随着中国种业创新力度的不断加大,人们对各类种质资源的保护和利用愈发重视,但是植物资源相当丰富,且植株普遍高大,全部保存成本会很高。因此,构建植物资源核心种质,是一种有效解决该问题的方法。核心种质的概念最早由Frankel 等[1]提出,即采用一定的方法选取最少的种质资源数量以最大程度地代表该类种质资源的遗传多样性。核心种质的构建主要有两种途径,分别是基于表型性状和基于分子标记基因型数据的构建方法。基于表型性状构建核心种质极易受环境的影响,因此误差较大,且需要对多种性状进行分析,工作量巨大。分子标记技术以信息量大、效率高、不受环境影响等特点[2],尤其是迅速发展的SSR标记具有稳定好、多态性高、共显性遗传、操作简便等优点[3],已逐渐成为构建核心种质的主要方法。目前,SSR 分子标记技术已经成功应用到多种作物的核心种质构建中,对资源保护和创新利用以及新品种的选育发挥了重要作用。如在蔬菜中,崔竣杰等[4]根据表型和SSR 标记构建了苦瓜的核心种质;李金龙等[5]利用SSR 分子标记技术研究甜荞种质资源的群体结构,进而构建甜荞的初级核心种质库;吴茵[6]在利用SRAP分子标记构建辣椒初级核心种质的基础上,利用SSR分子标记压缩初级核心种质进而构建了最终的核心种质;果树中鲁敏等[7]采用位点优先取样策略对收集的102 份自然种质构建核心种质;李冬波等[8]利用SSR分子标记对广西荔枝种质资源进行遗传多样性分析并构建核心种质,从而对资源保护发挥了重要作用;邹梁峰[9]基于表型性状和SSR 分子标记构建了毛花猕猴桃的核心种质,从而对中国广泛分布的这种特有的种质资源进行了很好的保护利用。刘娟等[10-11]分别基于表型性状和ISSR分子标记构建了野杏和南疆杏资源的核心种质,花卉中陈明堃等[12]统计建兰的表型并利用SSR标记筛选了核心种质,能够最大程度代表原始种质的遗传多样性。此外,林木中的杉木[13]、毛白杨[14]、刺槐[15]、核桃[16]、白蜡[17]、楸树[18]等都构建了核心种质,对于资源保护和创新利用有重要作用。
板栗(Castanea mollissima Bl.)为壳斗科栗属植物,果实味道鲜美,具有很高的营养价值和药用价值[19],早在6000 年前,古人就已经食用野生板栗[20]。中国板栗资源丰富,可分为华北、长江流域、西南、东南、西北与东北6 个品种群[21],现阶段约有300 个栽培品种(系)。同时,随着自然变异和人工选择,市场上逐渐出现新品种,同时淘汰一些过时的品种。因此,如果不及时发现和保护,就会造成优异种质资源的遗失和浪费,从而对今后资源的创新利用造成困难。所以,构建中国板栗品种的核心种质,对其保护和利用具有重要的意义。前人对板栗品种核心种质的构建仅在某一地区或所采集的板栗品种资源不够全面[22-23],不足以代表全国板栗品种资源的遗传多样性。笔者以国家板栗资源圃和江苏良种繁育基地收集的342 个品种(系)资源为材料,应用荧光SSR 分子标记技术获取片段信息,基于分层取样法、模拟退火算法和随机搜索算法构建全国板栗品种核心种质,从而为今后板栗新品种的创制、品种的种质鉴定和保存利用提供理论基础。
1 材料和方法
1.1 试验材料
试验材料为从山东省泰安市国家板栗资源圃和位于江苏板栗良种繁殖基地搜集的共342个板栗品种,它们原生地分布在16个省份。资源圃位于当地的丘陵山地地区,降水量较大,土壤适合板栗生长和栽培。
1.2 试验方法
1.2.1 DNA提取 板栗叶片DNA提取采用美基生物公司生产的广谱植物DNA 提取试剂盒。采用1.5%琼脂糖凝胶电泳检测DNA完整度,用超微量分光光度计测定DNA 浓度和纯度,将合格的DNA 保存在-20 ℃冰箱中。
1.2.2 荧光毛细管电泳SSR-PCR 扩增 SSR 引物采用本实验室之前所筛选得到的21 对高多态SSR标记进行PCR扩增[22]。PCR扩增采用20 μL反应体系:其中包含1 μL模板DNA(20~50 ng·μL-1),10 μL 2×Taq PCR Master Mixes(Takara, DaLian China),0.1 μL 正向引物(10 μmol·L-1),0.3 μL 反向引物(10 μmol·L-1),0.2 μL 荧光标记(FAM,HEX,ROX)的M13引物(10 μmol·L-1)(中国天津,擎科)和9.4 μL ddH2O。反应程序为:94 ℃预变性3 min,94 ℃变性30 s,56 ℃退火30 s,72 ℃延伸30 s,35 个循环,最后72 ℃延伸10 min,10 ℃保存(BIO-RAD PCR Thermal Cycler T100,USA)。
将3对荧光SSR引物的PCR扩增产物合并在一起,然后使用ABI 3730XL DNA 测序仪(Applied Biosystems, Foster City, CA, USA)通过毛细管电泳分析混合物中片段位置信息,并使用Gene Marker V 2.2.0 软件(Soft Genetics LLC, State College, PA,USA)读取等位位点具体的片段大小信息。
