欧李(Cerasus humilis)属于蔷薇科樱桃属植物[1],作为我国特有的矮生灌木果树资源,是实施“退耕还林”的重要树种之一。欧李树种适应性强,耐干旱、耐贫瘠、耐盐碱,对生长环境要求较低,具有短期内速生繁殖等优点[2]。目前,欧李在我国中西部干旱和半干旱地区以及一些退化工矿用地推广应用,不仅作用于改良土壤结构,还具有一定的经济效益[3]。
植物在生长发育过程中会受到非生物性胁迫和生物性胁迫的危害。在长期进化过程中,为了更好的生存,植物产生了一系列生理生化机制来适应、抵御不良环境的影响[4-5]。基因表达调控是一种重要的防御机制[6-7]。
NAC基因家族是植物中最大的基因家族之一,且是植物所特有的基因家族[8]。NAC转录因子的命名取自矮牵牛(Petunia hybrida)的NAM(no apical meristem)基因、拟南芥(Arabidopsis thaliana)的ATAF1/ATAF2 基因、以及CUC2(cup-shaped cotyledon)基因的首字母[9]。1996 年,Souer 等[10]从矮牵牛中克隆出第一个NAC家族成员NAM。NAC基因家族参与植物的根、茎、叶、花等器官的生长发育、果实成熟、激素调控[11-14]。同时,NAC基因也参与植物激素调控,如拟南芥ATAF1、水稻OsNAP,水稻SNAC2[15- 17]被证明参与ABA 信号转导途径;ANAC072 基因参与MeJA 信号转导途径,细胞分裂素、乙烯、赤霉素等的信号转导途径也有NAC 基因的参与[13,18-19]。NAC基因在植物抗干旱、冷害、高温、高盐等非生物胁迫中同样发挥着重要作用。Tran等[20]过表达3 个NAC 基因(ANAC019、ANAC055、ANAC072),使拟南芥的抗旱能力显著提高。Jeong等[21]发现干旱处理中OsNAC10 基因在水稻根和穗中的表达量显著提高,当该基因过表达时,可促进根生长,显著提高水稻产量。Lu等[22]将NAC家族中的ATAF1 基因敲除后,拟南芥ataf1-1 和ataf1-2 突变体在干旱响应试验中的恢复率相比于野生型,提高了6 倍左右。Hao等[23]发现大豆中的GmNAC11基因和GmNAC20 基因响应冷害胁迫,基因过表达植株对低温的耐受性显著提高,Hegedus等[24]在欧洲油菜中也证明NAC 基因作用于冷害胁迫。Mao 等[25]将小麦中的TaNAC2 基因在拟南芥中过表达,其与对照组相比抗低温能力显著提高。Dong 等[26]发现玉米NAC 家族中的NUT1 基因调控水分转运,从而影响玉米的高温表型,而NUT1 基因表达的时空特异性有利于农作物应对田间水分的不断变化,NUT1 基因可作为未来农作物抗高温胁迫的重要标靶。梁晓庆[27]对番茄进行高温胁迫,结果表明抑制SINAC1基因表达的转基因植株相比于正常植株抗高温能力显著降低,即证明SINAC1 基因正向调控番茄对高温的抗性。Li 等[28]在拟南芥和大豆毛状根中过表达GmNAC06 基因,植株均表现出较强的耐盐性,表明GmNAC06 基因在盐胁迫响应中发挥作用。Zheng等[29]研究发现,过表达OsNAC45基因增强了转基因水稻的耐盐性。近年来,NAC 基因家族在病害、虫害等生物性胁迫方面的研究也取得重要成果[30-36]。
基于已发表的欧李全基因组数据,笔者在本研究中采用生物信息学分析方法,对欧李NAC基因家族进行鉴定,并对其家族成员的理化性质、基因结构、染色体定位、共线性、启动子进行分析。同时,对不同胁迫处理下ChNAC 基因成员的表达量进行分析,旨在挖掘胁迫响应的关键ChNAC 基因,为欧李抗逆品种的选育奠定基础。
1 材料和方法
1.1 欧李NAC基因家族成员的鉴定及理化性质分析
欧李的全基因组数据来自Wang等[37]的报道,从pafm(http://pfam.xfam.org/)数据库中获得桃NAC基因的Hmmer文件,对欧李全基因组数据进行本地BLAST。并结合NCBI 在线网站(ttp://www.ncbi.nlm.nih.gov/)中桃、扁桃等相近物种的NAC 序列对欧李的NAC基因进行补充、完善并剔除重复。利用ExPASy 网站(https://web.expasy.org/protparam/)分析欧李NAC 基因的氨基酸数量、等电点、不稳定系数等理化性质。利用PSORT在线网站(https://psort.hgc.jp/form2.html)对亚细胞定位进行预测。利用SOPMA在线网站(https://prabi.ibcp.fr/htm/site/web)对蛋白质二级结构进行分析[38-39]。
1.2 欧李NAC基因家族分类
从TAIR 在线网站中(https://www.arabidopsis.org/)下载拟南芥NAC 基因家族的蛋白质序列与欧李NAC 基因家族的蛋白序列进行比对。使用MEGA7软件,将Bootstrap重复值设定为1000,其他参数均为默认值,采用邻接法构建系统进化树[38-39]。
1.3 欧李NAC基因家族基因结构分析
利用MEME 在线网站(https://meme-suite.org/meme)对NAC 蛋白的motif 基序进行预测,整合NAC 蛋白的位置信息,利用Tbtools 软件绘图并分析[38-39]。
1.4 欧李NAC基因家族染色体定位和共线性分析
对欧李的NAC 蛋白的位置信息和所处的染色体进行编号整理。利用Tbtools软件绘制图表[38-39]。
1.5 欧李NAC基因家族启动子分析
从欧李全基因组数据库中查找各NAC 转录因子ATG 密码子前2000 bp 的序列,利用PlantCARE在线网站(http://bioinformatics.psb.ugent.be/webTbtools/plantcare/html/)对其功能进行预测、整理。利用Origin软件绘制功能分布柱状图[38-39]。
1.6 欧李NAC基因组织特异性分析
采欧李品种农大4 号成熟期果实、叶片、根状茎、种仁及一年生根送往Biomarker 公司,提取各样品的总RNA,对RNA的质量进行检测,检测合格后构建cDNA文库,进行Illumina HiSeq平台高通量测序。获得不同组织基因的转录组文件后,利用注释信息对NAC基因进行筛选、整理。用Tbtool软件绘制基因表达热图[38-39]。
1.7 欧李NAC基因家族在胁迫诱导下的表达模式分析
以山西农业大学欧李种质资源圃欧李品种农大4号嫩枝扦插苗做材料。待扦插苗长至株高25 cm、具有10枚叶片时,选择根系发育旺盛的材料移至霍格兰溶液中进行水培。将植株分别置于加入了200 mmol·L-1 NaCl 溶液或20%PEG6000 溶液的霍格兰溶液中,25 ℃处理48 h 作为盐处理及干旱处理,将植株置于加入等量清水的霍格兰溶液中,25 ℃处理48 h,作为对照。将植株置于霍格兰溶液培养,分别进行4 ℃处理48 h做低温处理或40 ℃处理48 h 做高温处理,以25 ℃处理48 h,作为对照。每个处理设置5个生物学重复,48 h后分别在每株上采集植株中部3枚叶片,液氮处理后送往Biomarker公司,取各样品的总RNA,对RNA的质量进行检测,检测合格后构建cDNA文库,进行Illumina HiSeq平台高通量测序。获得不同组织基因的转录组文件后,利用注释信息对NAC基因进行筛选、整理。最后使用Tbtools软件绘制欧李NAC基因表达热图[40-42]。
2 结果与分析
2.1 欧李NAC基因家族的理化性质分析
