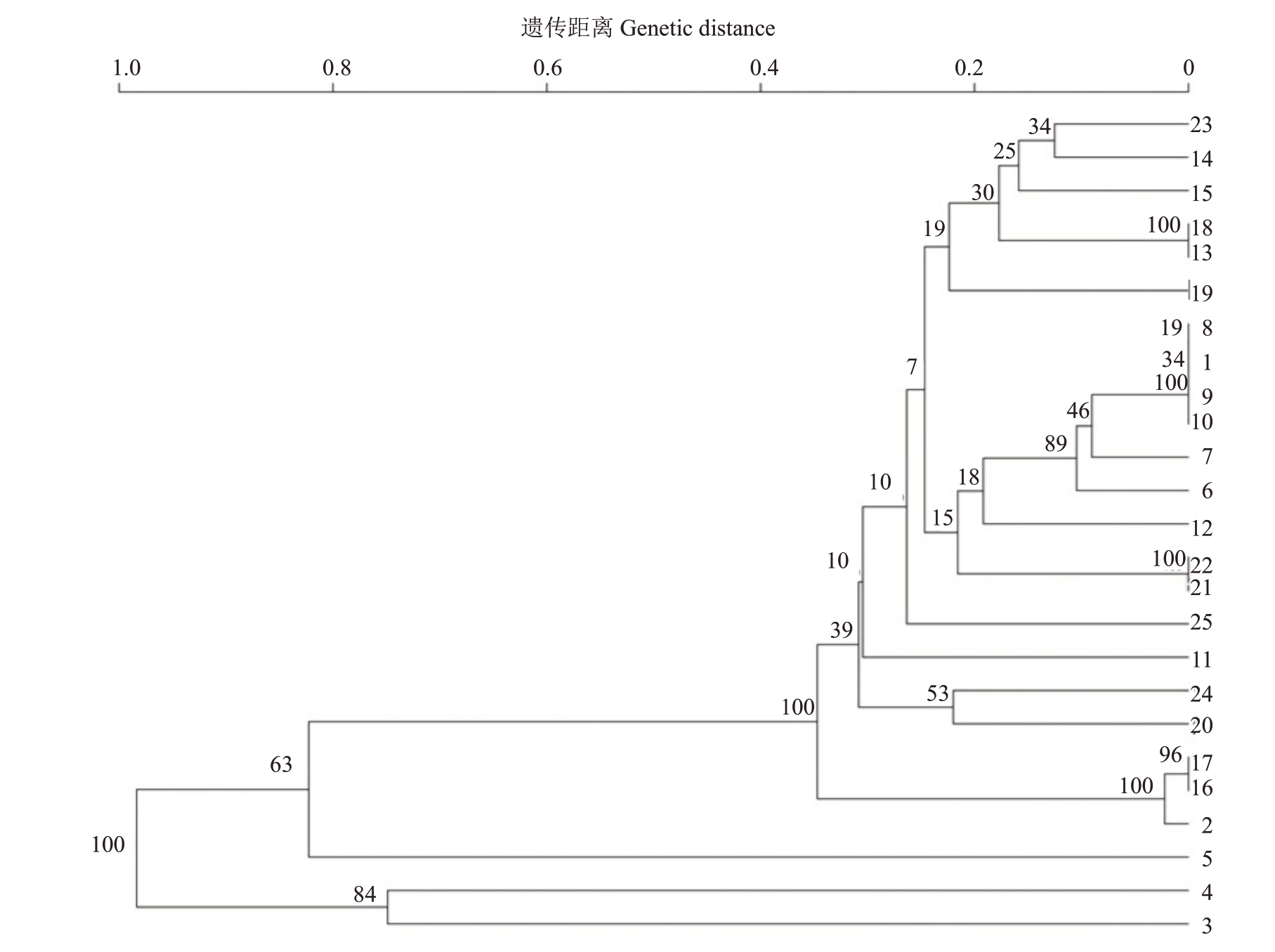
图2 基于SSR 标记的22 份柚类品种UPGMA 树状图
Fig.2 UPGMA tree map of 22 pomelo varieties based on SSR markers
3 讨 论
SSR 荧光标记法能精准识别目标DNA 片段大小,检测结果稳定、准确和高效,适用大批量品种的检测分析[11,17],已被广泛应用于品种鉴定和分子身份证的构建。李益等[16]利用SSR荧光标记筛选出21对核心引物,成功构建了包含500 份柑橘品种的DNA指纹图谱库,形成了基于荧光毛细管电泳平台的柑橘SSR分子标记品种鉴定体系。徐雷锋等[18]利用筛选的SSR引物,采用荧光测序技术结合新的带型编码方式构建96 份百合样本的分子身份证。朱田田等[19]利用荧光SSR 分子标记对甘肃省11 个当归(Angelica sinensis)品种(系)的194份当归样品进行遗传多样性和遗传结构分析,表明SSR荧光标记检测方法可以对目标DNA片段进行准确检测,结果可靠。关于柚类SSR标记的研究已有报道,刘冬峰等[20]利用4 对SSR 引物组合,区分18 份浙江柚类资源并构建了浙江地方柚类资源的种质鉴定图;刘勇等[21]利用具有多态性的33 对SSR 引物对来自中国的122份柚类资源进行了分析,分为7个组群,进而可细分为18 个亚组,主要以三大品种群组成:沙田柚品种群、文旦柚品种群及庞大的杂种柚品种群;王旭等[22]利用21对核心引物,筛选出17对具有高多态性的SSR 引物对68 份柚类种质资源亲缘关系进行分析,聚类分析结果与结构分析结果一致,表明垫江柚系列与长寿沙田柚具有很近的亲缘关系并有相同的基因型,从分子水平上说明了垫江柚系列可能属于沙田柚类型。国外对柚类也进行了遗传多样性研究,Kongsri等[23]对泰国优良商品品种,泰国中部、南部和北部的地方品种和国外的共53 个无性系进行评价,结果表明利用10个具有多态性SSR标记的引物,共产生33个等位基因,大多数柚品种,包括商业品种和杂交品种,都属于同一类群,而本地和国外品种比商业品种更多样化;Ahmed 等[24]利用60 个SSR标记对柚品种进行分子鉴定,发现了26 个多态性SSR 位点,具有77 个扩增等位基因,结果显示粉柚和白柚的遗传相似系数最大,其亲缘关系较近。