1.2.3 数据分析 按照PowerMarker V3.25[24]软件要求的格式,在Excel表格中将读取的SSR 原始“bp值型”数据转换成powermarker格式,缺失的数据赋值为“0”,并利用该软件及GenAlEx6.51[25-26]计算遗传多样性指标参数,包括等位基因数量(Na)、有效等位基因数(Ne)、观测杂合度(Ho)、期望杂合度(He)、多态信息含量(PIC)。
1.2.4 核心种质的构建(1)基于分层取样法筛选核心种质。基于最大等位基因数原则,根据聚类图采用逐步聚类留样,每一轮聚类,随机去除最低分类水平上两个个体中的一个,剩下的一个入选提取比例,第二轮提取比例为从上次提取的样品中聚类筛选,从而形成八个提取比例(100%、83.33%、66.67%、46.49%、31.58%、20.76%、13.74%、8.77%),并计算这8个提取比例的等位位点数量。
(2)基于模拟退火算法筛选核心种质。为构建合适的核心种质,再通过基于最大等位基因数(SAAN)的模拟退火算法来筛选和构建中国板栗品种的核心种质。在Power Marker v3.25 软件中建立一个梯度样本抓取数量方案来计算所选的不同容量的样品所保留的等位位点的数量,从而确定最终筛选的核心种质样品数量。为保证尽可能的保留等位位点数,采用5,10,15,20,25,30,35,40,45,55,60,65,70,75,80,85,90,95,100,105,110,115,120,125,130,135,140,145,150,155,160,165,170,175,180,185,190,195,200,205,210,215,220,225,230,235,240,245,250,255,260,265,270,275,280,285,290,295,300,305,310,315,320,325,330,335,340的梯度抓取方案来计算不同取样数量的等位位点数,从而确定等位位点最大时的抓取数量。
(3)基于随机搜索算法筛选核心种质。基于最大等位基因数(RS,AN,AN)的随机搜索算法来构建板栗品种的核心种质。该方法同样利用上述基于模拟退火算法的梯度抓取方案,比较不同取样数量的等位位点数量,从而确定抓取数量。
将上述3 种策略所获得的取样数量、等位位点数量等进行比较以确定最终的取样数量,进而来评估出3种核心种质构建策略中的最优策略。
最后通过Prism 软件[27]对核心种质和原始种质之间的Na、Ne、Ho、He 和PIC 值进行T 检验,从而验证所构建的核心种质的准确性、科学性和可靠性。
2 结果与分析
2.1 原始板栗品种(系)种质聚类分析
对所有资源进行遗传多样性分析,结果表明342份板栗品种资源共有212个等位位点,平均每对引物所保留的等位基因数(Na)为10.095,有效等位基因数(Ne)为3.488,观测杂合度为0.596,期望杂合度为0.658,多态信息含量为0.642。同时基于UPGMA方法进行聚类分析,结果如图1所示。全部资源可以划分为3个组,蓝色部分大多为北方品种,但混有31 份南方品种;红色部分大多为南方品种,但混有39 份北方品种;紫色部分共有42 份品种,其中大部分为山东资源,占78.57%,在地理位置上正好也为南北两大类群的交汇地带。由此推测,全部资源大致分为南北两个群体,但也存在较多的混杂,南北方板栗品种存在较多基因交流,山东地区为南北品种类群的混交地带。群体结构分析进一步证明,全部板栗资源大致分为两部分,但存在较多混杂。群体结构中混杂系数大于0.8 被认为存在较多基因交流,如北方品种群中混入的南方品种Y140系数大于0.8,因此会聚类到北方品种群中,表明该品种存在频繁的基因交流。

图1 全部板栗资源的UPGMA 系统树谱图(A)及群体结构图(B)
Fig.1 UPGMA dendrogram(A)and group structure(B)of all Castanea germplasm
2.2 核心种质构建与评价
2.2.1 构建必选种质库 种质资源库对于育种和基因挖掘具有重要作用。如图2 所示,根据板栗品种独特的成熟期、枝条长度和弯曲度、刺蓬色泽等,从全部资源中挑选出8个具有特殊性状的品种作为必选品种,而这些资源会在未来育种和遗传研究等方面发挥重要作用。其中,早熟型品种有京暑红、燕山早丰;短枝型品种有短枝1 号、短枝3 号和超短枝2号;垂枝型品种有垂枝2 号;雄花不育型品种有无花;红刺型品种有山东红栗。

图2 特殊种质资源
Fig.2 The special germplasm resources
2.2.2 核心种质构建 为了确定最合适的核心种质构建策略以及取样数量,笔者基于最大等位基因数,利用分层抽样法、模拟退火算法和随机搜索算法3种方法构建最优核心种质(图3)。

图3 3 种方法进行核心种质筛选对比
Fig.3 Comparison of three methods for core collection screening