通过本地BLAST 和NCBI 网站与桃等相近物种进行比对,利用NCBI预测其结构域,共筛选得到76 条NAC 基因,根据欧李的拉丁文种名(Cerasus humilis),按照基因位置信息依次编号为ChNAC01-ChNAC76(表1)。
表1 ChNAC 蛋白的理化性质分析
Table 1 Physicochemical properties of ChNAC
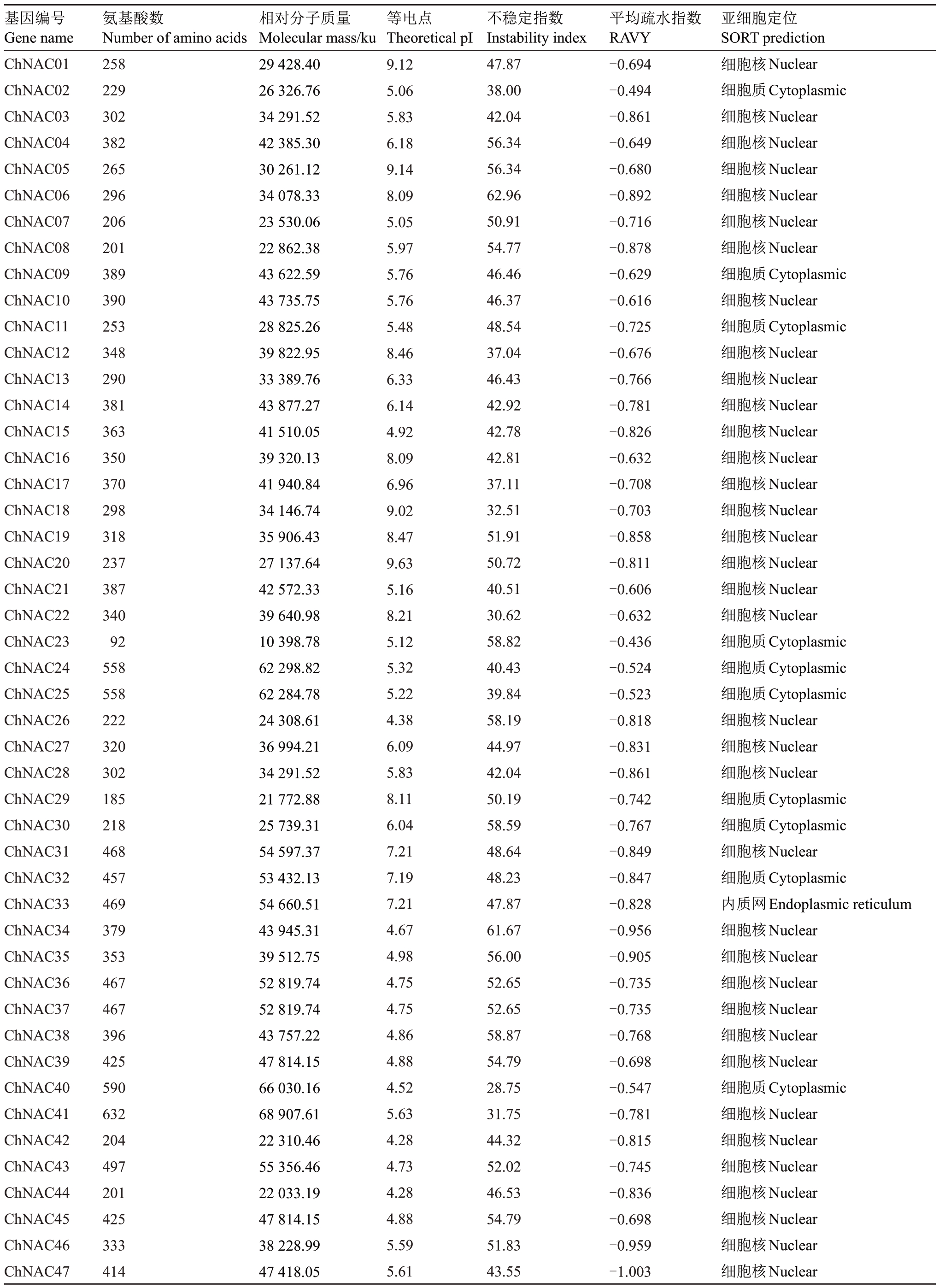
基因编号Gene name氨基酸数Number of amino acids相对分子质量Molecular mass/ku等电点Theoretical pI不稳定指数Instability index平均疏水指数RAVY亚细胞定位SORT prediction ChNAC01 ChNAC02 ChNAC03 ChNAC04 ChNAC05 ChNAC06 ChNAC07 ChNAC08 ChNAC09 ChNAC10 ChNAC11 ChNAC12 ChNAC13 ChNAC14 ChNAC15 ChNAC16 ChNAC17 ChNAC18 ChNAC19 ChNAC20 ChNAC21 ChNAC22 ChNAC23 ChNAC24 ChNAC25 ChNAC26 ChNAC27 ChNAC28 ChNAC29 ChNAC30 ChNAC31 ChNAC32 ChNAC33 ChNAC34 ChNAC35 ChNAC36 ChNAC37 ChNAC38 ChNAC39 ChNAC40 ChNAC41 ChNAC42 ChNAC43 ChNAC44 ChNAC45 ChNAC46 ChNAC47258229302 382265296206 201389390253 348290381363 350370298318 23738734092 558558222320 302185218468 457469379353 467467396425 590632204497 201425333414 29428.4026326.7634291.5242385.3030261.1234078.3323530.0622862.3843622.5943735.7528825.2639822.9533389.7643877.2741510.0539320.1341940.8434146.7435906.4327137.6442572.3339640.9810398.7862298.8262284.7824308.6136994.2134291.5221772.8825739.3154597.3753432.1354660.5143945.3139512.7552819.7452819.7443757.2247814.1566030.1668907.6122310.4655356.4622033.1947814.1538228.9947418.059.125.065.836.189.148.095.055.975.765.765.488.466.336.144.928.096.969.028.479.635.168.215.125.325.224.386.095.838.116.047.217.197.214.674.984.754.754.864.884.525.634.284.734.284.885.595.6147.8738.0042.0456.3456.3462.9650.9154.7746.4646.3748.5437.0446.4342.9242.7842.8137.1132.5151.9150.7240.5130.6258.8240.4339.8458.1944.9742.0450.1958.5948.6448.2347.8761.6756.0052.6552.6558.8754.7928.7531.7544.3252.0246.5354.7951.8343.55-0.694-0.494-0.861-0.649-0.680-0.892-0.716-0.878-0.629-0.616-0.725-0.676-0.766-0.781-0.826-0.632-0.708-0.703-0.858-0.811-0.606-0.632-0.436-0.524-0.523-0.818-0.831-0.861-0.742-0.767-0.849-0.847-0.828-0.956-0.905-0.735-0.735-0.768-0.698-0.547-0.781-0.815-0.745-0.836-0.698-0.959-1.003细胞核Nuclear细胞质Cytoplasmic细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞质Cytoplasmic细胞核Nuclear细胞质Cytoplasmic细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞质Cytoplasmic细胞质Cytoplasmic细胞质Cytoplasmic细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞质Cytoplasmic细胞质Cytoplasmic细胞核Nuclear细胞质Cytoplasmic内质网Endoplasmic reticulum细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞质Cytoplasmic细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear
表1 (续) Table 1(Continued)

基因编号Gene name氨基酸数Number of amino acids相对分子质量Molecular mass/ku等电点Theoretical pI不稳定指数Instability index平均疏水指数RAVY亚细胞定位SORT prediction ChNAC48 ChNAC49 ChNAC50 ChNAC51 ChNAC52 ChNAC53 ChNAC54 ChNAC55 ChNAC56 ChNAC57 ChNAC58 ChNAC59 ChNAC60 ChNAC61 ChNAC62 ChNAC63 ChNAC64 ChNAC65 ChNAC66 ChNAC67 ChNAC68 ChNAC69 ChNAC70 ChNAC71 ChNAC72 ChNAC73 ChNAC74 ChNAC75 ChNAC76346285335 347284271306 577432351351 378166306352 182262854362 344437483294 321454363295 37338338942.6532143.5938522.0438715.1432769.0530798.0135509.6465360.6948123.8939435.0739161.9242935.6417968.334936.3439567.1820939.4229584.1795216.8542308.9839123.1149328.4853683.9733023.5336523.0651744.8940353.3333935.4341068.5142910.395.066.324.957.745.648.766.174.904.585.015.397.015.798.006.616.447.964.936.315.216.356.986.086.016.417.787.048.458.2342.9461.3356.9443.4841.7435.7655.9846.1236.4331.3840.5938.4852.1651.1346.5249.0753.7548.1560.1640.8535.5246.6532.6241.0549.4046.5141.8938.9841.37-0.560-0.843-0.760-0.937-0.675-0.530-0.806-0.599-0.381-0.588-0.541-0.952-0.666-0.829-0.701-0.695-0.597-0.824-0.953-0.822-0.636-0.611-0.825-1.023-0.892-0.729-0.677-0.764-0.596细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞质Cytoplasmic细胞核Nuclear囊泡Vesicles