嫁接于园艺作物,尤其在园艺作物生产上应用广泛。在嫁接过程中选择根系相对发达的砧木,能够显著促进植株营养吸收、增强植株的抗性[1-2],而品质优良的接穗能改善植株活力、提高果实产量和品质[3-4]。因此,开展砧穗互作研究,对选择性状优良的砧木或接穗品种、获得优势互补的砧穗组合及促进嫁接技术在果树生产中的高效运用具有重要意义。
砧穗互作一直是园艺学领域研究的热点。在砧穗互作生理层面,前人主要从植物激素对嫁接体愈合的调控、砧木对植物抗性的影响以及砧木改善植株对水分和矿质离子的吸收等方面进行了探究。例如:植物内源激素如生长素(IAAs)、赤霉素(GAs)、茉莉酸(JAs)等作为信号物质参与嫁接过程愈伤组织的形成和嫁接体的生长发育[5-6];砧木能够通过调节抗氧化酶的活性,来缓解盐逆境对植株的损伤[7];在缺水条件下,根系发达的砧木能够显著提高植株对水分的吸收效率[8]。接穗对砧木的影响报道并不多,已有的研究主要集中在对根系各项生理指标的测定方面,如一般情况下柑橘的同一种砧木嫁接接穗后其根系活力、POD和SOD活性低于未嫁接接穗的砧木或与其差异不显著,此结果与葡萄上的研究相似[9]。在砧穗互作分子层面,前人主要从影响嫁接愈合相关基因的鉴定、砧穗间mRNA、siRNA等低分子的传递及其生物学功能等方面开展了研究。研究发现,烟草中的NbGH9B3基因可通过促进细胞壁形成影响嫁接组织创面的愈合,从而显著提高嫁接的成功率[10];植物在缺磷的条件下能将地上部合成的miR399 转运至根系调节根系磷酸盐的吸收和维持植株体内磷酸盐平衡[11];值得关注的是,越来越多的研究表明mRNA 等低分子可在砧穗间传递并调控相关农艺性状。
研究已证实拟南芥(Arabidopsis thaliana)、甘蓝(Brassica oleracea)、马铃薯(Solanum tuberosum)、杜梨(Pyrus betulaefolia)、葡萄(Vitis vinifera)等植物的韧皮部汁液中存在可移动的mRNA,它们所编码的蛋白质功能广泛[12-18]。但并非所有韧皮部中的mRNA 均能在嫁接体中长距离运输,目前已知的可在砧穗间相互移动的mRNA仍然较少。前人发现,拟南芥IAA18 和IAA28 基因在成熟叶片的维管束细胞转录后,其mRNA向下运输至根部从而抑制侧根的发生[19];拟南芥TCTP1/HSC70.1 等基因的mRNA经m5C 甲基化修饰后可移动到根部促进根系的发育[20];苹果GAI(GAINTENSIVE)基因的mRNA 能由根系长距离运输至叶片来调控植株矮化[21];苹果调节侧根形成的MpSLR/IAAI4基因的mRNA可由砧木转移至接穗进而影响其生长发育[22];梨PbWoxT1 基因的mRNA 能在砧木和接穗间双向移动并调控植株的形态[23]。