利用分层抽样策略,通过逐步聚类取样形成了8 个取样比例为100%、83.33%、66.67%、46.49%、31.58%、20.76%、13.74%、8.77%,并计算每个取样比例下的等位位点数量分别为212、207、202、196、185、179、163、152。随着取样比例的升高,所保留的等位位点数量也逐渐增多,当取样比例达到100%时,等位位点数量达到最大为212。
RS,AN,AN 为利用基于等位位点数量的随机搜索算法来构建核心种质。以5为梯度计算等位位点数量,结果表明,等位位点数量呈现波浪变化,当取样数量达到325份时所保留的等位位点数量达到最大值212。
SA,AN,AN 为利用基于等位位点数量的模拟退火算法来构建核心种质。结果表明当以5为梯度计算等位位点数量时,随着取样数量的增加等位位点数量也逐渐升高。当取样数量达到85份资源时,所保留的等位位点数达到最大值212,并且随着取样数量继续增加时,等位位点数量趋于平稳,不再升高或降低。
综上所述,通过对3 种方法在不同取样数量时的等位位点数比较分析,结果表明SA,AN,AN策略为构建核心种质的最优策略,且当种质资源数量达到85份时能够代表全部资源的等位基因数,即所保留的等位位点数为212。
2.2.3 核心种质评价及遗传多样性分析 为了验证这85 份核心种质资源的合理性、可靠性及科学性,首先,笔者对核心种质在全部资源的分布情况进行主坐标分析(PCoA)以及对核心种质进行群体结构分析。如图4 所示,Pop2 部分为核心种质,Pop1 部分为筛选之后的剩余种质,主坐标1和2分别解释了位点信息数据中34.36%和15.71%的变异,初步显示核心种质在全部种质资源中较为均匀的分布,从而表明所筛选结果的合理性。群体结构分析表明,85个核心种质资源与全部种质资源资源相同,大致分为两部分。
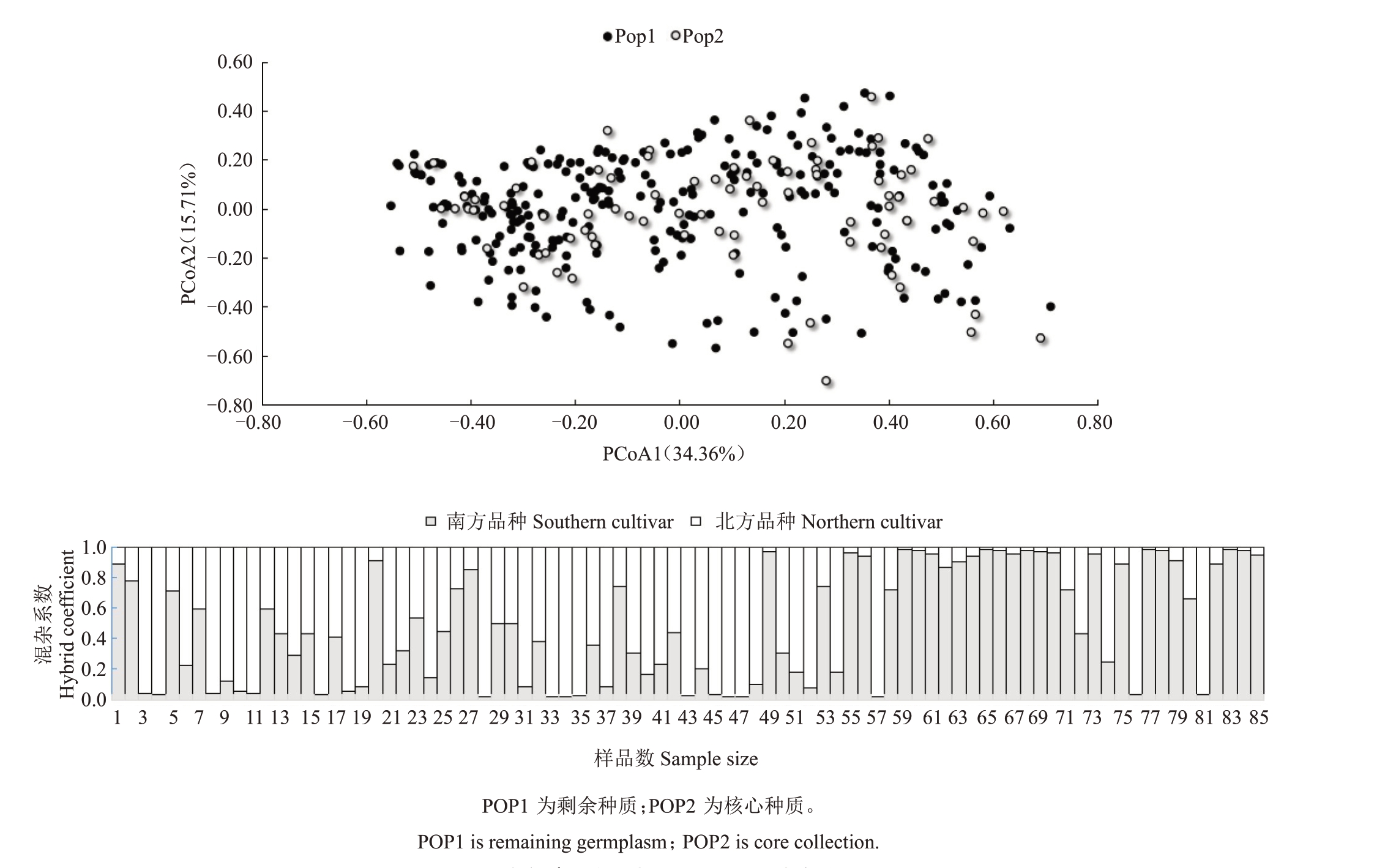
图4 核心种质主坐标分析及群体结构分析
Fig.4 Principal coordinate analysis(PCoA)and group structure of the core germplasm
随后笔者对所筛选的核心种质和原始种质之间的遗传多样性参数进行分析。如表1 所示,原始种质共342份,核心种质共85份,保留率为24.85%;核心种质的等位位点数量和原始种质的等位位点数相同,保留率为100%;等位基因数(Na)、有效等位基因数(Ne)、观测杂合度(Ho)、期望杂合度(He)、多态信息含量(PIC)的保留率分别为86.32%、85.46%、98.15%、90.12%和100.79%。
表1 核心种质筛选保留率
Table 1 Core germplasm screening retention rate
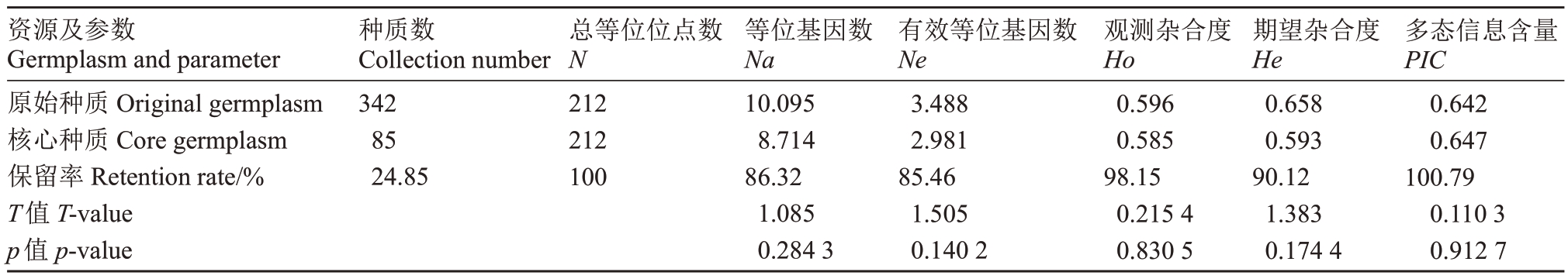
资源及参数Germplasm and parameter原始种质Original germplasm核心种质Core germplasm保留率Retention rate/%T值T-value p值p-value种质数Collection number 3428524.85总等位位点数N 212212100等位基因数Na 10.0958.71486.321.0850.2843有效等位基因数Ne 3.4882.98185.461.5050.1402观测杂合度Ho 0.5960.58598.150.21540.8305期望杂合度He 0.6580.59390.121.3830.1744多态信息含量PIC 0.6420.647100.790.11030.9127
对核心种质和原始种质的遗传多样性参数进行T 检验,结果显示所构建的核心种质各遗传多样性参数与原始种质之间没有显著差异,且其原生地涵盖11个省份,其他省份样品较少,分析中没有入选,表明所构建的85 个核心种质资源能够很好地保存原始种质资源的遗传多样性(表2)。
表2 筛选的85 个核心种质
Table 285 core germplasm screened

序号Number 123456789101112 13141516 17181920 21222324 25262728 29303132 33343536 37383940 414243品种Cultivars(lines)八月炸Bayuezha尖顶油栗A Jiandingyouli A大叶栗Dayeli燕山早丰Yanshanzaofeng确山10号Queshan 10 hao山东红栗Shandonghongli郯城3号Tancheng 3 hao西寨1号Xizhai 1 hao无花Wuhua大板红Dabanhong红栗6号Hongli 6 hao大栗青Daliqing鲁栗1号Luli 1 hao鲁栗3号Luli 3 hao鲁栗4号Luli 4 hao不育2号Buyu 2 hao威丰Weifeng XZ-1 N4-41美丰Meifeng圃308 Pu30821430682圃151 