of secretory system细胞质Cytoplasmic细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞质Cytoplasmic细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞质Cytoplasmic细胞核Nuclear细胞核Nuclear细胞核Nuclear细胞质Cytoplasmic细胞核Nuclear细胞核Nuclear细胞核Nuclear
欧李NAC 基因的理化性质和亚细胞定位结果表明,欧李NAC 基因中氨基酸最少的为ChNAC23 基因,仅有85 个氨基酸,其相对分子质量为10398.78 ku;最多的为ChNAC65 基因,有854 个氨基酸,其相对分子质量为95216.85 ku。55 个ChNAC 基因的氨基酸数量集中在200~400 个之间。ChNAC 蛋白的等电点值在4.28~9.63 之间,其中碱性蛋白33 个,酸性蛋白17 个,中性蛋白26 个。ChNAC 蛋白多为不稳定蛋白,仅有15 个NAC 蛋白的不稳定指数小于40,为稳定蛋白。其余61 个NAC 蛋白的不稳定指数均大于40 为不稳定蛋白。ChNAC蛋白的总平均疏水指数为-0.381~-1.023,均为亲水性蛋白。亚细胞定位结果显示:ChNAC33蛋白定位于内质网;ChNAC55 蛋白定位于囊泡中;ChNAC02、ChNAC09、ChNAC11 等15 个蛋白定位于细胞质中;其余ChNAC蛋白均定位于细胞核中。亚细胞定位结果的多样性,表明ChNAC蛋白在细胞的不同部位,不同细胞器中行使不同的功能,为植物体的正常生命活动提供保障。
为进一步明确ChNAC蛋白的空间结构,利用在线网站(https://npsa-prabi.ibcp.fr/cgi-bin/)分别对76条ChNAC蛋白的二级结构进行了预测,分析了其α-螺旋、延伸链、β-转角及无规则卷曲所占的比例。其中,α-螺旋所占比例在10.85%~45.05%之间;延伸链所占比例在9.69%~25.27%之间;β-转角所占比例在0~9.65%之间;无规程卷曲所占比例在38.29%~70.60%之间(表2)。
表2 ChNAC 蛋白的二级结构
Table 2 Secondary structure of ChNAC protein

基因编号Gene name基因编号Gene name ChNAC01 ChNAC02 ChNAC03 ChNAC04 ChNAC05 ChNAC06 ChNAC07 ChNAC08 ChNAC09 ChNAC10 ChNAC11 ChNAC12 ChNAC13 ChNAC14 ChNAC15 ChNAC16 ChNAC17 ChNAC18 ChNAC19 ChNAC20 ChNAC21 ChNAC22 ChNAC23 ChNAC24 ChNAC25 ChNAC26 ChNAC27 ChNAC28 ChNAC29 ChNAC30 ChNAC31 ChNAC32 ChNAC33 ChNAC34 ChNAC35 ChNAC36 ChNAC37 ChNAC38比例Ratio/%α-螺旋α-helix 10.8518.3411.5918.0616.9816.5520.3923.3822.8823.5915.0223.2824.8322.5721.7616.5725.6818.7915.7219.8318.3517.9426.0917.5623.3045.0519.3812.5819.4623.3923.9324.0727.0818.2127.2024.6324.6324.49延伸链Extended strand 18.9923.1417.8814.4017.7413.5114.5610.9513.6215.1316.6015.8010.3415.7517.6316.0012.9712.0823.9017.3015.2517.0620.6511.6513.6211.7113.1217.8817.3021.1017.0918.1618.3416.0912.4611.5611.5612.88 β-转角β-turn 5.814.802.983.403.022.033.883.484.635.133.164.023.794.466.894.864.593.026.604.224.392.650.002.513.944.952.813.645.414.593.213.723.843.173.684.074.073.54无规则卷曲Random coil 64.3453.7167.5564.1462.2667.9161.1762.1958.8756.1565.2256.9061.0357.2253.7262.5756.7666.1153.7758.6562.0262.3553.2668.2859.1438.2964.6965.8957.8450.9255.7754.0550.7562.5356.6659.7459.7459.09 ChNAC39 ChNAC40 ChNAC41 ChNAC42 ChNAC43 ChNAC44 ChNAC45 ChNAC46 ChNAC47 ChNAC48 ChNAC49 ChNAC50 ChNAC51 ChNAC52 ChNAC53 ChNAC54 ChNAC55 ChNAC56 ChNAC57 ChNAC58 ChNAC59 ChNAC60 ChNAC61 ChNAC62 ChNAC63 ChNAC64 ChNAC65 ChNAC66 ChNAC67 ChNAC68 ChNAC69 ChNAC70 ChNAC71 ChNAC72 ChNAC73 ChNAC74 ChNAC75 ChNAC76比例Ratio/%α-螺旋α-helix 21.4122.3727.8543.1429.5836.8221.4118.3217.1521.1020.7025.0720.7514.7916.6117.6528.6021.0624.2220.5117.2044.5816.8022.7318.6820.6124.9424.3114.5327.2316.9813.9518.6924.4513.5015.9319.0320.37延伸链Extended strand 14.5915.4218.8311.2711.8712.4414.5914.4117.1511.8511.9316.7214.9916.9017.3415.0310.9211.349.6914.2513.7610.8413.9310.8025.2720.6112.1814.0911.9213.509.7319.7319.0010.5714.6015.9313.9414.62 β-转角β-turn 3.534.759.655.883.426.973.534.504.114.053.165.076.344.935.173.273.123.943.993.994.762.414.103.128.244.201.993.873.204.812.697.827.174.632.752.033.494.18无规则卷曲Random coil 60.4757.4643.6739.7155.1343.7860.4762.7661.5963.0164.2153.1357.9363.3860.8964.0557.3763.6662.1161.2564.2942.1765.1663.3547.8054.5860.8957.7370.3554.4670.6058.5055.1460.3569.1566.1063.5460.84
