油脂是生物体能量代谢的主要来源,在生物膜合成、能量稳定、信号转导等方面发挥重要作用[1-2]。植物细胞中油脂以三酰甘油(Triacylglycerol,TAG)的形式合成于内质网,通过出芽的形式存储于特定细胞器油体(Lipid Droplet,LD)中。油体主要由三酰甘油(TAG)、磷脂(Phospholipid)和内嵌蛋白(Intrinsic proteins)组成。TAG 是植物种子中能量存储的有效形式之一,可以为种子萌发和幼苗初期生长提供必要的能量[3]。由亲水性的磷酸基团和疏水性的酰基团组成的磷脂是油体生物膜结构的主要组分,内嵌蛋白则主要负责维持油体的结构稳定性。油体表面的内嵌蛋白包括油体蛋白(Oleosin,OLEO),油体钙蛋白(Caleosin,CLO)、甾醇蛋白(Steroleosin,SLO)和油体相关蛋白(Lipid Droplet Associated Proteins,LDAP)等[4]。油体蛋白最早在绿藻中被发现,是油体含量最丰富的膜蛋白,约占种子总蛋白的8%[5-7]。随着分子生物学技术的快速发展,目前拟南芥(Arabidopsis thaliana)[8]、油菜(Brassica napus)[9]、大豆(Glycine max)[10]和芝麻(Sesamum indicum)[11]等物种中的油体蛋白均被研究。不同物种中oleosin 基因具有相似的理化性质:蛋白相对分子质量为15~26 ku,蛋白结构包含1 个疏水环区,具体由3 个脯氨酸残基和1 个丝氨酸残基(-PX5SPX3P-)组成的“脯氨酸结”[4],该结构域对油体蛋白靶向定位到油体至关重要[12-13]。
Oleosin是种子油体表面表达丰度最高的蛋白,另外发现oleosin 也定位于花粉的表面和茸毛层[14]。目前在拟南芥中鉴定到16个oleosin蛋白,其中5个主要在种子中表达,8 个在花药中表达,以及3 个同时在种子和花药中表达,其中在种子中表达的oleosin 蛋白主要功能是调节油体大小和影响种子萌发[2]。研究发现,oleosin 蛋白在种子干燥过程中对油体的稳定性和融合有一定的影响;另外在种子过冬阶段,oleosin 蛋白调控油体大小和种子活力,在种子萌发过程中oleosin蛋白调控脂质代谢[2]。拟南芥AtOle1 基因敲除后,突变体种子油体形态异常,油体异常积累,导致种子萌发延缓[15]。水稻中过表达黄豆oleosin基因,转基因水稻籽粒中油体蛋白数量增多、体积变小,种子含油量显著增加,但三酰甘油的脂肪酸组成并未发生变化,表明油体大小与总含油量呈负相关[16]。拟南芥ole1 ole2 双突变体中,发现种子细胞中具有不规则且增大的含油结构,同时,油体蛋白可通过防止低温自吸胀过程中油体的异常融合,提高种子细胞活力[17]。oleosin 基因可以维持油体的大小和结构的稳定性,影响脂类物质的积累,从而调节植物种子的含油量[18],并且种子活力可能与含油量相关[19]。由此可见,油体蛋白oleosin在种子生长发育过程中具有重要作用。
山核桃(Carya cathayensis),也称中国山核桃,属于胡桃科山核桃属,主要分布于浙江和安徽一带[20],已有500 多年的种植历史[21]。薄壳山核桃(Carya illinoinensis),也属于胡桃科山核桃属,是世界著名的胡桃科坚果树种,原产于美国[22]。山核桃与薄壳山核桃分别是东亚和北美洲东部的胡桃科山核桃属代表,也是胡桃科山核桃属中仅有的两种商业栽培的树种[21]。山核桃的坚果较小,外壳较硬,而且缺乏对非生物胁迫如高温、洪水、干旱和盐碱的耐受性,极大地限制了山核桃全世界范围内的商业化种植[21]。山核桃与薄壳山核桃含油量都高达70%,其中不饱和脂肪酸占90%以上[23],且具有补脑、降低血脂、预防心脑血管疾病等功效[24]。笔者以商业栽培山核桃属坚果树种的2 个代表(山核桃与美国山核桃)为研究对象,根据已公开发表的山核桃和薄壳山核桃基因组信息[21],通过生物信息学手段分析oleosin 基因家族,主要包括保守基序、系统进化、启动子区域顺式作用元件、表达模式及亚细胞定位研究等方面,为进一步筛选山核桃oleosin候选基因并进行功能分析提供一定的参考依据。
1 材料和方法
1.1 Oleosin基因鉴定
通过拟南芥数据库TAIR(https://www.arabidop-sis.org)获取拟南芥oleosin基因家族蛋白序列,作为参照序列,通过BioEdit进行本地BlastP从山核桃以及薄壳山核桃数据库(https://ngdc.cncb.ac.cn/databasecommons/database/id/4151)中获取oleosin 蛋白序列,利用Pfam 数据库(https://pfam.xfam.org)筛选包含oleosin结构域的蛋白序列。
1.2 Oleosin蛋白的理化性质分析
利 用 ProtParam(https://web.expasy.org/protparam)数据库分析山核桃和薄壳山核桃oleosin 蛋白理化性质,并且利用WoLF PSORT(https://wolfpsort.hgc.jp)在线工具进行亚细胞定位预测分析。
1.3 Oleosin蛋白保守基序和系统进化分析