建立“分子身份证”,对品种鉴定、种苗和农业生物产品的真伪鉴别具有重要意义。目前分子身份证的构建已在绿豆[25]、小麦[26]、水稻[27-28]、黍子[29]、核桃[30]等上应用。DNA 指纹图谱是构建分子身份证的基础,但指纹图谱区别于分子身份证。目前,构建分子身份证不同的品种采用不同的编码方法[18,31],笔者在本研究中采用扩增等位基因分子质量和结合图谱带型方式构建22份柚类资源分子身份证,在分子身份证上容易区分种质扩增的基因带型是纯合子还是杂合子,且这种编码方式需要的标记较少,形成的字符串不长。
目前,应用分子标记进行柑橘品种鉴定还处于研究阶段,对于亲缘关系很近的柑橘品种,比如本研究的桂柚1 号是沙田柚的变异单株,两者很难区分开;引物开发和设计的SSR 标记引物是在已知基因组上随机片段设计的,无法实现设计的引物在各条染色体上均匀分布,也不能代表整个基因组的信息。本研究形成的分子身份证能提供参考,但还不能全面的代表柚类品种的信息。因此需要结合全基因组测序等方法对品种进行更全面的鉴定。
4 结 论
本研究通过SSR荧光毛细管电泳检测,获得22份柚类的图谱带型和等位基因数据,得到了15份不同类型的分子身份证代码,其中12条为信息独特的身份证,为柚类及柑橘种质资源鉴定和品种保护提供依据。
[1] 张太平,彭少麟.柚的起源、演化及分布初探[J].生态学杂志,2000,19(5):58-61.
ZHANG Taiping,PENG Shaolin. Introduction to the origin and evolution of pomelo and its distribution in China[J]. Chinese Journal of Ecology,2000,19(5):58-61.
[2] 彭瑜,苏智先.部分柚类品种叶片性状分类研究[J].绵阳师范学院学报,2007,26(2):82-85.
PENG Yu,SU Zhixian. Study on leaf shape classification of some pomelo varieties[J]. Journal of Mianyang Teachers’College,2007,26(2):82-85.