Pu151大公书2号Dagongshu 2 hao栗园8号Liyuan 8 hao尖顶油栗B Jiandingyouli B东密坞无花Dongmiwuwuhua北味Beiwei超短枝2号Chaoduanzhi 2 hao短枝3号Duanzhi 3 hao短枝1号Duanzhi 1 hao青龙23号Qinglong 23 hao燕秋Yanqiu紫晶Zijing圣山栗Shengshanli燕山红栗Yanshanhongli垂枝2号Chuizhi 2 hao泰安薄壳Tai’anbaoke华丰Huafeng包丰Baofeng蒙山二早Mengshan’erzao京暑红Jingshuhong原生地Origin region河南确山Queshan,Henan山东郯城Tancheng,Shandong江苏溧阳Liyang,Jiangsu河北迁西Qianxi,Hebei河南信阳Xinyang,Henan山东泰安Tai’an,Shandong山东郯城Tancheng,Shandong河北迁西Qianxi,Hebei山东泰安Tai’an,Shandong河北宽城Kuancheng,Hebei山东泰安Tai’an,Shandong江苏溧阳Liyang,Jiangsu山东泰安Tai’an,Shandong山东泰安Tai’an,Shandong山东泰安Tai’an,Shandong山东泰安Tai’an,Shandong山东威海Weihai,Shandong山东泰安Tai’an,Shandong山东泰安Tai’an,Shandong山东Shandong山东泰安Tai’an,Shandong山东泰安Tai’an,Shandong山东泰安Tai’an,Shandong山东泰安Tai’an,Shandong山东莒南Junan,Shandong山东泰安Tai’an,Shandong山东郯城Tancheng,Shandong河北Hebei山东泰安Tai’an,Shandong山东泰安Tai’an,Shandong山东泰安Tai’an,Shandong山东泰安Tai’an,Shandong河北青龙Qinglong,Hebei山东莒南Junan,Shandong河北青龙Qinglong,Hebei山东泰安Tai’an,Shandong北京Beijing山东郯城Tancheng,Shandong山东泰安Tai’an,Shandong山东泰安Tai’an,Shandong山东Shandong山东蒙阴Mengyin,Shandong北京Beijing序号Number 44454647 48495051 52535455 56575859 60616263 64656667 68697071 72737475 76777879 80818283 8485品种Cultivars(lines)虎爪栗Huzhuali新选1号Xinxuan 1 hao迁西早红Qianxizaohong燕兴Yanxing辛庄2号Xinzhuang 2 hao高店1号Gaodian 1 hao灰拣Huijian柞14 Zuo14明拣Mingjian搓湾中栗Cuowanzhongli双季栗Shuangjili马齿青Machiqing开化双季栗Kaihuashuangjili广西玉林11号Guangxiyulin 11 hao镇巴双洁Zhenbashuangjie闭口红Bikouhong宜兴薄壳Yixingboke大金漆栗Dajinqili龙潭叶里藏Longtanyelicang油栗子Youlizi特早Tezao新杭迟栗Xinhangchili无名2 Wuming 2广果Guangguo六艳红Liuyanhong洋辣栗Yanglali红光油栗Hongguangyouli安徽1号Anhui 1 hao驴粪蛋Lvfendan六月爆Liuyuebao粘底板Zhandiban广德大红袍Guangdedahongpao双合大红袍Shuanghedahongpao焦刺替码Jiaocitima早庄B Zaozhuang B红毛早Hongmaozao上光栗Shangguangli Csb-1砖桥处暑红Zhuanqiaochushuhong林场优系Linchangyouxi乌山2号Wushan 2 hao东山早栗Dongshanzaoli原生地Origin region北京Beijing山东泰安Tai’an,Shandong河北迁西Qianxi,Hebei河北青龙Qinglong,Hebei北京Beijing河南信阳Xinyang,Henan陕西西安Xi’an,Shaanxi陕西柞水Zhashui,Shaanxi陕西西安Xi’an,Shaanxi江苏吴县Wuxian,Jiangsu湖南Hunan浙江上虞Shangyu,Zhejiang浙江Zhejiang广西Guangxi湖南Hunan湖北罗田Luotian,Hubei江苏宜兴Yixing,Jiangsu江苏东山Dongshan,Jiangsu江苏溧阳Liyang,Jiangsu安徽Anhui安徽Anhui安徽Anhui安徽Anhui安徽Anhui安徽Anhui安徽Anhui湖北Hubei安徽Anhui安徽Anhui湖北罗田Luotian,Hubei安徽舒城Shucheng,Anhui安徽Anhui安徽Anhui江苏Jiangsu江苏南京Nanjing,Jiangsu湖北京山Jingshan,Hubei浙江Zhejiang安徽Anhui安徽Anhui江苏溧阳Liyang,Jiangsu江苏Jiangsu江苏Jiangsu