2.2 欧李NAC基因系统进化分析
研究利用拟南芥的NAC基因序列做对照,参考Hisako 等[8]的研究结果将ChNAC 基因分为17 个亚族。不同的亚族在功能及进化关系上存在差异[43]。ONAC01 亚族中不含有ChNAC 基因,说明ONAC001 亚族中的拟南芥NAC 基因与ChNAC 基因无进化关系,其余亚族中均存在数量不等的ChNAC基因。ChNAC成员数量超过10个的有2个亚族。ANAC001 亚族中的家族成员数量最多,有15个家族成员。ONAC003亚族中含有11个家族成员。研究表明,ANAC001亚族作用于植物的生长发育及胁迫应答过程[44],ONAC003亚族参与调节开花时节[45],推测欧李NAC基因多具备以上功能。其余亚族中的成员数量均小于10 个:TIP 亚族中有8 个家族成员;NAC2亚族和OsNAC7亚族中均有7个家族成员;ONAC022 亚族中有6 个家族成员;NAP 亚族中有5 个家族成员;ANAC063 亚族中有3 个家族成员;ATAF亚族,NAM亚族,SENU5亚族,TERN亚族,ANAC011 亚族及OsNAC8 亚族中都有2 个家族成员。AtNAC3亚族,NAC1亚族中仅具有1个家族成员(图1)。

图1 ChNAC 基因系统进化树
Fig.1 Phylogenetic tree of ChNAC gene
2.3 欧李NAC基因结构及保守基序分析
ChNAC基因CDS片段数量在1~8个之间,多数CDS 片段数量为3~6 个,ChNAC15、ChNAC23 基因均为一整段的CDS 序列,其中没有内含子插入。ChNAC65基因由8个CDS片段组成,CDS片段数量最多(图2)。

图2 ChNAC 基因结构和保守基序
Fig. 2 Structure and conserved motif of ChNAC gene
使用MEME 在线网站将Motif 值设置为20,对欧李的NAC 基因进行显示分析。结果表明,ChNAC 基因的Motif 数量并不完全一致。Motif1、Motif2、Motif7出现的次数最多,且Motif2总出现在序列的最前端,有35 个ChNAC 基因均由Motif2、Motif8、Motif4、Motif5、Motif3、Motif6、Motif1、Motif7 组成,有11 个ChNAC 基因是在上述Motif 后端插入了其他一个或几个Motif组成,有10个ChNAC基因是在上述基础上少了一个或几个Motif 组成,ChNAC08 基因和ChNAC23 基因都仅含有2 个Motif,Motif13 只在ChNAC26 基因中出现了1 次,ChNAC43 基因中的Motif11、Motif18 连续出现了2次(图2)。
2.4 欧李NAC 基因家族染色体分布及种内共线性分析
利用ChNAC 基因的注释信息和Tbtools 绘图软件,可以清楚直观地了解ChNAC 基因在欧李8条染色体上的分布情况。如图3所示,ChNAC基因在染色体上的分布是不均匀的。第五条染色体上(Chr5)ChNAC基因的数量最多,有19个家族成员,且多集中于染色体的前端。其次是第一条染色体(Chr1)上,有12 个ChNAC 家族成员且分布较均匀。第三条染色体(Chr3)和第七条染色体(Chr7)上家族成员的数量是一致的,都有9 个家族成员。第四条染色体(Chr4)上家族成员数量最少,仅有4个家族成员。
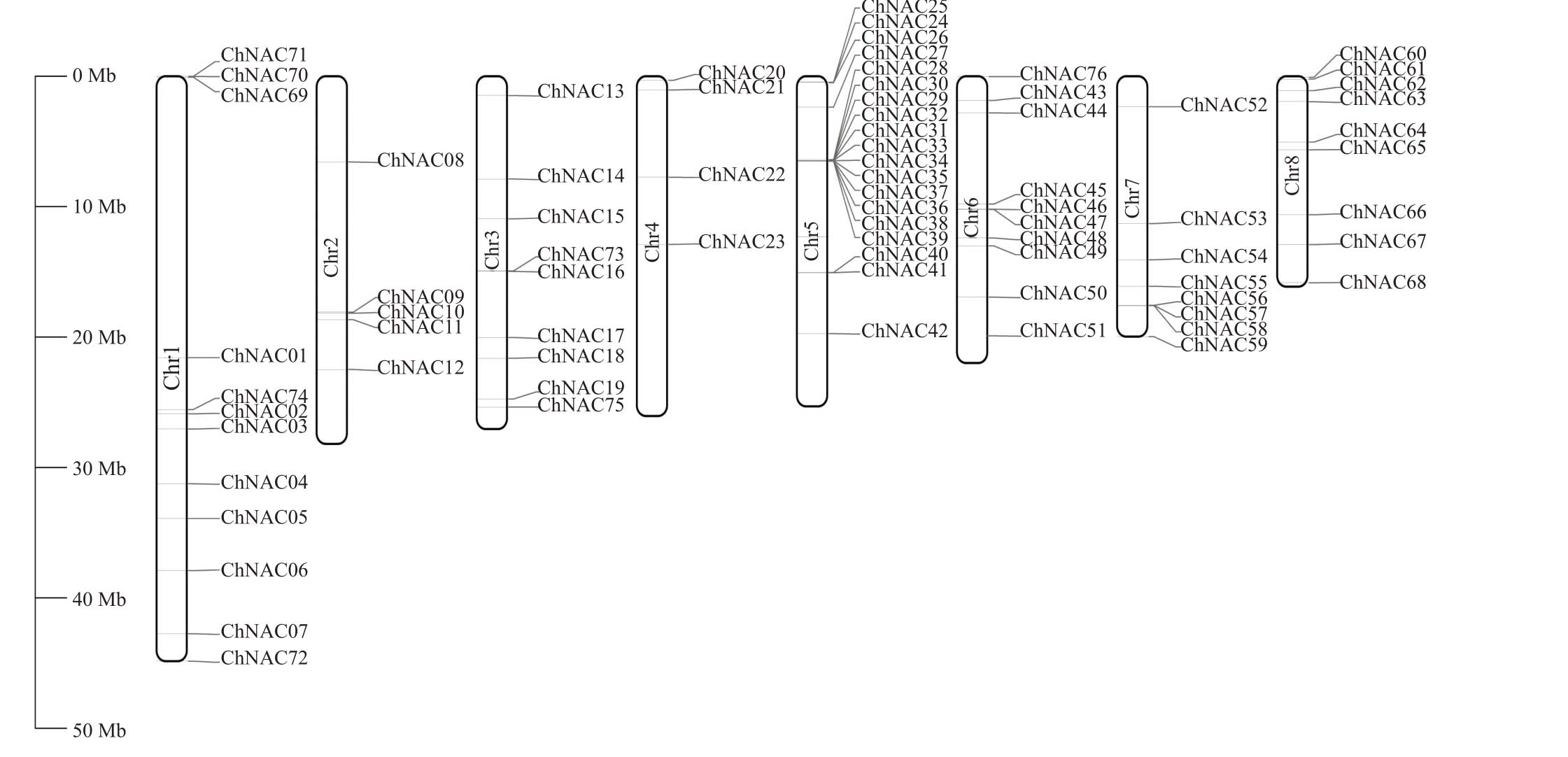
图3 ChNAC 基因染色体分布
Fig.3 Chromosomal distribution of ChNAC gene
为了进一步明确ChNAC 基因在染色体上的分布情况,对ChNAC基因的种内共线性进行了注释和分析,结果表明(图4),ChNAC基因家族中存在8对共线性基因:ChNAC12- ChNAC08、ChNAC11-ChNAC49、ChNAC73-ChNAC75、ChNAC20-ChNAC01、ChNAC20-ChNAC64、ChNAC01-ChNAC64、ChNAC25-ChNAC65、ChNAC27- ChNAC66,其中ChNAC01、ChNAC20、ChNAC64 这3 条基因是互为共线性,推测以上8 对基因可能是由染色体大片段复制而成的。
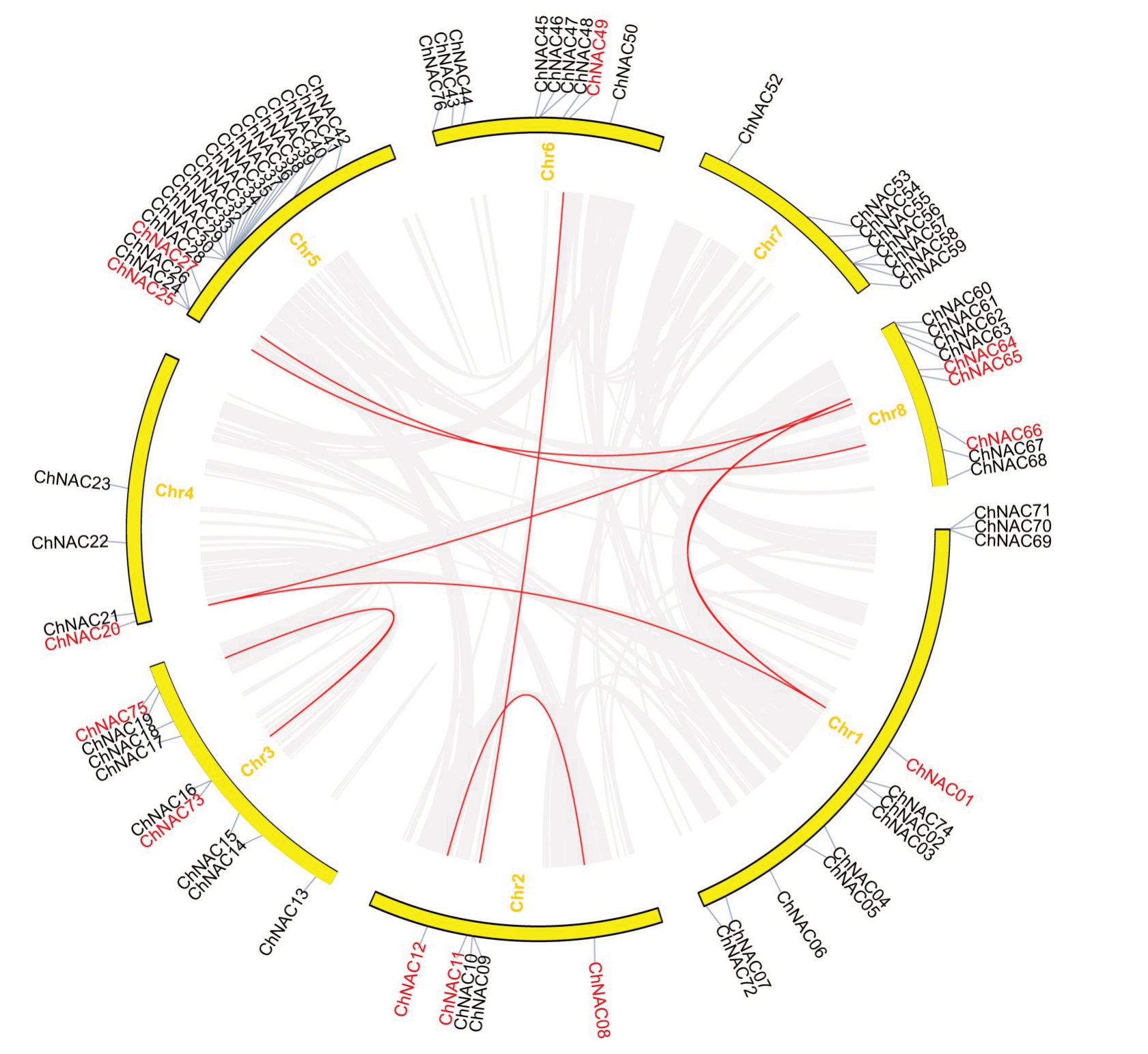
图4 ChNAC 基因共线性
Fig.4 ChNAC gene collinearity
2.5 欧李NAC基因启动子分析
为便于预测ChNAC 基因可能所具有的潜在功能,对ChNAC 基因起始密码子前2000 bp 的序列进行分析,并利用绘图软件进行整理并绘图。结果如图5 所示。ChNAC 基因顺式作用元件中除了CAAT、TATA等保证基因家族进行正常转录活动所需的组成型转录元件外,还含有植物激素类响应元件:TGA-element(生长素响应元件)、TCA-element(水杨酸响应元件)、P-box(赤霉素响应元件)、ABRE(脱落酸响应元件)、TGACG-motif(茉莉酸甲酯响应元件);植物生长发育响应元件:MSA-like(细胞周期响应元件)、circadian(昼夜节律控制响应元件)、RY-element(种子特异性响应元件)、O2-site(玉米醇溶蛋白代谢元件)、CAT-box(分生组织的表达元件)、GCN4-motif(胚乳发育元件)、HD-Zip 1(参与栅栏叶肉细胞分化的元素元件);结合位点MBS(MYB 结合位点元件)、HD-Zip 3(蛋白质结合位点元件)、Box 4(光响应元件)、LTR(低温胁迫响应元件)、ARE(厌氧诱导响应元件)、TC-rich repeats(参与防御和应激反应响应元件)等。其中,占比较大的是组成型转录元件、植物激素类响应元件和光响应元件。

图5 ChNAC 基因启动子分析
Fig. 5 Promoter analysis of ChNAC gene
2.6 欧李NAC基因组织特异性分析
欧李农大4号不同组织中,ChNAC基因的表达量存在差异(图6)。ChNAC01、ChNAC07、ChNAC08等16 个基因在果实中的表达量最高,且ChNAC07基因仅在果实中表达;ChNAC50、ChNAC53、ChNAC63 等15 个基因在叶片中的表达量最高,且ChNAC13、ChNAC53、ChNAC63基因仅在叶片中表达;ChNAC17、ChNAC66、ChNAC62等15个基因在根状茎中表达量最高,且ChNAC17、ChNAC59、ChNAC68 基因仅在根状茎中表达;ChNAC15、ChNAC46 等16 个基因在真根中的表达量最高;ChNAC36、ChNAC37 等10个基因在种仁中的表达量最高。 ChNAC23、ChNAC45、ChNAC47、ChNAC54 这4 个基因在上述组织中均不表达,推测其可能在花或其他器官中表达。ChNAC基因表达量在不同组织中存在差异,表明不同的ChNAC 基因调控不同组织的生长发育,即ChNAC基因的功能存在多样性,具有不同的生物学作用,ChNAC基因在欧李的生长发育中发挥着巨大作用。