山核桃与薄壳山核桃oleosin 蛋白保守基序分析参考徐胤等[25]的方法。利用MEME Suite 5.4.1(https://meme-suite.org/meme/doc/meme.html)对山核桃和薄壳山核桃oleosin蛋白保守基序进行分析,最后利用TBtools 软件进行优化。利用DNAMAN软件对oleosin 基因家族氨基酸序列进行多重比对分析。通过Uniprot(UniProt)数据库下载油菜、花生(Arachis hypogaea)和大豆的oleosin蛋白序列,结合已获得的拟南芥、山核桃、薄壳山核桃oleosin 蛋白序列,利用MEGA 7.0 软件通过邻接法(Neighbor-Joining,NJ)构建系统进化树,校对参数Bootstrap设置为1000,进一步利用Evolview(https://www.evolgenius.info/evolview-v2)在线工具进行优化。
1.4 Oleosin 基因家族启动子区域顺式作用元件分析
利用TBtools 软件从山核桃和薄壳山核桃基因组中导出oleosin 基因上游2000 bp 启动子区域序列,然后利用Plant CARE(https://bioinformatics.psb.ugent.be/webtools/plantcare/html)预测山核桃和薄壳山核桃oleosin蛋白启动子区域顺式作用元件。
1.5 山核桃oleosin 基因在胚发育不同阶段的表达模式分析
为了研究山核桃oleosin 基因在胚发育不同阶段的表达丰度,利用NCBI Short Read Archive database(SRA)(https://www.ncbi.nlm.nih.gov/sra/)获得山核桃胚发育不同阶段转录组数据(检索号为PRJNA687050)[26],获得oleosin 基因RPKM(Reads Per Kilobase Per Million Mapped Reads)数值,利用MEV4.9.0绘制热图。
1.6 低温胁迫及ABA处理下山核桃oleosin基因的表达模式分析
研究材料为浙江农林大学张启香教授课题组已成功诱导的山核桃愈伤组织。每个愈伤培养皿直径为90 mm,高度为20 mm,培养皿中愈伤个数平均为9个。3个愈伤培养皿为1个重复。每个处理3个重复。对生长状况良好的山核桃愈伤组织进行4 ℃低温和100 μmol·L-1 ABA 处理,并分别在处理0、2、4、6、8 和12 h 后取0.5 g 愈伤组织,液氮速冻,利用Trizol 法[27]提取RNA,并反转录合成cDNA,用于后续qRT-PCR 检测。反转录试剂盒(PrimeScriptTMRTreagent Kit,RR047A)和 TB green 试剂(RR820A)购于TaKaRa公司。
利用Primer Primer 5软件设计山核桃oleosin基因序列的特异性定量引物,由生工生物工程(上海)股份有限公司合成,以山核桃CCARF基因为内参基因(具体引物序列如下表1)。基因相对表达量计算采用2-ΔΔCt方法。
表1 荧光定量PCR 引物序列
Table 1 primer sequence of qPCR

基因名称Gene name CCA0791S0002 CCA0514S0208 CCA0520S0025 CCA0656S0074 CCA0674S0096 CCA0779S0183 CCARF正向引物(5’-3’)Forward Primer(5’-3’)CACTCTCGTACCCGTCTTCG ATCCTTGGGTTCTTGGCGTC GTCCAAAGTCCCAGAGAGGC ATTCAGTCCTGTTCTCGTCCC CGATCCTAGTCCCGGTAGGT GGTCCAAAGTCCCAAGGAGG TGTGGTTGAAGCTAGGGATG反向引物(5’-3’)Reverse Primer(5’-3’)GCTTTGTGTGCTCGAGTTGC CCTGTGCACCAGAGATGCTG CCGAGCTCAAAAATCCAGCG ATTTGTGCCGCCCCAGG CACAAACTGCCCGTACTCCT GAAGACCCGGGATAGCGATG GCGTTGACGGAGTGAGTG
1.7 山核桃oleosin蛋白亚细胞定位分析
载体与试剂:pBI221-UBQ10-GFP 载体由笔者所在实验室保存;DNA 聚合酶(Phanta Max Super-Fidelity DNA Polymerase,P505)购于南京诺唯赞生物科技有限公司;琼脂糖凝胶纯化回收试剂盒(E.Z.N.A.®Gel Extraction Kit,OMEGA)购于南京百思禾生物科技有限公司;质粒小提试剂盒(TIANGEN®Mini Plasmid Kit)购于天根生化科技(北京)有限公司;限制性内切酶QuickCut™Xba Ⅰ和QuickCut™BamH Ⅰ与T4 DNA 连接酶均购于TaKaRa 公司;质粒测序由北京擎科生物技术有限公司完成。