[3] 陈振光,赖钟雄.中国柚的种质资源及其研究[J].福建农学院学报(自然科学版),1993,22(3):290-295.
CHEN Zhenguang,LAI Zhongxiong. Introduction and research of pummelo germplasm in China[J].Journal of Fujian Agricultural University(Natural Science Edition),1993,22(3):290-295.
[4] 武晓晓,唐艳,邓崇岭.柑桔孢粉学研究进展[J].中国南方果树,2017,46(5):148-153.
WU Xiaoxiao,TANG Yan,DENG Chongling.Advances in studies of citrus palynology[J].South China Fruits,2017,46(5):148-153.
[5] 武晓晓,唐艳,邓崇岭,刘冰浩,陈传武,牛英.柑橘不同种属花粉形态观察[J].果树学报,2018,35(7):794-801.
WU Xiaoxiao,TANG Yan,DENG Chongling,LIU Binghao,CHEN Chuanwu,NIU Ying. Observation of citrus pollen morphology by scanning electron microscopy[J]. Journal of Fruit Science,2018,35(7):794-801.
[6] 姜成东,彭振坤.两个沙田柚品系同工酶分析[J].湖北民族学院学报(自然科学版),2004,20(4):1-4.
JIANG Chengdong,PENG Zhenkun.Analysis of isodynamic enzyme for two breeds of shatian pomelo[J]. Journal of Hubei Minzu University(Natural Science Edition),2004,20(4):1-4.
[7] SAMARINA L S,KULYAN R V,KONINSKAYA N G,GORSHKOV V M,RYNDIN A V,HANKE M V,FLACHOWSKY H,REIM S. Genetic diversity and phylogenetic relationships among citrus germplasm in the Western Caucasus assessed with SSR and organelle DNA markers[J]. Scientia Horticulturae,2021,288(4):110355.
[8] GOLEIN B,BIGONAH M,AZADVAR M,GOLMOHAMMADI M.Analysis of genetic relationship between‘Bakraee’(Citrus sp.) and some known Citrus genotypes through SSR and PCR-RFLP markers[J]. Scientia Horticulturae,2012,148(1):147-153.
[9] NIE X H,WANG Z H,LIU N W,SONG L,YAN B Q,XING Y,ZHANG Q,FANG K F,ZHAO Y L,CHEN X,WANG G P,QIN L,CAO Q Q. Fingerprinting 146 Chinese chestnut (Castanea mollissima Blume) accessions and selecting a core collection using SSR markers[J]. Journal of Integrative Agriculture,2021,20(5):1277-1286.
[10] SCHLAUTMAN B,BOLIVAR-MEDINA J,HODAPP S,ZALAPA J. Cranberry SSR multiplexing panels for DNA horticultural fingerprinting and genetic studies[J]. Scientia Horticulturae,2017,219:280-286.
[11] 郝晨阳,王兰芬,贾继增,董玉琛,张学勇.SSR 荧光标记和银染技术的比较分析[J].作物学报,2005,31(2):144-149.
HAO Chenyang,WANG Lanfen,JIA Jizeng,DONG Yuchen,ZHANG Xueyong. Comparison of fluorescence and silverstaining detection systems of microsatellite markers[J]. Acta Agronomica Sinica,2005,31(2):144-149.
[12] 刘伟,李淼,李桂祥,董晓民,高晓兰,张安宁.应用SSR 荧光标记法构建山东地方桃种质资源分子身份证[J].山东农业科学,2022,54(2):6-13.
LIU Wei,LI Miao,LI Guixiang,DONG Xiaomin,GAO Xiaolan,ZHANG Anning. Using fluorescent labeled SSR markers to establish molecular ID of pench germplasm resources from Shandong province[J]. Shandong Agricultural Sciences,2022,54(2):6-13.
[13] 苑克俊,牛庆霖,葛福荣,王江勇,王培久.利用荧光SSR 标记构建杏新品系的分子身份证[J].北方园艺,2018(4):34-40.
YUAN Kejun,NIU Qinglin,GE Furong,WANG Jiangyong,WANG Peijiu. Using the fluorescent labeled SSR markers to establish molecular identity of apricot germplasms[J]. Northern Horticulture,2018(4):34-40.
[14] 张小双,曹玉芬,齐丹,张莹,田路明,董星光,霍宏亮,徐家玉,刘超.秋子梨基于SSR 荧光标记的分子身份证构建及亲缘关系分析[J].中国南方果树,2018,47(4):92-98.