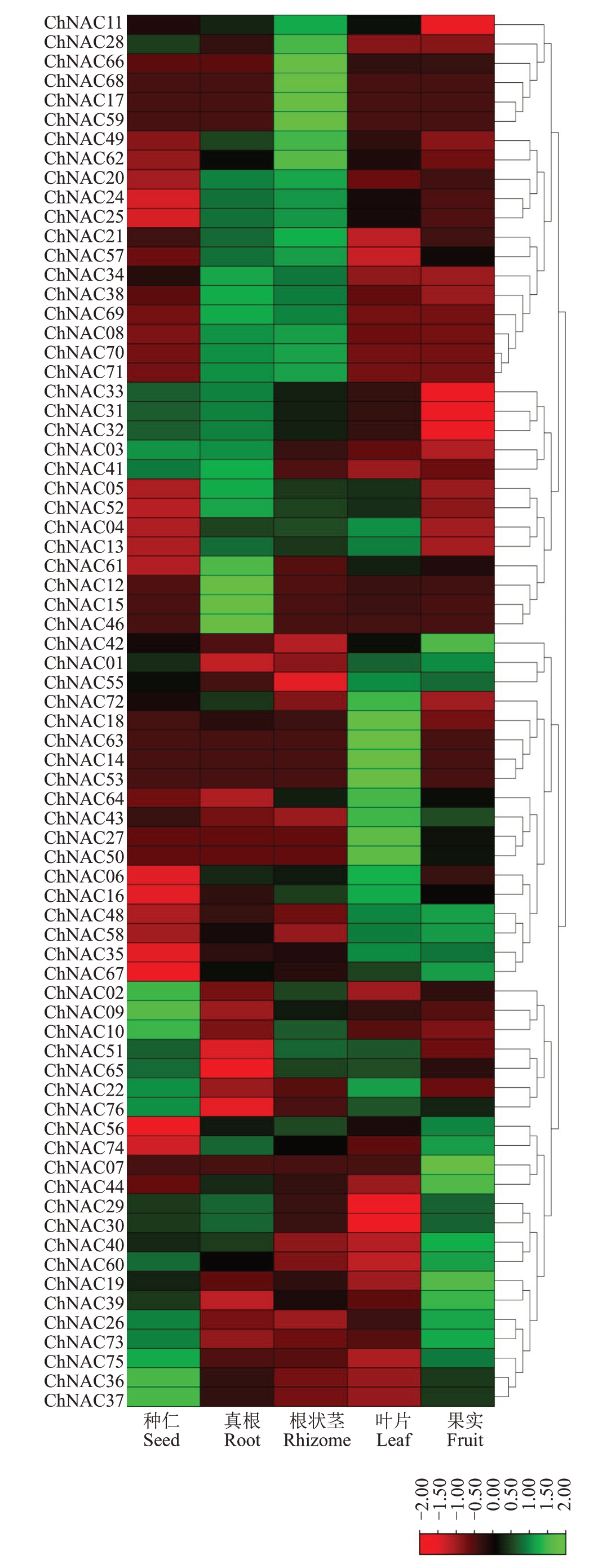
图6 不同组织中ChNAC 基因表达图谱
Fig.6 Expression map of ChNAC gene in different tissues
2.7 欧李NAC基因在非生物胁迫诱导下的表达模式分析
对60 个ChNAC 基因在胁迫中的表达量进行分析,结果表明(图7):在盐胁迫中,ChNAC15、ChNAC65、ChNAC74 等40 个基因的表达量上调,ChNAC24、ChNAC26、ChNAC27等14 个基因的表达量下降,ChNAC36、ChNAC37、ChNAC39等6个基因的表达量处理前后均差距不大;在模拟干旱胁迫中ChNAC01、ChNAC03、ChNAC18等38 个基因的表达量上调,ChNAC24、ChNAC11、ChNAC13 等13 个基因的表达量下降,ChNAC09、ChNAC10、ChNAC34等9个基因的表达量处理前后差距不大;在高温胁迫中,ChNAC08、ChNAC09、ChNAC10等38 个基因的表达量上调,ChNAC01、ChNAC13、ChNAC25 等19 个基因的表达量下降。ChNAC41、ChNAC62、ChNAC63 这3 个基因的表达量处理前后差距不大;在低温胁迫中,ChNAC24、ChNAC25、ChNAC35等40 个基因的表达量上调,ChNAC01、ChNAC03、ChNAC13 等18 个基因的表达量下调,ChNAC15、ChNAC63 这2 个基因的表达量在处理前后差距不大。综上,大多数NAC基因在胁迫条件下,通过正向调控来激活抗逆机制的运转,进而维持植物正常的生长发育。

图7 不同胁迫处理中ChNAC 基因表达图谱
Fig.7 Expression map of ChNAC gene under different stress
在胁迫处理下,一些基因的表达模式基本相似,如:ChNAC24 基因和ChNAC25 基因在低温胁迫下均为上调表达;在盐胁迫、模拟干旱及高温胁迫下均下调表达。ChNAC31 基因、ChNAC32 基因和ChNAC33基因在上述4种胁迫下表达量均上调。
ChNAC49 基因在上述胁迫下表达量均显著升高。在盐胁迫下,ChNAC49基因表达量与对照组相比上调了477 倍;在模拟干旱胁迫下,ChNAC49 基因表达量与对照组相比上调了124 倍;在低温胁迫下,ChNAC49 基因与对照组相比表达量上调了28 倍。ChNAC08基因在高温胁迫下的表达量上调值最大,与对照组相比上调了70.6 倍。ChNAC49 基因在高温胁迫下表达量的上调值仅次于ChNAC08基因,与对照组相比其上调了27.95倍。ChNAC64基因在盐胁迫下的表达量下降幅度最多,仅为对照组的14.29%;ChNAC17基因在模拟干旱胁迫下表达量的下降幅度最多,仅为对照组的15.96%;ChNAC01 基因的表达量在高温胁迫下的下降幅度最多,表达量仅为对照组的16.01%;ChNAC11基因的表达量在高温胁迫下的下降幅度最多,表达量仅为对照组的26.14%。说明这几个基因在上述4种胁迫下发挥着更重要的作用,是保证胁迫环境中植株正常生长发育的关键,对其表达量进行胁迫诱导处理,可能会获得所需的抗性植株。
3 讨论
通过生物信息学分析首次从欧李基因组中筛选出76个NAC基因家族成员。ChNAC基因家族成员数量低于水稻(151)[8]、核桃(121)[41]、甘蔗(85)[46]、龙眼(114)[47]、西瓜(104)[48]等,高于黑麦草(72)[49]等,综合来看,ChNAC 家族成员数量相比于其他物种较少,可能是基因组中没有大规模的片段复制且相对保守造成的[50]。
Ooka 等[8]首次根据NAC 基因的结构与功能将水稻和拟南芥的NAC 基因家族分为17 个亚族,大多数学者多用此方法作为NAC 基因家族的分类标准。笔者在本研究中以拟南芥NAC 基因作为参照将ChNAC 基因家族分为17 个亚族,各亚族所包含的ChNAC家族成员数量不同,其功能也可能存在差异。Clercq 等[51]证明拟南芥NAC2 亚族基因参与植物氧化的应激反应。Fujiwara 等[45]认为ONAC003亚族基因参与开花调节。依据系统进化树的同源关系,可以推测ChNAC对应亚族中的基因可能具有相似功能。NAM亚族及NAC1亚族中的拟南芥基因,被证明与拟南芥的形态发生有关[52],由此推测ChNAC52 基因、ChNAC62 基因、ChNAC68 基因中的一种或者几种,可能作用于欧李的形态发生过程中。ATAF、AtNAC3亚族参与拟南芥的应激反应[53],与其同源性高的ChNAC 基因,可能在欧李的抗旱、抗盐碱方面发挥着重要的作用。
启动子是RNA聚合酶识别、结合和开始转录的一段DNA 序列,它含有RNA 聚合酶特异性结合和转录起始所需的保守序列,多数位于结构基因转录起始点的上游,关系到基因的时空表达,是基因的开关,影响基因的表达程度。对启动子元件进行分析,可以推测其潜在功能[54-58]。ChNAC68、ChNAC72、ChNAC76等基因存在种子特异性作用元件,推测其可能在欧李种子胚乳的发育中具有重要作用;ChNAC03、ChNAC05 等基因存在胚乳表达元件,提示这些基因可能作用于种子表达或者种子萌发时期;ChNAC04、ChNAC05、ChNAC06等基因含有分生组织表达的相关作用元件,其可能作用于植物分生组织的形成;ChNAC03、ChNAC72 等基因的作用元件表明其可能诱导参与栅栏叶肉细胞分化的元素形成;O2-site 是一种代谢网络调控有关的作用元件,ChNAC03、ChNAC04、ChNAC05 等基因存在这种启动子元件说明这些基因可能参与玉米醇溶蛋白代谢途径;ChNAC基因的启动子中含有许多响应激素调控的作用元件(TGA-elemet响应生长素调控,P-box响应赤霉素调控,TCA-elemet 响应水杨酸调控,ABRE 响应脱落酸调控,TGACG-motif 响应茉莉酸甲酯调控)防御基因和应激反应作用元件提示基因在应对胁迫方面有重大作用,如ChNAC03、ChNAC04、ChNAC05等基因。启动子分析结果表明ChNAC 基因家族作用于欧李的生长、发育、组织分化、抗逆等多个方面,这与其他物种中NAC 基因家族的研究结果相一致[59-61]。
当植物感受到外界刺激时,通过信号传导途径传递给胁迫应答因子,从而激活植物的抗逆反应,减少植物受到的损伤,NAC基因在这个过程中扮演着重要角色。盐胁迫被认为是一种渗透胁迫[62],有学者认为,NAC 基因在盐胁迫时作用于植物ABA、乙烯、GA 等代谢途径,促进这些激素的合成,从而提高植物的抗盐性[63-64];也有学者认为,盐胁迫时可通过NA+、K+离子的改变,或可溶性糖、游离脯氨酸等低分子渗透调节物的增加来维持体内的渗透压,进而抵御盐胁迫,维持植物正常生长发育[65-66]。后续可对盐胁迫下欧李NAC 转基因植株中上述物质的含量进行测定,进而来验证这一猜想。温度胁迫时,NAC 基因主要通过调节温度应激基因启动子的表达及合成,进而来激发植物的抗逆机制的运作[67-68]。干旱胁迫时,一类NAC 转录因子与MYC-like 元件结合,在胁迫应答中起作用[20],另一类NAC基因[69]主要在气孔的保卫组织中表达,当植物感受到外界刺激时,会促进气孔关闭,从而使植株的抗旱性提高。Wang 等[70]在拟南芥中对ChNAC06 基因过表达,发现其在干旱胁迫下表达量升高,根系中ABA敏感性升高,抗干旱能力显著提高。笔者在干旱胁迫下所获得的结果与之相一致。尤其是ChNAC49基因,在盐胁迫、干旱胁迫、低温胁迫下的表达量上调幅度均最大,在高温胁迫下的表达量上调幅度仅次于ChNAC08 基因。推测其可能在更多的胁迫下同样发挥着重要的作用,对欧李的抗性研究可能是一种“关键基因”。且该基因在叶片、根状茎、根中均表达。可对该基因进行基因过表达处理,获得转基因植物,以期挖掘出ChNAC 基因家族更多的潜在功能,为欧李抗性植株的培育做贡献,也可根据不同品种ChNAC基因家族的表达量,初步筛选出具有特殊抗性的欧李品种。