3 讨论
3.1 板栗品种遗传多样性分析
基于SSR 标记研究种群遗传多样性已成为最方便快捷的方法,前人[28-33]均利用SSR 技术研究种群遗传多样性。而关于板栗遗传多样性的研究也已有报道。张馨方等[34]利用SSR 分子标记技术对279份板栗资源进行遗传多样性分析,平均每对引物检测到3.38个等位位点,有效等位基因数为1.53,聚类结果表明群体间遗传关系和地理来源有一定联系。此外,江锡兵等[35]、张馨方等[36]均采用SSR标记对部分地区板栗资源进行遗传多样性分析。笔者对全国板栗品种资源进行广泛收集,并且基于SSR荧光分子标记技术对其进行遗传多样性分析,总等位位点为212 个,平均每对引物检测到的等位基因数为10.095,有效等位基因数为3.488,明显高于前人的研究结果。因此,笔者在本研究中的21对SSR分子标记能够反映出板栗品种的遗传多样性。此外,对342 份板栗品种资源进行聚类分析,结果表明中国板栗品种资源大致分为南北两个群体。但是存在大部分南北混杂品种,对于其混杂原因还有待进一步研究分析。另外,选取聚类分析中得到的南北方板栗品种的含水量、可溶性糖含量、直链淀粉含量、支链淀粉含量、总淀粉含量等19个表型性状进行差异显著性分析,结果显示,南北方板栗品种的可溶性糖含量、直链淀粉含量、支链淀粉含量、刺苞厚、坚果厚、坚果宽呈显著性差异;总苞质量、苞质量、刺苞宽、刺苞高、苞肉厚、单粒质量呈极显著差异;含水量、总淀粉含量、出实率等其他指标无显著差异。此外,上述指标中除了直链淀粉含量、出实率和果形指数外,其余南方品种的表型指标值均大于北方品种,如含水量、可溶性糖含量、单粒质量等指标值,推测可能与南北方的环境差异有关。通过表型分析,进一步证明板栗品种(系)大致分为南北两个群体。
3.2 板栗品种核心种质构建分析
种质资源的收集和保存对物种的遗传多样性保护和育种具有重要作用,但是一个物种的种质资源往往比较丰富,不方便进行遗传育种研究[12]。因此,选择一定方法构建核心种质对深入挖掘优异种质资源,提高资源利用价值和效率具有重要意义[1]。构建策略的选择是核心种质的重点,不同的取样策略对核心种质样品的选择具有直接的影响。此外,等位基因的数量是多样性的反映[37],保留更多的等位基因对育种有重要作用[38]。李金龙等[5]基于聚类和表型,构建了甜荞530份种质;李洪果等[39]基于等位基因最大化原则构建了杜仲189 份种质;王春梅等[40]、陈天青等[41]、何建文等[42]都用了一种方法构建了玉米、小麦、辣椒等的核心种质。因此,为了使核心种质更具有代表性和可靠性,笔者在本研究中采用3种方法进行核心种质构建,同时比较3种方法所保留的等位位点数量,进而筛选出最少的种质资源以最大程度地代表原始种质的遗传多样性。
中国板栗品种丰富,资源众多,不易保存,因此构建核心种质尤为重要。前人对于板栗核心种质也有研究,如张馨方等[23]采用多次聚类的方法构建了燕山板栗的核心种质,不足以代表全国范围内的板栗品种资源;Nie等[22]基于最大遗传多样性的模拟退火算法构建了45份板栗品种的核心种质,但是只运用一种取样策略且研究的样品较少,具有局限性。因此亟需构建涵盖全国板栗品种资源遗传多样性的核心种质。
笔者通过SA,AN,AN、RS,AN,AN和分层抽样3 种方法构建核心种质,最终选择SA,AN,AN 作为最优策略并且构建的85 份资源约占原始种质库的24.85%,能够充分代表原始种质的遗传多样性,符合前人关于核心种质取样比例在5%~40%的研究结论[43]。基于等位基因数的模拟退火算法相比另外两种构建策略而言,能够以最少的资源代表整个原始种群的遗传多样性,且能够更好地保存资源。随着市场的需要及科技的发展,会不断出现新的优异板栗品种资源,因此构建核心种质是一个动态的过程,在今后的研究中需要不断完善核心种质。
4 结论
笔者在本研究中基于SSR 分子标记,揭示了南北方板栗品种(系)存在较多基因交流,同时利用SA,AN,AN、RS,AN,AN 和分层抽样3 种策略构建核心种质,确定基于等位基因数的模拟退火算法为最优取样策略,最终构建85份核心种质。
[1] FRANKEL O H,BROWN A H D.Plant genetic resources today:A critical appraisal[M]//HOLDEN J H W,WILLIAMS J T.Crop genetic resources:conservation and evaluation. George Allan and Unwin,London,1984:249-257.