柑橘是我国栽培面积最大的果树,但由于其童期较长的特点,故生产中需运用嫁接技术缩短生长和育种周期。迄今柑橘砧穗互作的研究主要停留在生理层面,诸如砧穗间mRNA传递的分子层面的研究鲜有报道。目前鉴定mRNA 运输性采用的主要方法有:根据候选基因的SNPs 位点设计dCAPS引物,然后将SNP 位点转化为dCAPS 标记进行酶切验证。但由于SNP-dCAPS 敏感度较低,故无法验证SNP 位点是否杂交存在一定缺陷;研究表明,在拟南芥中大部分可传递mRNA 具有修饰碱基5-甲基胞嘧啶(m5C)的特点,因此可以根据该特点初步鉴定砧穗mRNA 的运输性;环剥树体侧枝后对韧皮部汁液进行检测,可以发现韧皮部中一些基因的信号消失了,从而判断其mRNA 可能由其他部位转移而来。由于红夏橙与枳壳的韧皮部汁液较难提取,故笔者在本研究中通过分析比对柑橘红夏橙接穗(Citrus sinensis)和枳砧木(Poncirus trifoli-ate)的特征性单核苷酸多态性(single nucleotide polymorphisms,SNPs),筛选出了可在柑橘砧穗间传递的mRNA 分子,进一步采用SNP-PCR 对这些mRNA 进行了验证。笔者在本研究中鉴定了柑橘砧穗间传递的mRNA,为深入探究这些mRNA 的功能、推进砧穗互作分子机制研究提供了候选基因。
1 材料和方法
1.1 材料
以红夏橙(Citrus sinensis Osbeck)和枳(Poncirus trifoliata L.Raf)的实生苗为主要材料,选取生长2年且生长量一致的枳作砧木,红夏橙芽和枳芽作接穗,分别将红夏橙和枳接穗芽接到枳砧木上(图1)。
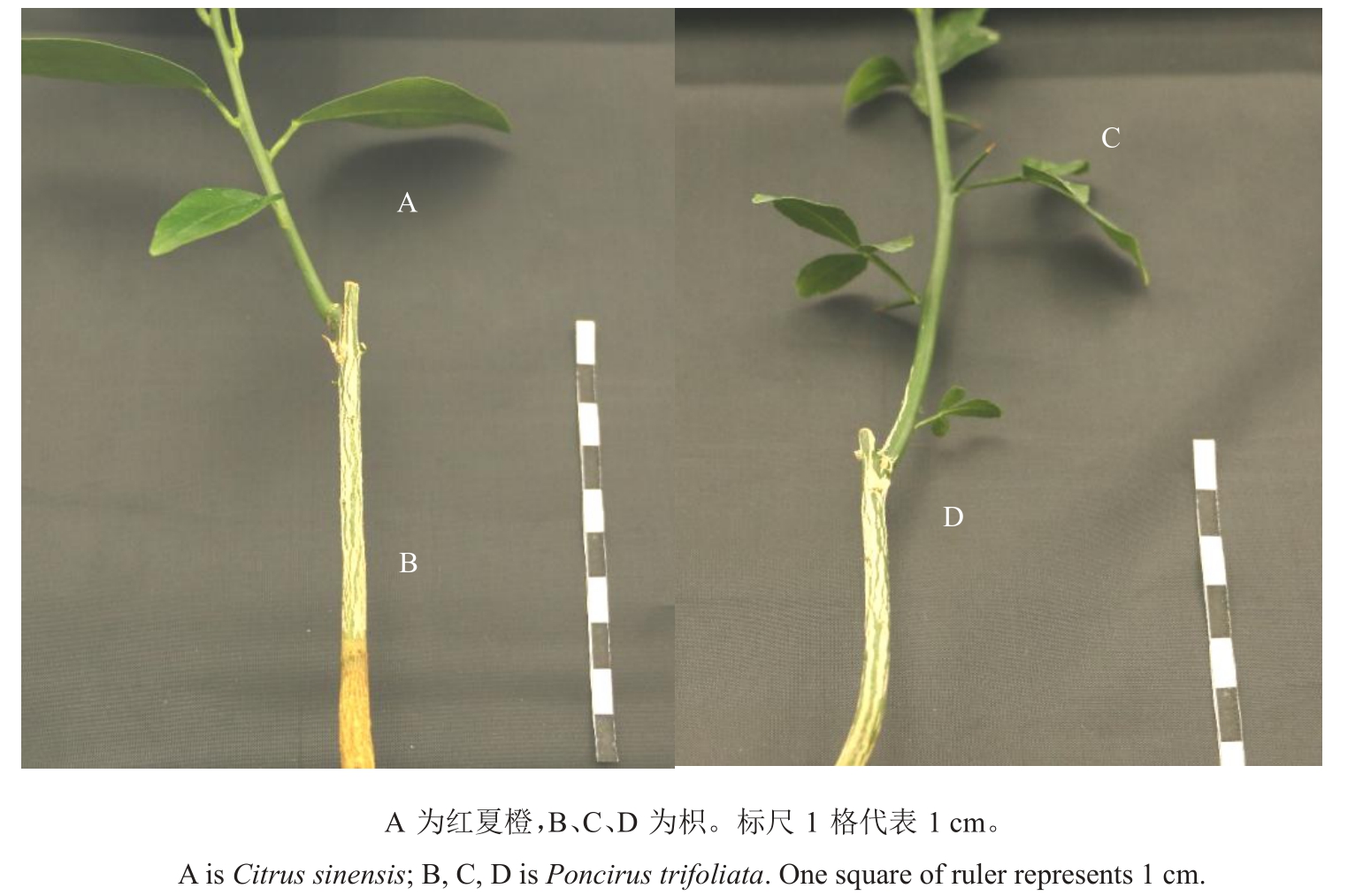
图1 红夏橙/枳嫁接组合(左)、枳/枳的嫁接组合(右)
Fig.1 The grafted combination of Citrus sinensis/Poncirus trifoliata(on the left)and the grafted combination of Poncirus trifoliata/Poncirus trifoliata(on the right)
1.2 取样及预处理
2016年5月在华中农业大学果树标本园内采集2种嫁接组合的长势一致的苗木清洗干净后置于保温袋中带回实验室,然后对每个样品的接穗、交界面和砧木3个部位进行采集,每个部位采3个样品并保证每个样品长度为1 cm,液氮速冻后放于-80 ℃保存。
1.3 柑橘组织RNA的提取