4 结论
笔者在本研究中首次鉴定并分析了76 个欧李NAC 基因家族成员,明确了胁迫条件下各ChNAC基因的表达模式,发现ChNAC49基因在各胁迫处理下表达量的变化幅度显著高于其他基因,后期可对ChNAC49基因进行克隆,培育出抗性更强的欧李转基因株系,为ChNAC基因家族的功能验证奠定了理论基础。
[1] 俞德浚.中国果树分类学[M].北京:农业出版社,1979.YU Dejun. Chinese fruit tree taxonomy[M]. Beijing:Agricultural Press,1979.
[2] 张东为,贾天会,舒乔生.水保优良树种欧李的研究进展及今后研究方向[J].中国水土保持,2012(1):45-47.ZHANG Dongwei,JIA Tianhui,SHU Qiaosheng.Research progress and future research direction of Cerasus humilis[J].Soil and Water Conservation in China,2012(1):45-47.
[3] 吴亚芳,李晓艳,郭金丽,李连国,李东方,张薇,邵郅胜.欧李适应非生物胁迫的研究进展及应用前景[J].特产研究,2022,44(1):104-110.WU Yafang,LI Xiaoyan,GUO Jinli,LI Lianguo,LI Dongfang,ZHANG Wei,SHAO Zhisheng. Research progress and application prospect of cerasus humilis adaptation to abiotic stress[J].Special Wild Economic Animal and Plant Research,2022,44(1):104-110.
[4] SHINOZAKI K,YAMAGUCHI-SHINOZAKI K,SEKI M.Regulatory network of gene expression in the drought and cold stress responses[J]. Current Opinion in Plant Biology,2003,6(5):410-417.
[5] SINGH K B,FOLEY R C,OÑATE-SÁNCHEZ L.Transcription factors in plant defense and stress responses[J].Current Opinion in Plant Biology,2002,5(5):430-436.
[6] HUANG G T,MA S L,BAI L P,ZHANG L,MA H,JIA P,LIU J,ZHONG M,GUO Z F. Signal transduction during cold,salt,and drought stresses in plants[J]. Molecular Biology Reports,2012,39(2):969-987.
[7] CHEN W J,ZHU T.Networks of transcription factors with roles inenvironmental stress response[J]. Trends in Plant Science,2004,9(12):591-596.
[8] OOKA H,SATOH K,DOI K,NAGATA T,OTOMO Y,MURAKMI K,MATSUBARA K,OASTO N,KAWAI J,CARNINCI P,HAYASHIZAKI Y,SUZUKI K,KOJIMA K,TAKAHARA Y,YAMAMOTO K,KIKUCHI S.Comprehensive analysis of NAC Family Genes in Oryza sativa and Arabidopsis thaliana[J].Dna Research,2003,10(6):239-247.
[9] AIDA M,ISHIDA T,FUKAKI H,FUJISAWA H,TASAKA M.Genes involved in organ separation in Arabidopsis:An analysis of the cupshaped cotyledon mutant[J]. The Plant Cell,1997,9(6):841-857.
[10] SOUER E,VAN HOUWELINGEN A,KLOOS D,MOL J,KOES R.The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries[J]. Cell,1996,85(2):159-170.
[11] 柳展基,邵凤霞,唐桂英.植物NAC 转录因子的结构功能及其表达调控研究进展[J]. 西北植物学报,2007,27(9):1915-1920.LIU Zhanji,SHAO Fengxia,TANG Guiying.The research progress of structure,function and regulation of plant NAC transcription factors[J].Acta Botanica Boreali-Occidentalia Sinica,2007,27(9):1915-1920.
[12] XIE Q,FRUGIS G,COLGAN D,CHUA N H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promotelateral root development[J]. Genes & Development,2000,14(23):3024-3036.
[13] MAO C J,HE J M,LIU L N,DENG Q M,YAO X F,LIU C M,QIAO Y L,LI P,MING F.OsNAC2 integrates auxin and cytokinin pathways to modulate rice root development[J]. Plant Biotechnology Journal,2020,18(2):429-442.
[14] GUO Y F,GAN S S.At NAP,a NAC family transcription factor,has an important role in leaf senescence[J]. The Plant Journal,2006,46(4):601-612.