ZHANG Xiaoshuang,CAO Yufen,QI Dan,ZHANG Ying,TIAN Luming,DONG Xingguang,HUO Hongliang,XU Jiayu,LIU Chao.Analysis of genetic relationship and establish molecular ID of Pyrus ussuriensis based on SSR molecular marker[J].South China Fruits,2018,47(4):92-98.
[15] 李慧峰,王涛,冉昆.利用SSR 荧光标记构建41 份山东省苹果资源分子身份证[J].沈阳农业大学学报,2020,51(1):70-77.
LI Huifeng,WANG Tao,RAN Kun. Using the fluorescent labeled SSR markers to establish the molecular identity of 41 Malus germplasms in Shandong province[J]. Journal of Shenyang Agricultural University,2020,51(1):70-77.
[16] 李益,马先锋,唐浩,李娜,江东,龙桂友,李大志,牛英,韩瑞玺,邓子牛.柑橘品种鉴定的SSR 标记开发和指纹图谱库构建[J].中国农业科学,2018,51(15):149-159.
LI Yi,MA Xianfeng,TANG Hao,LI Na,JIANG Dong,LONG Guiyou,LI Dazhi,NIU Ying,HAN Ruixi,DENG Ziniu. SSR markers screening for identification of citrus cultivar and construction of DNA fingerprinting library[J]. Scientia Agricultura Sinica,2018,51(15):149-159.
[17] CMEJLOVA J,REJLOVA M,PAPRSTEIN F,CMEJLA R. A new one-tube reaction kit for the SSR genotyping of apple (Malus×domestica Borkh.)[J].Plant Science,2021,303:110768.
[18] 徐雷锋,葛亮,袁素霞,任君芳,袁迎迎,李雅男,刘春,明军.利用荧光标记SSR 构建百合种质资源分子身份证[J]. 园艺学报,2014,41(10):2055-2064.
XU Leifeng,GE Liang,YUAN Suxia,REN Junfang,YUAN Yingying,LI Ya’nan,LIU Chun,MING Jun. Using the fluorescent labeled SSR markers to establish molecular identity of lily germplasms[J]. Acta Horticulturae Sinica,2014,41(10):2055-2064.
[19] 朱田田,张明惠,王富胜,王圆圆,栗孟飞,晋玲.基于SSR 荧光标记的不同品种(系)当归遗传关系分析及分子身份证构建[J].中草药,2022,53(12):3774-3783.
ZHU Tiantian,ZHANG Minghui,WANG Fusheng,WANG Yuanyuan,LI Mengfei,JIN Ling. Genetic relationship analysis and molecular ID codes construction of different cultivars(lines) of Angelica sinensis based on fluorescent labeled SSR markers[J]. Chinese Traditional and Herbal Drugs,2022,53(12):3774-3783.
[20] 刘冬峰,陈巍,林绍生,徐文荣,郭秀珠,黄品湖.基于SSR 标记的浙江地方柚类种质资源遗传关系分析[J]. 果树学报,2017,34(2):166-174.
LIU Dongfeng,CHEN Wei,LIN Shaosheng,XU Wenrong,GUO Xiuzhu,HUANG Pinhu. Analysis of genetic relationship of pummelo germplasms by SSR markers in Zhejiang province[J].Journal of Fruit Science,2017,34(2):166-174.
[21] 刘勇,刘德春,吴波,孙中海.利用SSR 标记对中国柚类资源及近缘种遗传多样性研究[J]. 农业生物技术学报,2006,14(1):90-95.
LIU Yong,LIU Dechun,WU Bo,SUN Zhonghai.Genetic diversity of pummelo and their relatives based on SSR markers[J].Journal of Agricultural Biotechnology,2006,14(1):90-95.
[22] 王旭,彭洁,朱延松,杨胜男,张晓楠,余洪,江东,梁大成.基于SSR 分子标记的68 份柚类种质资源亲缘关系分析[J].安徽农业科学,2021,49(4):100-103.
WANG Xu,PENG Jie,ZHU Yansong,YANG Shengnan,ZHANG Xiaonan,YU Hong,JIANG Dong,LIANG Dacheng.Analysis of genetic relationship of 68 pummelo germplasm resources based on SSR molecular marker[J]. Journal of Anhui Agricultural Sciences,2021,49(4):100-103.