用SnapGene 软件分别在山核桃oleosin 基因CCA0520S0025 和CCA0791S0002 的CDS 序列两端设计引物(CCA0520S0025-F:GGGTCTAGAATGTCTGATCACTCAAGGCCATTG、CCA0520S0025-R:GGGGGATCCTGTCCTCCTTCCCTCTTGGGC、CCA07-91S0002-F:GGGTCTAGAATGGCCGAGAACCAACAGCA 和CCA0791S0002-R:GGGGGATCCGCCCTGTGCACCAGAGACAT)。引物下划线的为酶切位点Xba Ⅰ(TCTAGA)和BamH Ⅰ(GGATCC)。利用Trizol法[27]提取山核桃胚RNA,利用反转录试剂盒(TRAN,北京,P30702)获得cDNA。以cDNA 为模板,用CCA0520S0025-F 与CCA0520S0025-R 和CCA0791S0002-F 与CCA0791S0002-R 引物对山核桃oleosin基因CCA0520S0025和CCA0791S0002的CDS 序列进行PCR 扩增。PCR 反应体系50 μL:2×Phanta Max Buffer 25 μL,10 μmol·L-1正反向引物各2 μL,10 mmol·L-1 dNTP 1 μL,模板cDNA 4 μL,ddH2O 15 μL,Phanta Max Super-Fidelity DNA Polymerase 1 μL。PCR 反应程序:95 ℃预变性5 min,95 ℃变性15 s,58 ℃退火15 s,72 ℃延伸1 min,30个循环,72 ℃延伸5 min。利用琼脂糖凝胶纯化回收试剂盒回收目的基因片段。利用限制性内切酶Xba Ⅰ和BamH Ⅰ对pBI221-UBQ10-GFP 载体与回收的目的基因片段进行双酶切。通过酶切连接的方法,构建表达载体pBI221-UBQ10-CCA0520S0025-GFP 和pBI221-UBQ10-CCA0791S0002-GFP,经测序验证正确后,提取质粒。
拟南芥悬浮细胞瞬时转化体系参考Zeng 等[28]的方法。将培养5 d 的拟南芥悬浮细胞转至50 mL离心管中,1000 r·min-1离心2 min,弃上清液,加入酶解液置于25 ℃摇床解离2 h 后,750 r·min-1离心10 min,弃上清液,加入2 倍的EP Buffer 重悬细胞,750 r·min-1离心10 min,2 次重复,吸取500 μL 拟南芥悬浮细胞至电击杯,加入一定浓度的质粒,如pBI221- UBQ10- CCA0520S0025- GFP 和pBI221-UBQ10-CCA0791S0002-GFP 分别与AtOLEO2-RFP(拟南芥oleosin基因)共转化,静置5~10 min,以130 V和1000 μF电压电击后,加入2 mL TEX Buffer重悬细胞,倒入35 mm培养皿置于25 ℃培养12 h。利用激光共聚焦显微镜(Leica TCS SP8)观察亚细胞定位情况,具体分析山核桃oleosin基因与拟南芥oleosin标志基因AtOLEO2的亚细胞共定位情况,绿色荧光(GFP)接收波长范围为488~510 nm,红色荧光(RFP)接收波长范围为532~588 nm。试验所用的拟南芥oleosin 基因(AtOLEO2-RFP)质粒由笔者所在实验室保存。
2 结果与分析
2.1 Oleosin基因家族成员鉴定及理化性质分析
以拟南芥oleosin 基因[29]作为参照,在山核桃和薄壳山核桃基因组中分别鉴定到7 个和8 个具有脯氨酸发卡结构域的oleosin 基因,进一步利用Prot-Param 软件对15 个oleosin 蛋白的理化性质进行分析。结果显示(表2),山核桃和薄壳山核桃oleosin基因编码139~166 个氨基酸;蛋白质分子质量为14.54~17.51 ku;等电点范围为9.33~10.36,蛋白不稳定系数介于28.71~63.13 之间,脂肪系数为88.86~112.11,平均亲水性介于0.098~0.467 之间。对亚细胞定位进行预测,结果发现,CCA0520S0025、CCA0779S0183、CIL1212S0020、CIL0938S0150 可能定位于叶绿体,CCA0514S0208 和CIL0009S0029可能定位于液泡膜,其余9 个山核桃oleosin 蛋白可能定位于细胞质膜。综上,山核桃与薄壳山核桃的蛋白质分子质量均小于25 ku,具有较高的等电点和脂肪系数,即山核桃和薄壳山核桃具有相似的理化性质。
表2 山核桃和薄壳山核桃oleosin 基因家族成员理化特征分析
Table 2 Analysis of protein characterization of oleosin genes family members in Carya cathayensis and Carya illinoinensis

物种Species山核桃Carya cathayensis基因ID Gene ID CCA0674S0096氨基酸长度Amino acid length/aa 147分子质量Molecular weight/ku 15.675 40等电点PI 9.57不稳定系数Instability index 29.04脂肪系数Aliphatic index 112.11平均亲水性Grand average of hydropathi-city 0.467 CCA0514S020814815.722 509.5141.45107.500.411 CCA0656S007213914.580 119.8338.59107.410.422 CCA0656S007413914.592 169.8337.98110.220.459 CCA0791S000215616.192 8610.2734.7694.550.263 CCA0520S002515416.223 0010.2855.85105.840.147 CCA0779S018314014.536 0410.1362.88102.500.224薄壳山核桃Carya illinoinensis