[15] JENSEN M K,LINDEMOSE S,DE MASI F,REIMER J J,NIELSEN M,PERERA V,WORKMAN C T,TURCK F,GRANT M R,MUNDY J,PETERSEN M,SKRIVER K.ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana[J]. FEBS Open Bio,2013,3:321-327.
[16] CHEN X,WANG Y F,LÜ B,LI J,LUO L Q,LU S C,ZHANG X,MA H,MING F.The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway[J].Plant and Cell Physiology,2014,55(3):604-619.
[17] HU H H,YOU J,FANG Y J,ZHU X Y,QI Z Y,XIONG L Z.Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice[J].Plant Molecular Biology,2008,67(1):169-181.
[18] CHEN X,LU S C,WANG Y F,ZHANG X,LÜ B,LUO L Q,XI D D,SHEN J B,MA H,MING F.OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice[J].The Plant Journal,2015,82(2):302-314.
[19] HE X J,MU R L,CAO W H,ZHANG Z G,ZHANG J S,CHEN S Y.At NAC2,a transcription factor downstream of ethylene and auxin signaling pathways,is involved in salt stress response and lateral root development[J].The Plant Journal,2005,44(6):903-916.
[20] TRAN L P,NAKASHIMA K,SAKUMA Y,SINPSON S D,FUJITA Y,MARUYAMA K,FUJITA M,SEKI M,SHINOZAKI K,YAMAGUCHI- SHINOZAKI K. Isolation and functional analysis of Arabidopsis stress in ducible NAC transcription factors that bind to a drought respon sive cis-element in the early responsive to dehydration stress l promoter[J]. The Plant Cell,2004,16(9):2481-2498.
[21] JEONG J S,KIM Y S,BAEK K H,JUNG H,HA S H,CHOI Y D,KIM M,REUZEAU C,KIM J K.Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions[J]. Plant Physiology,2010,153(1):185-197.
[22] LU P L,CHEN N Z,AN R,SU Z,QI B S,REN F,CHEN J,WANG X C.A novel drought-inducible gene,ATAF1,encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis[J].Plant Molecular Biology,2007,63(2):289-305.
[23] HAO Y J,WEI W,SONG Q X,CHEN H W,ZHANG Y Q,WANG F,ZOU H F,GANG L,TIAN A G,ZHANG W K,MA B,ZHANG J S,CHEN S Y. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants[J].The Plant Journal,2011,68(2):302-313.
[24] HEGEDUS D,YU M,BALDWIN D,GRUBER M,SHARPE A,PARKIN I,WHITWILL S,LYDIATE D. Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress[J].Plant Molecular Biology,2003,53(3):383-397.
[25] MAO X G,ZHANG H Y,QIAN X Y,LI A,ZHAO G Y,JING R L.TaNAC2,a NAC-type wheat transcription factor conferring enhanced mult-iple abiotic stress tolerances in Arabidopsis[J].Journal of Experimental Botany,2012,63(8):2933-2946.
[26] DONG Z B,XU Z N,XU L,XU L,GALLI M,GALLAVOTTI A,DOONER H K,CHUCK G. Necrotic upper tips1 mimics heat and drought stress and encodes a protoxylem-specific transcription factor in maize[J]. Proceedings of the National Academy of Sciences,2020,117(34):20908-20919.
[27] 梁晓庆.高温胁迫下番茄SlNAC1 基因的功能分析[D].泰安:山东农业大学,2015.LIANG Xiaoqing. Functional analysis of the tomato SlNAC1 gene under heat stress[D]. Tai’an:Shandong Agricultural University,2015.
[28] LI M,CHEN R,JIANG Q Y,SUN X J,ZHANG H,HU Z.Gm-NAC06,a NAC domain transcription factor enhances salt stress tolerance in soybean[J].Plant Molecular Biology,2021,105(3):333-345.
[29] ZHENG X N,CHEN B,LU G J,HAN B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance[J]. Biochemical and Biophysical Research Communications,2009,379(4):985-989.
[30] COLLINGE M,BOLLER T. Differential induction of two potato genes,Stprx2 and StNAC in response to infection by Phytophthora infestans and to wounding[J]. Plant Molecular Biology,2001,46(5):521-529.
[31] JENSEN M K,RUNG J H,GREGERSEN P L,GJETTING T,FUGLSANG A T,HANSEN M,JOEHNK N,LYNGKJAER M F,COLLINGE D B. The HvNAC6 transcription factor:a positive regulator of penetrationresistance in barley and Arabidopsis[J].Plant Molecular Biology,2007,65(1):137-150.
[32] DELESSERT C,KAZAN K,WILSON I W,STRAETEN D V D,MANNERS J,DENNIS E S,DOLFERUS R. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis[J].The Plant Journal,2005,43(5):745-757.
[33] BU Q Y,JIANG H L,LI C B,ZHAI Q Z,ZHANG J,WU X Y,SUN J Q,XIE Q,LI C Y.Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses[J]. Cell Research,2008,18(7):756-767.
[34] LIN R M,ZHAO W S,MENG X B,WANG M,PENG Y L.Rice gene OsNAC19 encodes a novel NAC-domain transcription factor and responds to infection by Magnaporthe grisea[J].Plant Science,2007,172(1):120-130.
[35] REN T,QU F,MORRIS T J. HRT gene function requires interac-tion between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus[J].The Plant Cell,2000,12(10):1917-1925.
[36] SELTH L A,DOGRA S C,RASHEED M S,HEALY H,RANDLES J W,REZAIAN M A. A NAC domainprotein interacts with Tomato leaf curl virus replicationaccessory protein and enhances viral repli-cation[J]. The Plant Cell,2005,17(1):311-325.
[37] WANG P F,YI S K,MU X P,ZHANG J C,DU J J. Chromosome-level genome assembly of Cerasus humilis using pacbio and Hi-C technologies[J].Frontiers in Genetics,2020,11:956.
[38] 张晓旭,郭彩珍,穆霄鹏,郭夕雯,王鹏飞,汪泽文,曹琴,杜俊杰.欧李AP2 基因家族的鉴定与表达分析[J].分子植物育种,2022,20(7):2162-2169.ZHANG Xiaoxu,GUO Caizhen,MU Xiaopeng,GUO Xiwen,WANG Pengfei,WANG Zewen,CAO Qin,DU Junjie. Identification and expression analysis of AP2 gene family in chinese dwarf cherry [Cerasus humilis (Bge.) Sok][J]. Molecular Plant Breeding,2022,20(7):2162-2169.
[39] 汪泽文,杨依维,王鹏飞,张建成,穆霄鹏,付鸿博,杜俊杰.欧李DREB 基因家族的鉴定与分析[J].植物生理学报,2020,56(3):413-422.WANG Zewen,YANG Yiwei,WANG Pengfei,ZHANG Jiancheng,MU Xiaopeng,FU Hongbo,DU Junjie. Identification and analysis of DREB gene family in Cerasus humilis[J].Plant Physiology Journal,2020,56(3):413-422.
[40] 钱禛锋,谷书杰,赵雪婷,饶席兵,曾丹,沈庆庆,张蓉琼,陈疏影,何丽莲,李富生.基于转录组的蔗茅CML 基因家族鉴定及冷胁迫表达分析[J]. 农业生物技术学报,2022,30(5):885-895.QIAN Zhenfeng,GU Shujie,ZHAO Xueting,RAO Xibing,ZENG Dan,SHEN Qingqing,ZHANG Rongqiong,CHEN Shuying,HE Liliang,LI Fusheng. Identification and cold stress expression analysis of CML gene family in Erianthus fulvus based on transcriptome[J].Chinese journal of agricultural biotechnology,2022,30(5):885-895.
[41] 亢超,郭彩华,张雪蒙,刘金明,袁星,全绍文,牛建新. 核桃NAC 基因家族的全基因组鉴定与分析[J].果树学报,2021,38(9):1444-1458.KANG Chao,GUO Caihua,ZHANG Xuemeng,LIU Jinming,YUAN Xing,QUAN Shaowen,NIU Jianxin. Genome- wide identification and analysis of NAC gene family in walnut (Juglans regia L.)[J]. Journal of Fruit Science,2021,38(9):1444-1458.
[42] 郭禄芹,赵世豪,朱华玉,胡建斌,孙守如,马长生,杨路明.167份西瓜种质材料的遗传多样性分析[J]. 中国瓜菜,2018,31(1):5-11.GUO Luqin,ZHAO Shihao,ZHU Huayu,HU Jianbin,SUN Shouru,MA Changsheng,YANG Luming.The genetic diversity analysis of 167 watermelon germplasms[J].China Cucurbits and Vegetables,2018,31(1):5-11.
[43] ZHU T T,NEVO E,SUN D F,PENG J H. Phylogenetic analyses unravel the evolutionary history of NAC proteins in plant[J].Evolution,2012,66(6):1833-1848.