[2] 艾叶,陈璐,谢泰祥,陈娟,兰思仁,彭东辉.基于SSR 荧光标记构建建兰品种核心种质[J]. 园艺学报,2019,46(10):1999-2008.AI Ye,CHEN Lu,XIE Taixiang,CHEN Juan,LAN Siren,PENG Donghui. Construction of core collection of Cymbidium ensifolium cultivars based on SSR fluorescent makers[J]. Acta Horticulture Sinica,2019,46(10):1999-2008.
[3] HILDE N.Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants[J]. Molecular Ecology,2004,13(5):1143-1155.
[4] 崔竣杰,程蛟文,曹毅,胡开林.基于SSR 标记和表型性状构建苦瓜核心种质的研究[J].中国蔬菜,2022(2):25-32.CUI Junjie,CHENG Jiaowen,CAO Yi,HU Kailin. Studies on constructure of bitter gourd core collection based on SSR markers and phenotypictraits[J].China Vegetables,2022(2):25-32.
[5] 李金龙,范昱,赵梦雨,康珍,杨克理,张凯旋,周美亮.基于表型性状和SSR 分子标记构建甜荞初级核心种质[J].植物遗传资源学报,2021,22(5):1240-1247.LI Jinlong,FAN Yu,ZHAO Mengyu,KANG Zhen,YANG Keli,ZHANG Kaixuan,ZHOU Meiliang. Constructure of primary of core collection of buckwheat germplasm resources based on phenotypic traits and SSR[J]. Journal of Plant Genetic Resources,2021,22(5):1240-1247.
[6] 吴茵.基于SRAP、SSR 标记的辣椒种质遗传多样性分析与核心种质构建[D].南昌:江西农业大学,2017.WU Yin. Genetic diversity analysis and establishment of the core collection of pepper germplasm resuorces based on SRAP and SSR markers[D]. Nanchang:Jiangxi Agricultural University,2017.
[7] 鲁敏,张怀山,白静,安华明.基于果实品质与EST-SSR 标记的刺梨居群遗传多样性分析及核心种质构建[J].分子植物育种,2020,18(9):3098-3106.LU Min,ZHANG Huaishan,BAI Jing,AN Huaming. Genetic diversity analysis and core collection in Rosa roxburghii based on fruits quality and EST- SSR markers[J]. Molecular Plant Breeding,2020,18(9):3098-3106.
[8] 李冬波,徐宁,秦献泉,李鸿莉,侯延杰,邱宏业,张树伟,朱建华,彭宏祥.广西原产和引种荔枝种质资源的遗传多样性分析及核心种质构建[J].南方农业学报,2020,51(7):1537-1544.LI Dongbo,XU Ning,QIN Xianquan,LI Hongli,HOU Yanjie,QIU Hongye,ZHANG Shuwei,ZHU Jianhua,PENG Hongxiang.Genetic diversity and construction of core collections of litchi(Litchi chinensis Sonn.)germplasm originated and introduced in Guangxi[J]. Journal of Southern Agriculture,2020,51(7):1537-1544.
[9] 邹梁峰. 毛花猕猴桃雄株核心种质构建及遗传多样性分析[D].南昌:江西农业大学,2019.ZOU Liangfeng. Establishment of the core collection of male germplasm resources of Actinidia eriantha and analysis of its genetic diversity[D]. Nanchang:Jiangxi Agricultural University,2019.
[10] 刘娟,廖康,曹倩,孙琪,刘欢,贾杨.利用表型性状构建新疆野杏种质资源核心种质[J].果树学报,2015,32(5):787-796.LIU Juan,LIAO Kang,CAO Qian,SUN Qi,LIU Huan,JIA Yang. Establishment of wild apricot core collection based on phenotypic characters[J]. Journal of Fruit Science.2015,32(5):787-796.
[11] 刘娟,廖康,曼苏尔·那斯尔,曹倩,江振斌,贾杨.利用ISSR分子标记构建南疆杏种质资源核心种质[J].果树学报,2015,32(3):374-384.LIU Juan,LIAO Kang,Mansur·Nasir,CAO Qian,JIANG Zhenbin,JIA Yang. Core-germplasm construction of apricot collections in South of Xinjiang by ISSR molecular markers[J]. Journal of Fruit Science,2015,32(3):374-384.
[12] 陈明堃,陈璐,孙维红,马山虎,兰思仁,彭东辉,刘仲健,艾叶.建兰种质资源遗传多样性分析及核心种质构建[J].园艺学报,2022,49(1):175-186.CHEN Mingkun,CHEN Lu,SUN Weihong,MA Shanhu,LAN Siren,PENG Donghui,LIU Zhongjian,AI Ye. Genetic diversity analysis and core collection of Cymbidium ensifolium germplasm resources[J]. Acta Horticulture Sinica,2022,49(1):175-186.
[13] 李魁鹏,陈仕昌,程琳,贺锦锋,唐红亮,谭文婧,余代渊,王斌,莫宗恒,黄开勇.基于SSR 标记构建广西杉木核心种质[J].广西科学,2021,28(5):511-519.LI Kuipeng,CHEN Shichang,CHENG Lin,HE Jinfeng,TANG Hongliang,TAN Wenjing,YU Daiyuan,WANG Bin,MO Zongheng,HUANG Kaiyong. Construction of core germplasm of Cunninghamia lanceolata in Guangxi based on SSR markers[J].Guangxi Sciences,2021,28(5):511-519.
[14] 毛秀红,朱士利,李善文,华辉,田书勇,仲伟国,董玉峰,安新民.基于荧光SSR 标记的毛白杨核心种质构建[J].北京林业大学学报,2020,42(7):40-47.MAO Xiuhong,ZHU Shili,LI Shanwen,HUA Hui,TIAN Shuyong,ZHONG Weiguo,DONG Yufeng,AN Xinmin. Core germplasm construction of Populus tomentosa based on the fluorescent SSR markers[J]. Journal of Beijing Forestry University,2020,42(7):40-47.