CIL1212S002015616.948 699.3341.5498.910.408 CIL0212S001514715.728 489.4728.71105.510.401 CIL0252S000915616.163 8410.3640.1692.120.204 CIL0009S002914014.646 019.9836.90106.000.341 CIL1494S004515416.237 9810.2850.19102.010.131 CIL0940S011913914.558 049.8032.75107.410.389 CIL0938S015016617.505 379.9340.7088.860.098 CIL0922S019014014.583 1810.3663.13109.500.274亚细胞定位Subcellular localization细胞质膜Plasma membrane液泡膜Vacular membrane细胞质膜Plasma membrane细胞质膜Plasma membrane细胞质膜Plasma membrane叶绿体Chloroplast叶绿体Chloroplast叶绿体Chloroplast细胞质膜Plasma membrane细胞质膜Plasma membrane液泡膜Vacular membrane细胞质膜Plasma membrane细胞质膜Plasma membrane叶绿体Chloroplast细胞质膜Plasma membrane
2.2 Oleosin 蛋白质保守结构域及启动子顺式作用元件分析
通过氨基酸多重序列比对,发现山核桃和薄壳山核桃oleosin 蛋白都具有一个典型的且高度保守的“脯氨酸结”基序(图1-A),即包含1个丝氨酸残基和3个脯氨酸残基,可简化为PX5SPX3P,该结构是油体中特有的结构。对山核桃与薄壳山核桃oleosin基因结构分析,发现oleosin基因家族成员均含有一个外显子,无内含子。山核桃与薄壳山核桃oleosin蛋白保守基序分析显示,山核桃和薄壳山核桃15个oleosin基因均包含motif 1、motif 2、motif 3和motif 4基序,推测该4 个基序在oleosin 蛋白序列中高度保守;从基序结构和基序位置来看,CIL0938S0150、CCA0791S0002 与CIL0252S0009 的基序位置相似,而CIL0938S0150 蛋白序列存在额外的motif 15 基序,可能具有特异性的功能;类似的,CCA0656S0072、CCA0656S0074 与CIL094S0119 3个蛋白具有一致的保守基序,暗示蛋白结构和功能可能相似(图1-B)。

图1 山核桃和薄壳山核桃oleosin 蛋白保守基序和启动子顺式作用元件分析
Fig.1 Analysis of conserved motif and the promoter cis-acting elements of oleosin protein in Carya cathayensis and Carya illinoinensis
通过PlantCARE在线工具对山核桃和薄壳山核桃oleosin 基因启动子顺式作用元件进行功能预测。结果发现山核桃和薄壳山核桃oleosin 基因启动子均含有激素响应和胁迫响应元件。在山核桃和薄壳山核桃中分别有5个和7个oleosin基因启动子中含有MeJA响应元件、分别有5个和4个oleosin基因启动子中含有GA 响应元件、分别有5 个和6 个oleosin 基因启动子中含有ABA 响应元件。除了CCA0520S0025、CIL0922S0190 和CIL0252S0009外,其余oleosin基因均含有光周期响应元件。同时发现CCA0779S0183、CIL0009S0029、CIL1212S0020、CIL0212S0015 与CIL0252S0009 启动子中含有干旱响应元件。CCA0779S0183、CIL0009S0029 与CIL1494S0045启动子中含有低温响应元件(图1-C)。在15个oleosin 基因启动子中均检测到应对环境变化和激素信号相关的顺式作用元件,因此推测山核桃和薄壳山核桃oleosin 基因响应逆境及激素信号而参与调控植物生长发育。
2.3 Oleosin基因家族系统发育进化分析
利用MEGA7.0 软件对山核桃(7 个)和薄壳山核桃(8 个)、拟南芥(17 个)、油菜(17 个)、花生(17个)和大豆(14 个)的oleosin 基因进行系统进化分析。结果表明,以上80个基因可划分为4个亚家族,包括T-Oleosin、U-Oleosin、SH-Oleosin和SL-Oleosin(图2)。山核桃与薄壳山核桃15个oleosin基因分布在U-Oleosin,SH-Oleosin,SL-Oleosin 3 个亚家族。SL-Oleosin 是最大亚族分支,约占山核桃和薄壳山核桃oleosin 基因成员的1/2。在SL-Oleosin 亚家族中,山核桃和薄壳山核桃oleosin基因分布在2个分支中,CCA0791S0002、CIL0938S0150与CIL0252S0009处于同一分支,亲缘关系最近。另一分支中CCA0520S0025 与CIL1494S0045、CCA0779S0183和CIL0922S0190汇聚。在U-Oleosin 亚家族中,山核桃和薄壳山核桃oleosin 基因同样分布在2 个分支中,CCA0674S0096 和CIL0212S0015 聚在同一分支,亲缘关系最近;CIL1212S0020 分布在另一分支中,未发现亲缘关系较近的山核桃oleosin基因。在SH-Oleosin 分支中,山核桃与薄壳山核桃oleosin 基因聚在同一分支,亲缘关系较近,先与花生和大豆oleosin 基因汇聚在一个分支,最后与十字花科的油菜和拟南芥oleosin 基因汇聚在一起。