[44] 康桂娟,黎瑜,曾日中.巴西橡胶树HbNTL2 基因的克隆和表达分析[J].湖南农业大学学报(自然科学版),2015,41(6):602-609.KANG Guijuan,LI Yu,ZENG Rizhong. Clone and expression analysis on gene HbNTL2 from Hevea brasiliensis Muell.Arg.[J].Journal of Hunan Agricultural University (Natural Sciences),2015,41(6):602-609.
[45] FUJIWARA S,MITSUDA N.ANAC075,a putative regulator of VASCULAR-RELATED NAC-DOMAIN7,is a repressor of flowering[J].Plant Biotechnology,2016,33(4):255-265.
[46] RAMASWAMY M,NARAYANAN J,MANICKAVACHAGAM G,ATHIAPPAN S,ARUN M,GOMATHI R,RAM B.Genome wide analysis of NAC gene family′sequences′in sugarcane and its comparative phylogenetic relationship with rice,sorghum,maize and Arabidopsis for prediction of stress associated NAC genes[J].Agri Gene,2017,3:1-11.
[47] MUNIR N,CHEN Y K,CHEN X H,NAWAZ M A,IFTIKHAR J,RIZWAN H M,SHEN X,LIN Y L,XU X H,LAI Z X. Genome-wide identification and comprehensive analyses of NAC transcription factor gene family and expression patterns during somatic embryogenesis in Dimocarpus longan Lour.[J]. Plant Physiology and Biochemistry,2020,157:169-184.
[48] LÜ X L,LAN S R,GUY K M,YANG J H,ZHANG M F,HU Z Y.Global expressions landscape of NAC transcription factor family and their responses to abiotic stresses in Citrullus lanatus[J].Scientific Reports,2016,6:30574.
[49] NIE G,YANG X Y,YANG Z F,ZHONG M Y,ZHU Y Q,ZHOU J,APPIAH C,LIAO Z C,FENG G Y,ZHANG X Q. Genomewide investigation of the NAC transcript factor family inperennial ryegrass (Lolium perenne L.) and expression analysis under various abiotic stressor[J].Genomics,2020,112(6):4224-4231.
[50] SONG J,GAO Z H,HUO X M,SUN H L,XU Y S,SHI T,NI Z J. Genome-wide identification of the auxin response factor(ARF) gene family and expression analysis of its role associated with pistil development in Japanese apricot (Prunus mume Sieb.et Zucc.)[J].Acta Physiologiae Plantarum,2015,37(8):145.
[51] CLERCQ I D,VERNEIRSSEN V,AKEN O V,VANDEPOELE K,MURCHA M W,LAW S R,INZE A,NG S,IVANOVA A,ROMBAUT D,COTTE B V D,JASPERS P,PEER Y V D,KANGASJARVI J,WHELAN J,BREUSEGEM F V. The mem-branebound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis[J]. The Plant Cell,2013,25(9):3472-3490.
[52] AIDA M,ISHIDA T,FUAFAKI H,TASAKA M. Genes involved in organ separation in Arabidopsis:an analysis of the cupshaped cotyledon mutant[J].The Plant Cell,1997,9(6):841-857.
[53] COLLINGE M,BOLLER T. Differential induction of two potato genes,Stprx2 and StNAC,in response to infection by Phytophthora infestans and to wounding[J].2001,46(5):521-529.
[54] 苏炳茜,张舒婷,刘蒲东,李晓斐,戚峰,林玉玲,赖钟雄.龙眼TMK 基因家族成员的全基因组鉴定及表达分析[J].果树学报,2021,38(10):1668-1680.SU Bingxi,ZHANG Shuting,LIU Pudong,LI Xiaofei,QI Feng,LIN Yuling,LAI Zhongxiong. Genome-wide identification and expression analysis of TMK gene family in Dimocarpus longan Lour.[J].Journal of Fruit Science,2021,38(10):1668-1680.
[55] 杨杰,陈蓉,胡文娟,吴巧玲,佟晓楠,李兴涛.枳PP2C 基因家族的鉴定及表达分析[J].果树学报,2022,39(4):532-547.YANG Jie,CHEN Rong,HU Wenjuan,WU Qiaoling,TONG Xiaonan,LI Xingtao. Identification and expression analysis of PP2C gene family in Poncirus trifoliata[J].Journal of Fruit Science,2022,39(4):532-547.
[56] 谭彬,魏鹏程,栗焕楠,连晓东,陈谭星,郑先波,程钧,王伟,王小贝,冯建灿.桃PEBP 基因家族全基因组鉴定及桃TFL1 基因功能分析[J].果树学报,2020,37(10):1443-1454.TAN Bin,WEI Pengcheng,LI Huannan,LIAN Xiaodong,CHEN Tanxing,ZHENG Xianbo,CHENG Jun,WANG Wei,WANG Xiaobai,FENG Jiancan. Genome-wide identification of PEBP gene family and functional analysis of TFL1 gene in peach (Prunus persica)[J]. Journal of Fruit Science,2020,37(10):1443-1454.
[57] 周婉莹,曲东,张晓娟,张文慧,刘艳飞,张羽.中华猕猴桃fasciclin 基因家族生物信息学分析及克隆[J].果树学报,2020,37(8):1144-1155.ZHOU Wanying,QU Dong,ZHANG Xiaojuan,ZHANG Wenhui,LIU Yanfei,ZHANG Yu. Bioinformatics analysis and cloning of fasciclin gene family in kiwifruit[J]. Journal of Fruit Science,2020,37(8):1144-1155.
[58] 陈迪飞,魏秀清,许玲,许家辉,曾黎辉.莲雾PG 基因家族全基因组鉴定及表达分析[J].果树学报,2022,39(4):548-563.CHEN Difei,WEI Xiuqing,XU Ling,XU Jiahui,ZENG Lihui.Genome-wide identification and expression analysis of PG gene family in wax apple[Syzygium samarangense(Bl.)Merr.et Perry][J].Journal of Fruit Science,2022,39(4):548-563.
[59] LIU J,CHEN T Y,YUAN Y,ZHOU J H,ZHAO Y Y,HUANG L Q. Bioinformatics and expression analysis of NAC transcription factor genes in Scutellaria baicalensis[J]. World Journal of Traditional Chinese Medicine,2018,4(2):37-42.
[60] LING L,SONG L L,WANG Y J,GUO C H.Genome-wide analysis and expression patterns of the NAC transcription factor family in Medicago truncatula[J]. Physiology and Molecular Biology of Plants:An International Journal of Functional Plant Biology,2017,23(2):343-356.
[61] SAIDI M N,MERGBY D,BRINI F. Identification and expression analysis of the NAC transcription factor family in durum wheat (Triticum turgidum L. ssp. durum)[J]. Plant Physiology and Biochemistry,2017,112:117-128.
[62] ZHU J K. Abiotic stress signaling and responses in plants[J].Cell,2016,167(2):313-324.
[63] YANG R,DENG C,QUYANG B,YE Z. Molecular analysis of two salt-responsive NAC-family genes and their expression analysis in tomato[J].Molecular Biology Reports,2011,38(2):857-863.
[64] LIU Q L,XU K D,ZHAO L J,PAN Y Z,JIANG B B,ZHANG H Q,LIU G L.Overexpression of a novel chrysanthemum NAC transcription factor gene enhances salt tolerance in tobacco[J].Biotechnology Letters,2011,33(10):2073-2082.
[65] 张丹,马玉花.NAC 转录因子在植物响应非生物胁迫中的作用[J].生物技术通报,2019,35(12):144-151.ZHANG Dan,MA Yuhua.The role of NAC transcription factors in plant response to abiotic stress[J]. Biotechnology Bulletin,2019,35(12):144-151.
[66] SONG S Y,CHEN Y,CHEN J,DAI X Y,ZHANG W H.Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress[J].Planta,2011,234(2):331-345.
[67] KNIGHT M R,KNIGHT H. Low-temperature perception leading to gene expression and cold tolerance in higher plants[J].New Phytologist,2012,195(4):737-751.
[68] GUAN Q M,YUE X L,ZENG H T,ZHU J H.The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress- responsive gene regulation and thermotolerance in Arabidopsis[J].The Plant Cell,2014,26(1):438-453.
[69] HU H H,DAI M Q,YAO J L,XIAO B Z,LI X H,ZHANG Q F,XIONG L Z. Overexpressing a NAM,ATAF,and CUC(NAC)transcription factor enhances drought resistance and salt tolerance in rice[J]. Proceedings of the National Academy of Sciences,2006,103(35):12987-12992.
[70] WANG F,WANG J W,SUN L J,SONG X S. The molecular cloning and functional characterization of ChNAC1,a NACtranscription factor in Cerasus humilis[J].Plant Growth Regulation,2019,89(3):331-343.