[15] 杨欣超,张凯权,王静,贾汉森,李云,段劼,马履一.基于SSR分子标记的刺槐遗传多样性分析及核心种质的构建[J].分子植物育种,2020,18(9):3086-3097.YANG Xinchao,ZHANG Kaiquan,WANG Jing,JIA Hansen,LI Yun,DUAN Jie,MA Lüyi. Analysis genetic diversity and construction of core collection of black locust based on SSR markers[J].Molecular Plant Breeding,2020,18(9):3086-3097.
[16] 吴涛,陈少瑜,肖良俊,宁德鲁,潘莉,贺娜,朱云凤.基于SSR标记的云南省核桃种质资源遗传多样性及核心种质构建[J].植物遗传资源学报,2020,21(3):767-774.WU Tao,CHEN Shaoyu,XIAO Liangjun,NING Delu,,PAN Li,HE Na,ZHU Yunfeng.Genetic diversity analysis and core collection construction of walnut germplasm in Yunnan province[J].Journal of Plant Genetic Resources,2020,21(3):767-774.
[17] 燕丽萍,吴德军,毛秀红,姚俊修,任飞,李善文,王开芳,王因花,刘翠兰.基于SSR 荧光标记的白蜡核心种质构建[J].中南林业科技大学学报,2019,39(7):1-9.YAN Liping,WU Dejun,MAO Xiuhong,YAO Junxiu,REN Fei,LI Shanwen,WANG Kaifang,WANG Yinhua,LIU Cuilan.Construction of core collection of Fraxinus based on SSR molecular marker[J].Journal of Central South University of Forestry and Technology,2019,39(7):1-9.
[18] 方乐成,夏慧敏,麻文俊,张新叶.基于SSR 标记的楸树遗传多样性及核心种质构建[J].东北林业大学学报,2017,45(8):1-5.FANG Yuecheng,XIA Huimin,MA Wenjun,ZHANG Xinye.Genetic diversity analysis and primary core collection of Catalpa bungei germplasm with SSR markers[J]. Journal of Northeast Forestry University,2017,45(8):1-5.
[19] 徐志祥,高绘菊.板栗营养价值及其养生保健功能[J].食品研究与开发,2004(5):118-119.XU Zhixiang,GAO Huiju. Nutritional value and health care function of chestnut[J]. Food Research and Development,2004(5):118-119.
[20] 易哲.板栗市场天地宽[J].河南林业,2001(1):63.YI Zhe.Wide chestnut market[J].Henan Forestry,2001(1):63.
[21] 杨阳,郭燕,张树航,李颖,张馨方,王广鹏.中国栗属植物起源演化和分类研究进展[J].河北农业科学,2017,21(2):25-28.YANG Yang,GUO Yan,ZHANG Shuhang,LI Ying,ZHANG Xinfang,WANG Guangpeng.The origin,evolution and classification of Chestnut germplasm resources in China[J]. Journal of Hebei Agricultural Sciences,2017,21(02):25-28.
[22] NIE X H,WANG Z H,LIU N W,SONG L,YAN B Q,XING Y,ZHANG Q,FANG K F,ZHAO Y F,CHEN X,WANG G P,QIN L,CAO Q P. Fingerprinting 146 Chinese chestnut (Castanea mollissima Blume.) accessions and selecting a core collection using SSR markers[J]. Journal of Integrative Agriculture,2021,20(5):1277-1286.
[23] 张馨方,张树航,李颖,郭燕,王广鹏.基于SSR 标记构建燕山板栗核心种质[J].华北农学报,2021,36(S1):31-38.ZHANG Xinfang,ZHANG Shuhang,LI Ying,GUO Yan,WANG Guangpeng. Construction of core collection of Yanshan chestnut germplasm based on SSR markers[J]. Acta Agriculturae Boreali-Sinica,2021,36(S1):31-38.
[24] LIU K J,MUSE S V. Powermarker:an integrated analysis environment for genetic marker analysis[J].Bioinformatics(Oxford,England),2005,21(9):2128-2129.
[25] PEAKALL R,SMOUSE P E. GenAlEx 6.5:genetic analysis in Excel. Population genetic software for teaching and research--an update[J]. Bioinformatics (Oxford,England),2012,28(19):2537-2539.
[26] PEAKALL R,SMOUSE P E. GENALEX 6:Genetic analysis in Excel. Population genetic software for teaching and research[J].Molecular Ecology Notes,2006,6(1):288-295.
[27] 王浩,侯惠民.科学图形计算软件GraphPad Prism 3.0[J].中国医药工业杂志,2001,32(4):184-187.WANG Hao,HOU Huimin. Scientific graph software GraphPad Prism3.0[J]. Chinese journal of Pharmaceuticals,2001,32(4):184-187.
[28] 刘冬峰,陈巍,林绍生,徐文荣,郭秀珠,黄品湖.基于SSR 标记的浙江地方柚类种质资源遗传关系分析[J]. 果树学报,2017,34(2):166-174.LIU Dongfeng,CHEN Wei,LIN Shaosheng,XU Wenrong,GUO Xiuzhu,HUANG Pinhu. Analysis of genetic relationship of pummelo germplasms by SSR markers in Zhejiang province[J].Journal of Fruit Science,2017,34(2):166-174.
[29] 赵爽,苏淑钗,陈志钢,盛淑艳,王忠.基于SSR 的辽宁铁岭地区平榛遗传多样性与居群遗传结构分析[J].果树学报,2016,33(1):24-33.ZHAO Shuang,SU Shuchai,CHEN Zhigang,SHENG Shuyan,WANG Zhong.An assessment of the genetic diversity of population genetic structure concerning the Corylus heterophylla Fisch,grown in the tieling district of Liaoning provence ,using SSR markers[J].Journal of Fruit Science,2016,33(1):24-33.
[30] 高源,王大江,王昆,丛佩华,李连文,朴继成.基于荧光SSR分析中国原产苹果属植物17 个种的遗传多样性和遗传结构[J].果树学报,2020,37(11):1611-1622.GAO Yuan,WANG Dajiang,WANG Kun,CONG Peihua,LI Lianwen,PU Jicheng.Genetic diversity and population structure of 17 species of Malus Mill. Native to china based on fluorescent SSR analysis[J]. Journal of Fruit Science,2020,37(11):1611-1622.