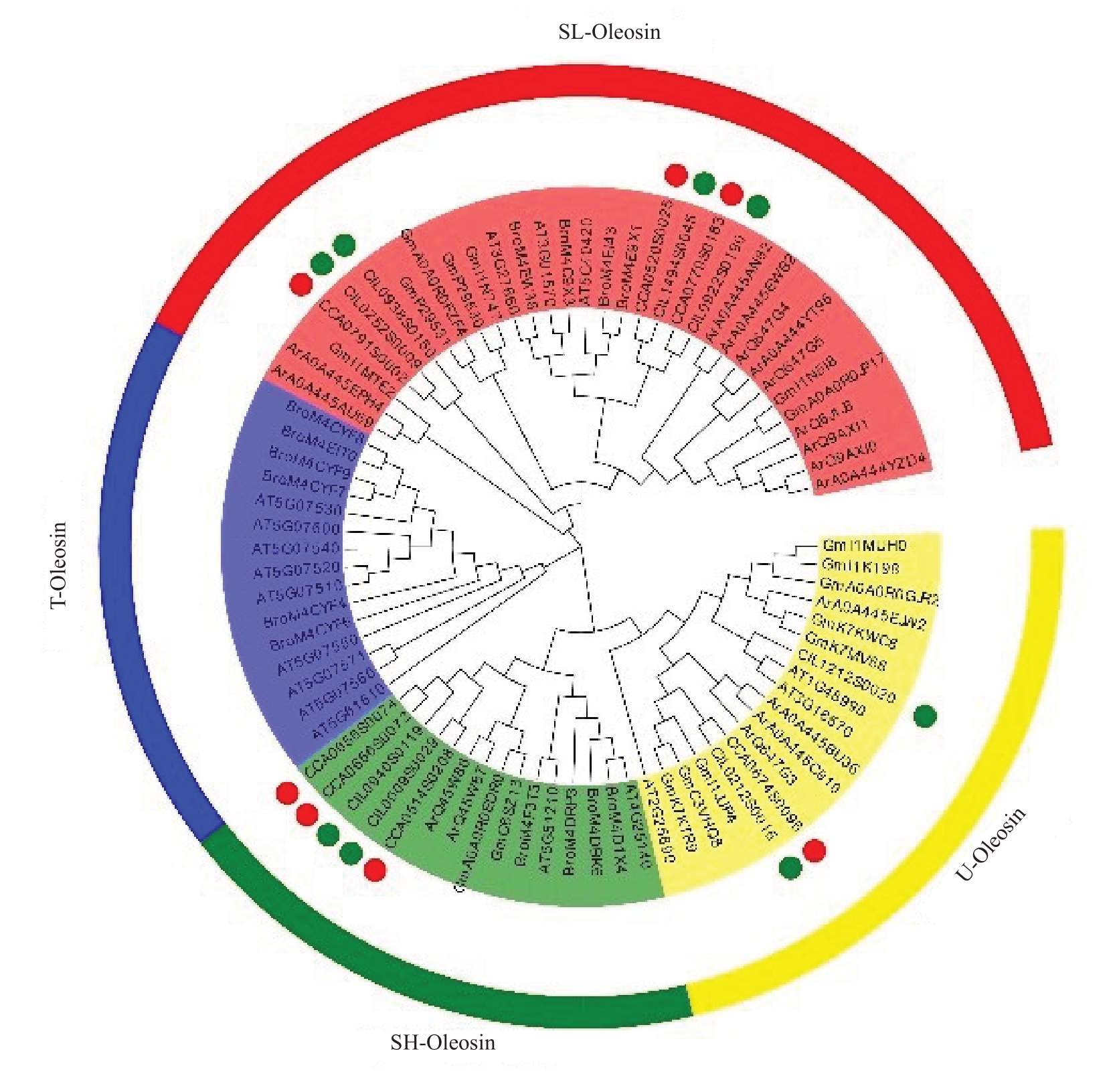
图2 山核桃、薄壳山核桃、拟南芥、大豆、花生和油菜oleosin 基因系统进化分析
Fig.2 Phylogenetic analysis of oleosin genes family in Carya cathayensis,C.illinoinensis,Arabidopsis thaliana,Glycine max,Arachis hypogaea and Brassica napus
2.4 山核桃oleosin 基因在胚发育不同阶段的表达模式分析
利用山核桃胚发育不同阶段转录组数据库,分析oleosin 基因在胚发育不同阶段(S1、S2、S3、S4、S5)的表达模式(图3)。从oleosin 基因表达水平分析,CCA0674S0096、CCA0779S0183、CCA0514S0208与CCA0520S0025 表达趋势一致可聚为一类;CCA0791S0002、CCA0656S0072 与CCA0656S0074表达趋势一致可聚为一类。从山核桃胚发育阶段分析,oleosin基因表达模式在胚发育的S1、S2和S3阶段可聚为一类,S4和S5阶段可聚为一类。
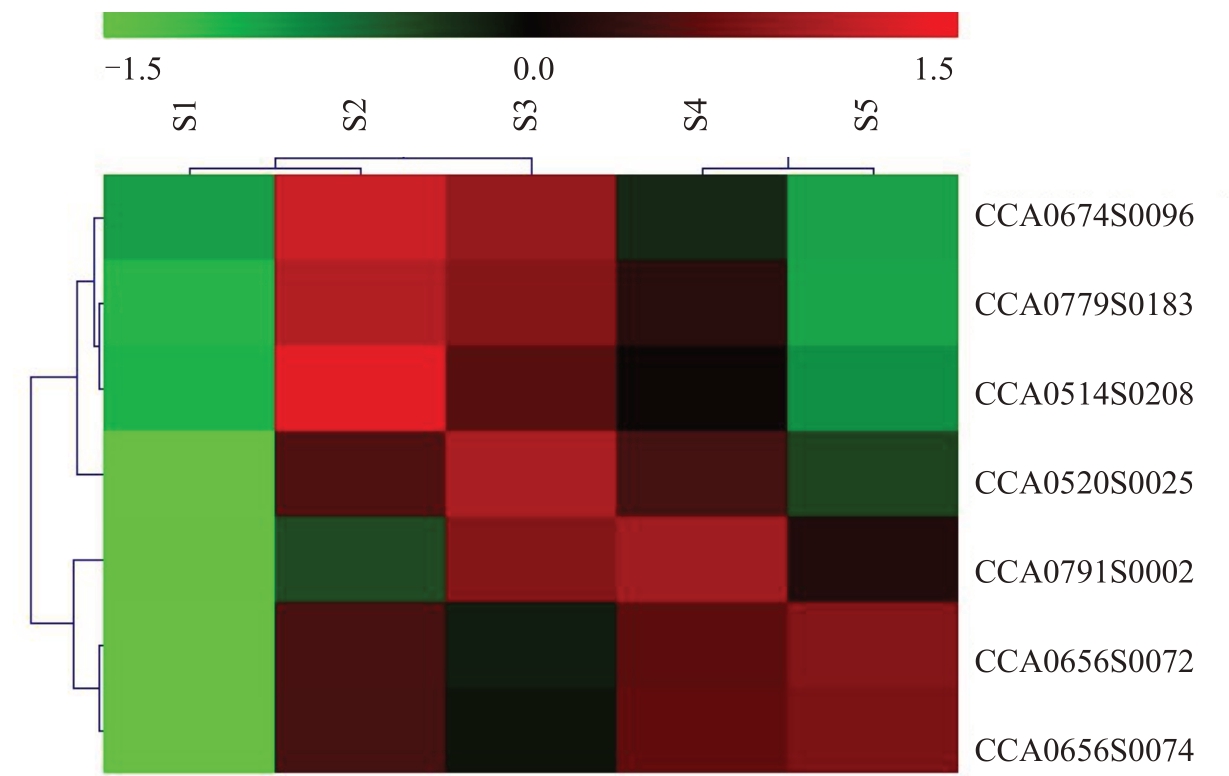
图3 山核桃oleosin 基因在胚不同发育阶段的表达模式
Fig.3 Expression pattern of oleosin genes family in the embryo during different developmental stages of Carya cathayensis
相对S1时期,山核桃oleosin基因在胚发育不同阶段均呈现不同程度的上调表达;在CCA0656S0072 与CCA0656S0074 中oleosin 基因在S5 阶段显著上调表达;S2 到S3 阶段,CCA0520S0025 和CCA0791S0002 中oleosin 基因表达量显著增加,因此推测oleosin基因在胚发育不同阶段具有功能多样性。
2.5 山核桃oleosin基因在ABA处理与低温胁迫下的表达模式分析
山核桃oleosin 基因启动子区域含有1 个至多个不等的ABA 响应元件和低温胁迫响应相关元件。挑选山核桃6 个oleosin 基因,利用qRT-PCR分析ABA 和低温胁迫条件下的基因表达模式。结果显示(图4-A),在ABA 处理条件下,山核桃oleosin 基因均呈现出上升表达趋势。CCA0791S0002、CCA050S0025、CCA0514S0208 和CCA0674S0096 基因表达模式相近,呈现上升表达趋势,CCA0791S0002和CCA050S0025均在ABA处理2 h 达到表达峰值;CCA0656S0074 在ABA 处理后呈现持续上升表达趋势,表达量在处理12 h后达到最高值;CCA0779S0183 在ABA 处理4 h 后出现下降表达趋势,表达量在2 h达到表达峰值。

图4 山核桃oleosin 基因在ABA 和低温胁迫处理下的表达模式
Fig.4 Expression patterns of Carya cathayensis oleosin genes under ABA and low-temperature treatments
在低温胁迫处理条件下,山核桃oleosin基因表达呈现出3种不同的表达模式(图4-B),分别呈现上升表达趋势(CCA0791S0002和CCA0520S0025)、先下降后上升再下降的双峰表达趋势(CCA0514S0208)以及下降表达趋势(CCA0656S0074、CCA0674S0096 和CCA0779S0183)。
2.6 Oleosin蛋白亚细胞定位分析
为进一步研究oleosin 蛋白的亚细胞定位,分别构建融合GFP 标签的oleosin 蛋白CCA0520S0025和CCA0791S0002 瞬时转化表达载体(pBI221-UBQ10- CCA0520S0025- GFP 和pBI221- UBQ10-CCA0791S0002-GFP)(图5)。利用拟南芥悬浮细胞瞬时转化体系,以拟南芥AtOLEO2-RFP 为阳性对照,观察oleosin蛋白的亚细胞定位情况。结果发现CCA0520S0025-GFP 和CCA0791S0002-GFP 均与AtOLEO2-RFP 共定位,且都定位于油体,表明CCA0520S0025和CCA0791S0002均是油体蛋白。

图5 CCA0520S0025 和CCA0791S0002 亚细胞定位
Fig.5 Subcellular localization of CCA0520S0025 and CCA0791S0002 in the Carya cathayensis
3 讨 论