[23] KONGSRI S,BOONPRAKOB U. Assessment of genetic relationships among pummelo cultivars [Citrus maxima (Burm.)Merrill] using simple sequence repeat markers[J]. Maejo Internaional Journal of Science and Technology,2016,10(2):209-219.
[24] AHMED S,RATTANPAL H S,SINGH G. Diversity,characterization and evaluation in pummelo (Citrus maxima Merr.) cultivars using SSR markers and quality parameters[J]. Indian Journal of Genetics and Plant Breeding,2019,79(3):594-605.
[25] ZHAO Y N,WANG Y,WANG L X,ZHANG D J. Molecular identification of mung bean accessions (Vigna radiata L.) from Northeast China using capillary electrophoresis with fluorescence-labeled SSR markers[J].Food and Energy Security,2020,9(1):1-29.
[26] 白晓倩,陈于,张仕杰,赵玉强,王武,朱灿灿.基于表型性状和SSR 标记的板栗品种遗传多样性分析及分子身份证构建[J].植物遗传资源学报,2022,23(4):972-984.
BAI Xiaoqian,CHEN Yu,ZHANG Shijie,ZHAO Yuqiang,WANG Wu,ZHU Cancan.Genetic diversity analysis and fingerprinting of chestnut varieties based on phenotypic traits and SSR markers[J]. Journal of Plant Genetic Resources,2022,23(4):972-984.
[27] 陆徐忠,倪金龙,李莉,汪秀峰,马卉,张小娟,杨剑波. 利用SSR 分子指纹和商品信息构建水稻品种身份证[J].作物学报,2014,40(5):823-829.
LU Xuzhong,NI Jinlong,LI Li,WANG Xiufeng,MA Hui,ZHANG Xiaojuan,YANG Jianbo. Construction of rice variety indentity using SSR fingerpring and commodity information[J].Acta Agronomica Sinica,2014,40(5):823-829.
[28] 管俊娇,杨晓洪,王江民,张鹏,黄清梅,白洁,余志慧,张建华.云粳系列水稻品种分子身份证的构建[J]. 江苏农业学报,2018,34(6):1201-1206.
GUAN Junjiao,YANG Xiaohong,WANG Jiangmin,ZHANG Peng,HUANG Qingmei,BAI Jie,YU Zhihui,ZHANG Jianhua.Establishment of molecular identity for rice variety of Yunjing series[J]. Jiangsu Journal of Agricultural Sciences,2018,34(6):1201-1206.
[29] 丁艺冰,丁雨格,陈凌,王海岗,陈喜明,王瑞云,乔治军.基于高基元SSR 构建黍子DNA 分子身份证[J]. 山西农业科学,2022,50(5):620-629.
DING Yibing,DING Yuge,CHEN Ling,WANG Haigang,CHEN Ximing,WANG Ruiyun,QIAO Zhijun. Construction of DNA molecular ID card of Panicum miliaceum based on highmotif SSR[J]. Journal of Shanxi Agricultural Sciences,2022,50(5):620-629.
[30] 梁燕,韩传明,周继磊,孙超,王翠香,李春明,王静,闵旭峰,公庆党,孟晓烨,杨绪强.山东核桃良种SSR 指纹图谱及分子身份证的构建:基于毛细管电泳分析[J].中国农学通报,2022,38(15):113-121.
LIANG Yan,HAN Chuanming,ZHOU Jilei,SUN Chao,WANG Cuixiang,LI Chunming,WANG Jing,MIN Xufeng,GONG Qingdang,MENG Xiaoye,YANG Xuqiang.Construction of SSR fingerprint and molecular identity card of Shandong elite Julans regia cultivars:Based on capillary electrophoresis analysis[J].Chinese Agricultural Science Bulletin,2022,38(15):115-121.
[31] 马旭丹,孙辽,李元元,肖本泽.部分水稻不育系的指纹图谱构建和遗传多样性分析[J].杂交水稻,2015,30(6):64-70.
MA Xudan,SUN Liao,LI Yuanyuan,XIAO Benze.Analysis of genetic polymorphism and fingerprinting of some male sterile lines in rice[J].Hybrid Rice,2015,30(6):64-70.