[31] 钟敏,廖光联,李章云,邹梁峰,黄清,陈璐,黄春辉,陶俊杰,朱博,徐小彪.野生毛花猕猴桃雄花花器性状及SSR 遗传多样性研究[J].果树学报,2018,35(6):658-667.ZHONG Min,LIAO Guanglian,LI Zhangyun,ZOU Liangfeng,HUANG Qing,CHEN Lu,HUANG Chunhui,TAO Junjie,ZHU Bo,XU Xiaobiao. Genetic diversity of wild male kiwifruit (Actinidia eriantha Benth.) germplasm based on SSR and morphological markers[J]. Journal of Fruit Science,2018,35(6):658-667.
[32] 马凯,赵钰,张恒,韩立群,赵国庆,王继勋.基于SSR 标记的中亚生态区核桃(Juglans regia L.)遗传多样性与种群结构分析[J].果树学报,2021,38(11):1854-1867.MA Kai,ZHAO Yu,ZHANG Heng,HAN Liqun,ZHAO Guoqing,WANG Jixun.Analysis of genetic diversity and population structure of walnut (Juglans regia L.)in central Asia ecological region based on SSR markers[J].Journal of Fruit Science,2021,38(11):1854-1867.
[33] 郭禄芹,赵世豪,朱华玉,胡建斌,孙守如,马长生,杨路明.167份西瓜种质材料的遗传多样性分析[J]. 中国瓜菜,2018,31(1):5-11.GUO Luqin,ZHAO Shihao,ZHU Huayu,HU Jianbin,SUN Shouru,MA Changsheng,YANG Luming.The genetic diversity analysis of 167 watermelon germplasms[J]. China Cucurbits and Vegetables,2018,31(1):5-11.
[34] 张馨方,张树航,李颖,郭燕,王广鹏.基于SSR 标记的板栗种质资源遗传多样性分析[J].分子植物育种,2020,18(15):5164-5175.ZHANG Xinfang,ZHANG Shuhang,LI Ying,GUO Yan,WANG Guangpeng. Genetic diversity analysis of Castanea mollissima germplasm resources based on SSR markers[J]. Molecular Plant Breeding,2020,18(15):5164-5175.
[35] 江锡兵,汤丹,龚榜初,赖俊声.基于SSR 标记的板栗地方品种遗传多样性与关联分析[J].园艺学报,2015,42(12):2478-2488.JIANG Xibing,TANG Dan,GONG Bangchu,LAI Junsheng.Genetic diversity and association analysis of local cultivars of Chinese chestnut based on SSR markers[J]. Acta Horticulturae Sinica,2015,42(12):2478-2488.
[36] 张馨方,张树航,李颖,郭燕,王广鹏.基于SSR 标记的燕山板栗种质资源遗传多样性分析[J].中国农业大学学报,2020,25(4):61-71.ZHANG Xinfang,ZAHNG Shuhang,LI Ying,GUO Yan,WANG Guangpeng.Genetic diversity analysis of chestnut germplasm in Yanshan region based on SSR markers[J]. Journal of China Agricultural University,2020,25(4):61-71.
[37] BATAILLON T M,DAVID J L,SCHOEN D J. Neutral genetic markers and conservation genetics:simulated germplasm collections[J].Genetics,1996,144(1).
[38] ZHAO W G,CHO G T,MA K H,CHUNG J W,GWAG J G,PARK Y J. Development of an allele-mining set in rice using a heuristic algorithm and SSR genotype data with least redundancy for the post-genomic era[J]. Molecular Breeding,2010,26(4):639-651.
[39] 李洪果,许基煌,杜红岩,乌云塔娜,刘攀峰,杜庆鑫.基于等位基因最大化法初步构建杜仲核心种质[J].林业科学,2018,54(2):42-51.LI Hongguo,XU Jihuang,DU Hongyan,WUYUN Tana,LIU Panfeng,DU Qingxin. Preliminary construction of core collection of Eucommia ulmoides based on allele number maximization strategy[J].Scientia Silvae Sinicae,2018,54(2):42-51.
[40] 王春梅,任洪,沈建华,赵晓燕,曹绍书,王竹.贵州玉米种质资源遗传多样性及核心种质库构建[J].西南农业学报,2016,29(5):1018-1022.WANG Chunmei,REN Hong,SHEN Jianhua,ZHAO Xiaoyan,CAO Shaoshu,WANG Zhu.Molecular evaluation of Guizhou local maize genetic diversity and construction of core collection[J].Southwest China Journal of Agricultural Sciences,2016,29(5):1018-1022.
[41] 陈天青,隋建枢,张立异,王伟,田世飞,杨康林,何庆才.贵州小麦育种核心种质的遗传多样性分析[J]. 西南农业学报,2015,28(5):1879-1887.CHEN Tianqing,SUI Jianshu,ZHANG Liyi,WANG Wei,TIAN Shifei,YANG Kanglin,HE Qingcai. Genetic diversity of core wheat breeding germplasm in Guizhou[J]. Southwest China Journal of Agricultural Sciences,2015,28(5):1879-1887.
[42] 何建文,韩世玉.基于SSR 标记不同距离聚类与抽样方法构建辣椒核心种质库[J].西南农业学报,2015,28(5):2199-2204.HE Jianwen,HAN Shiyu. Construction of core Capsicum germplasm bank based on different distance clustering and sampling method of SSR marker[J]. Southwest China Journal of Agricultural Sciences,2015,28(5):2199-2204.
[43] LI G S,ZHANG L J,BAI C K. Chinese Cornus officinalis:genetic resources,genetic diversity and core collection[J]. Genetic Resources and Crop Evolution,2012,59(8):1659-1671.