本研究从山核桃和薄壳山核桃基因组中分别鉴定到7 个和8 个含有完整开放阅读框的oleosin 基因。目前已从拟南芥和油菜基因组中分别鉴定到了17 个和48 个oleosin 基因[29]。在四倍体棉种陆地棉(Gossypium hirsutum)和巴巴登棉(Gossypium barbadense)中分别鉴定出25个和24个oleosin基因,而在二倍体棉种树木棉(Gossypium arboreum)和雷蒙地棉(Gossypium raimondii)中分别鉴定出12 个和13 个oleosin 基因[30]。相对于油菜oleosin 蛋白序列末端的高度可变性,导致其理化性质存在一定差异性,属于两亲性蛋白[29]。通过理化性质分析发现,山核桃和薄壳山核桃oleosin 蛋白属于碱性蛋白。源自于不同物种的油体蛋白可划分为6 大类谱系:POleosin,U-Oleosin,SL-Oleosin,SH-Oleosin,T-Oleosin 和M-Oleosin[4, 6]。其中M-Oleosin 存在于花椰菜[31-32],T-Oleosin 仅存在于十字花科物种,P-Oleosin主要分布于绿藻,是U-Oleosin 的来源,P-Oleosin 可进一步进化为SL-Oleosin 和SH-Oleosin[4]。通过系统进化分析,CIL12125S0020、AT3G18570 和AT1G48990 聚在同一分支,亲缘关系较近,推测具有相似的生物学功能。同时发现,CCA0656S0072与CCA0656S0074、CCA0520S0025 与CIL149480045、CCA0779S0183 与CIL092280190 的蛋白序列、基因结构及进化上较为相近,分别可归类为山核桃与薄壳山核桃的同源蛋白。
油体蛋白oleosin 是存在于油体表面的一类重要蛋白,在油体形成、油体形态稳定性、种子萌发及响应逆境胁迫等方面具有重要调控作用[33]。油体蛋白oleosin调控油体形成。研究发现,墨西哥鳄梨中油体蛋白PaOle1 是中果皮特有的,与果实成熟相关,在青果期高表达,并且与果实含油量相关[34]。在山核桃胚发育过程中,胚含油量在S1 到S5 阶段不断增加,其中S2到S3阶段为快速积累阶段[26]。分析发现山核桃oleosin 基因在胚发育不同阶段也呈现上调表达趋势,其中CCA0520S0025 和CCA07-91S0002在S2到S3阶段的表达量显著上调,推测在胚发育含油量快速积累阶段发挥重要作用。油体蛋白oleosin 调控油体大小与脂肪酸含量。在烟草中过表达油用芍药PoOle17.5 基因,烟草种子百粒质量和脂肪酸含量显著增加[18];在拟南芥中超表达油菜oleosin 基因(BnaOLE1- BnaOLE4),除BnaOLE1外,转基因拟南芥种子的油体均显著大于野生型;同时发现转基因拟南芥株系脂肪酸组成改变,主要表现为亚油酸含量增加而花生酸含量降低[29]。
油体蛋白oleosin响应逆境胁迫。研究发现,甘蓝型油菜经30 μmol·L-1 JA处理后,oleosin基因表达量增高[35];在干旱状态下,野生型拟南芥经ABA 和山梨醇处理后,oleosin 基因表达量增高[36];高粱(Sorghum bicolor)[37]oleosin 基因通过调节膜渗透性,可参与ABA响应;0.5 μmol·L-1 ABA处理拟南芥AtSAG 突变体时,OLE2 基因的表达量显著上升[38]。油体蛋白能维持低温胁迫下油体的完整性,进而提高种子的抗寒力,保证种子经过低温胁迫后仍然能正常萌发[17]。研究发现,在油体蛋白缺失的突变体中,油体蛋白水平影响了种子的发芽和耐冻性;拟南芥单突变体种子ole1 和ole2 在低温处理后,萌发率降低;双突变体种子ole1ole2和ole1ole3经过4 ℃低温处理后加快了油体的融合,使种子出现致死现象[16]。在拟南芥oleosin1 突变体中超表达饲用高粱oleosin基因(SbOle)并进行冷冻处理,结果发现超表达SbOle基因植株提高了拟南芥的发芽率[39]。
综上所述,油体蛋白oleosin 在油体形成、种子萌发以及逆境胁迫中发挥重要作用,对于山核桃oleosin 蛋白功能分析是下一步重要工作内容,特别是CCA0520S0025 和CCA0791S0002 响应山核桃油脂积累,响应ABA 以及低温诱导,可能参与调控山核桃种子形成含油量积累、种子萌发等生物学过程。
4 结 论
对山核桃和薄壳山核桃基因组中oleosin 基因家族成员进行了基因结构分析;系统进化分析,发现oleosin 基因家族成员主要分布在U-Oleosin、SLOleosin 和SH-Oleosin 3 个亚家族;顺式作用元件分析,结果显示oleosin基因家族成员可能参与逆境胁迫和激素调控。荧光定量PCR结果表明山核桃oleosin 基因CCA0520S0025 和CCA0791S0002 响应ABA 和低温胁迫。基于转录组数据分析表明oleosin 基因家族成员在山核桃胚发育过程中发挥重要作用。亚细胞定位分析进一步表明CCA0520S0025和CCA0791S0002 蛋白均定位于油体,可能在油脂合成过程中发挥重要作用。
[1]陈镇,李秀丽,陈法志.植物油体合成及功能研究进展[J].世界科技研究与发展,2021,43(2):182-191.CHEN Zhen,LI Xiuli,CHEN Fazhi. Research progress on biological synthesis and biological function in plant oil body[J].World Sic-Tech R&D,2021,43(2):182-191.
[2]SHAO Q,LIU X F,SU T,MA C L,WANG P P.New insights into the role of seed oil body proteins in metabolism and plant development[J].Front in Plant Science,2019,10:1568.
[3]CHAPMAN K D,DYER J M,MULLEN R T. Biogenesis and functions of lipid droplets in plants:thematic review series:Lipid droplet synthesis and metabolism:from yeast to man[J]. Journal of Lipid Research,2012,53(2):215-226.
[4]HUANG A.Plant lipid droplets and their associated proteins:Potential for rapid advances[J]. Plant Physiology,2018,176(3):1894-1918.
[5]CAPUANO F,BEAUDOIN F,NAPIER J A,SHEWRY P R.Properties and exploitation of oleosins[J]. Biotechnology Advances,2007,25(2):203-206.
[6]HUANG M D,HUANG A H. Bioinformatics reveal five lineages of oleosins and the mechanism of lineage evolution related to structure/function from green algae to seed plants[J].Plant Physiology,2015,169(1):453-470.
[7]DERUYFFELAERE C,BOUCHEZ I,MORIN H,GUILLOT A,MIQUEL M,FROISSARD M,CHARDOT T,D′ANDREA S.Ubiquitin-mediated proteasomal degradation of oleosins is involved in oil body mobilization during post-germinative seedling growth in Arabidopsis[J]. Plant Cell Physiology,2015,56(7):1374-1387.
[8]ROUX E,BAUMBERGER S,AXELOS M A,CHARDOT T.Oleosins of Arabidopsis thaliana:expression in Escherichia coli,purification,and functional properties[J]. Journal of Agricultural and Food Chemistry,2004,52(16):5245-5249.
[9]KEDDIE J S,HYUBNER G,SLOCOMBEl S P,JARVIS R P,CUMMINS I,EDWARDS E W,SHAW C H,MURPHY D J.Cloning and characterisation of an oleosin gene from Brassica napus[J].Plant Molecular Biology,1992,19(3):443-453.
[10]XU M Y,LIU D H,LI G Q. Cloning of soybean 24 kDa oleosin gene and its transient expression as a carrier for forgeign protein[J].Agricultural Sciences in China,2004,3(5):321-329.
[11]TAI S S,CHEN M C,PENG C C,TAEN J T. Gene family of oleosin isoforms and their structural stabilization in sesame seed oil bodies[J]. Bioscience,Biotechnology,and Biochemistry,2002,66(10):2146-2153.
[12]眭顺照,祝钦泷,郑丽,郭余龙,李名扬,裴炎.植物蛋白oleosin 及其在植物基因工程中的应用[J]. 中国生物工程杂志,2003(6):17-21.SUI Shunzhao,ZHU Qinlong,ZHENG Li,GUO Yulong,LI Mingyang,PEI Yan. Plant oleosin and their application on plant gene engineerings[J].China Biotechnology,2003(6):17-21.
[13]VAN ROOIJEN G J H,MOTONEY M M. Plant seed oil-bodies as carriers for foreign proteins[J]. Bio-Technolgy,1995,13(1):72-77.
[14]KIM H U,HSIEH K,RATNAYAKE C,HUANG A H.A novel group of oleosins is present inside the pollen of Arabidopsis[J].The Journal of Biological Chemistry,2002,277(25):22677-22684.
[15]SILOTO R,FINDLAY K,LOPEZ-VILLALOBOS A,YEUNG E,CORY N,MOLONEY M. The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis[J]. The Plant Cell,2006,18(8):1961-1974.
[16]LIU W X,LIU H L,QU L Q. Embryo-specific expression of soybean oleosin altered oil body morphogenesis and increased lipid content in transgenic rice seeds[J].Theoretical and Applied Genetics,2013,126(9):2289-2297.
[17]SHIMADA T L,SHIMADA T,TAKAHASHI H,FUKAO Y,HARA-NISHIMURA I.A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana[J].The Plant Journal,2008,55(5):798-809.
[18]ZHAO D Q,LI T T,LI Z Y,SUN J,TAO J. Characteristics of Paeonia ostii seed oil body and OLE17.5 determining oil body morphology[J].Food Chemistry,2020,319:126548.
[19]GU J W,HOU D L,LI Y H,CHAO H B,ZHANG K,WANG H,XIANG J,RABOANATAHIRY N,WANG B S,LI M T.Integration of proteomic and genomic approaches to dissect seed germination vigor in Brassica napus seeds differing in oil content[J].BMC Plant Biology,2019,19(1):21.
[20]邓杨勇,高龙军.山核桃产业发展现状及对策[J].现代农业科技,2020(8):91-92.DENG Yangyong,GAO Longjun. Development situation and countermeasures of hickory industry[J]. Modern Agricultural Science and Technology,2020(8):91-92.
[21]HUANG Y J,XIAN L H,ZHANG Z R,ZHANG R,WANG Z J,HUANG C Y,HUANG R,LUAN Y M,FAN T Q,WANG J H,SHEN C,ZHANG S M,WANG X W,RANDALL J,ZHENG B S,WU J S,ZHANG Q X,XIA G H,XU C M,CHEN M,ZHANG L S,JIANG W K,GAO L Z,CHEN Z D,LESLIE C A,GRAUKE L J,HUANG J Q. The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition[J]. Gigascience,2019,8(5):giz036.
[22]吕秋菊,沈月琴,高宇列,黄坚钦.山核桃产业的发展过程、动因及展望[J].浙江农林大学学报,2012,29(1):97-103.LÜ Qiuju,SHEN Yueqin,GAO Yulie,HUANG Jianqin. Development process,agents and prospect of hickory industry[J].Journal of Zhejiang A&F University,2012,29(1):97-103.
[23]黄春颖,黄有军,吴建峰,黄仁,栾雨濛,张深梅,王正加,张启香,黄坚钦.SAD 和FAD 家族基因调控山核桃不饱和脂肪酸组分配比[J].园艺学报,2018,45(2):250-260.HUANG Chunying,HUANG Yuojun,WU Jianfeng,HUANG Ren,LUAN Yumeng,ZHANG Shenmei,WANG Zhengjia,ZHANG Qixiang,HUANG Jianqin. SAD and FAD genes regulate the ratio of unsaturated fatty acid components in Carya cathayensis[J].Acta Horticulturae Sinica,2018,45(2):250-260.
[24]郭传友,黄坚钦,方炎明.山核桃研究综述及展望[J].经济林研究,2004(1):61-63.GUO Chuanyou,HUANG Jianqin,FANG Yanming.Review and perspective of research on Carya cathayensis[J].Non-wood Forest Research,2004(1):61-63.
[25]徐胤,胡秋涛,侯丹,鲁海雯,林新春.毛竹NCED 基因家族的全基因组鉴定及表达分析[J]. 农业生物技术学报,2021,29(6):1061-1072.XU Ying,HU Qiutao,HOU Dan,LU Haiwen,LIN Xinchun.Genome- wide identification and expression analysis of NCED gene family in Phyllostachys edulis[J]. Journal of Agricultural Biotechnology,2021,29(6):1061-1072.
[26]HUANG C Y,LI Y,WANG K T,XI J W,XU Y F,SI X L,PEI D,LYU S H,XIA G H,WANG J H,LI P P,YE H Y,XING Y L,WANG Y G,HUANG J Q.Analysis of lipidomics profile of Carya cathayensis nuts and lipid dynamic changes during embryonic development[J].Food Chemistry,2022,370:130975.
[27]CONNOLLY M A,CLAUSEN P A,LAZAR J G.Preparation of RNA from plant tissue using trizol[J].Cold Spring Harbor Protocols,2006(1):pdb.prot4105.
[28]ZENG Y L,JI C Y,LIN Y S,JIANG L W.Transient expression of fluorescent fusion proteins in Arabidopsis protoplasts[J].Methods in Molecular Biology,2021,2200:157-165.
[29]CHEN K,YIN Y T,LIU S,GUO Z Y,ZHANG K,LIANG Y,ZHANG L N,ZHAO W G,CHAO H B,LI M T. Genome-wide identification and functional analysis of oleosin genes in Brassica napus L.[J].BMC Plant Biology,2019,19(1):294.
[30]YUAN Y C,CAO X Z,ZHANG H J,LIU C Y,ZHANG Y X,SONG X L,GAI S P. Genome-wide identification and analysis of oleosin gene family in four cotton species and its involvement in oil accumulation and germination[J]. BMC Plant Biology,2021,21(1):569.
[31]HUANG C Y,HUANG A H C. Unique motifs and length of hairpin in oleosin target the cytosolic side of endoplasmic reticulum and budding lipid droplet[J]. Plant Physiology,2017,174(4):2248-2260.
[32]FANG Y,ZHU R L,MISHLER B. Evolution of oleosin in land plants[J].PloS One,2014,9(8):e103806.
[33]HUANG A H C.Oil bodies and oleosins in seeds[J].Annual Review of Plant Physiology,1992,43:177-200.
[34]SANCHEZ-ALBARRCHEZ F,SUAREZ-RODRIGUEZ L M,RUIZ-HERRERA L F,LOPEZ-MEZA J E,LOPEZ-GOMEZ R.Two oleosins expressed in the mesocarp of native Mexican Avocado,key genes in the oil content[J]. Plant Foods Human Nutrition,2021,76(1):20-25.
[35]HAYS D B,WILEN R W,SHENG C,MOLONEY M M,PHARIS R P. Embryo-specific gene expression in microspore-derived embryos of Brassica napus.An interaction between abscisic acid and jasmonic acid1,2[J].Plant Physiology,1999,119(3):1065-1072.
[36]TAKAHASHI S,KATAGIRI T,YAMAGUCHI-SHINOZAKI K,SHINOZAKI K.An Arabidopsis gene encoding a Ca2+-binding protein is induced by abscisic acid during dehydration[J].Plant Cell Physiology,2000,41(7):898-903.
[37]BUCHANAN C D,LIM S,SALZMAN R A,KAGIAMPAKIS I,MORISHIGE D T,WEERS B D,KLEIN R R,PRATT L H,CORDONNIER-PRATT M M,KLEIN P E,MULLET J E. Sorghum bicolor’s transcriptome response to dehydration,high salinity and ABA[J]. Plant Molecular Biology,2005,58(5):699-720.
[38]CHEN C T,WU C A,MIAO J M,LEI Y X,ZHAO D X,SUN D,YANG G D,HUANG J G,ZHENG C C. Arabidopsis SAG protein containing the MDN1 domain participates in seed germination and seedling development by negatively regulating ABI3 and ABI5[J].Journal of Experimental Botany,2014,65(1):35-45.
[39]OJHA R,KAUR S,SINHA K,CHAWLA K,KAUR S,JADHAV H,KAUR M,BHUNIA R K. Characterization of oleosin genes from forage sorghum in Arabidopsis and yeast reveals their role in storage lipid stability[J].Planta,2021,254(5):97.