部位(接穗、砧木、交界面)采集样本各1 cm,将样品立即放入液氮彻底研磨,然后分装到1.5 mL离心管中,保证每管包含0.1 g 样品;每管加入1 mL Trizol试剂迅速混匀后室温静置10 min;加入200 μL氯仿,盖严离心管盖剧烈振荡15 s 后室温静置10 min,4 ℃,12 000 r·min-1条件下离心15 min,将离心后上清液转移至新的1.5 mL RNase free的离心管中,加入0.5 mL 异丙醇后室温放置10 min;4 ℃,12 000 r·min-1条件下离心10 min,弃上清液后加入1 mL 75%乙醇溶液,短暂涡旋;4 ℃,7500 r·min-1的条件下离心5 min;弃上清液后室温静置5~10 min,使RNA 沉淀干燥,加入30 μL 无RNase 水溶解后60 ℃水浴10~15 min;取1 μL RNA进行琼脂凝胶电泳检测RNA 完整性,同时检测RNA 浓度,将剩余RNA保存在-80 ℃冰箱内。
1.4 无参转录组的组装
对2 个不同组合的柑橘嫁接组合的3 个部位的样品进行转录组测序,得到转录组数据后对所有测序数据用拼接软件Trinity 进行组装,具体参数见附录(Trinity组装的命令)。
1.5 红夏橙与枳的特征性SNP 分析及可传递mRNA判断
使用GATK 处理组装好的枳转录组,同时结合NCBI 中红夏橙的基因组数据筛选鉴定二者的特征性SNP,具体参数见附录(鉴定SNP 的命令)。对比红夏橙和枳各自的特征性SNP 位点,可区分红夏橙和枳的转录子,即在红夏橙该位点为纯合子且不同于枳的类型。若在红夏橙接穗样品中检测到杂合的特征性SNP 位点且在3 个生物学重复的样本中均出现此情况,即证明该转录子能够由砧木转移至接穗;同理在枳砧木样品中检测到杂合特征性SNP 位点,即证明该转录子能够由接穗转移至砧木。
1.6 基于特征性SNP位点的RT-PCR验证
选取包含特征性SNP 位点的候选基因片段,使用SnapGene软件设计引物,候选可传递mRNA引物序列(表1)。以柑橘异源嫁接组合的cDNA 为模板进行扩增,纯化后测序,每一候选基因选取3个不同单株样本进行多次验证。
表1 SNP-RT-PCR 引物
Table 1 Primer sequences of SNP-RT-PCR

基因Gene Cs2g16070.1条带大小Size of fragment/bp 311 Pt2g028010.4344 Pt3g020940.1引物序列(5’-3’)Primer sequence(5’-3’)F:GCAGATGCAGTTCTACTCAAGTG R:AGAGACCTCAGACCCAAAGTAGA F:AGTTGGCTGCTAGGATATCTAGG R:CAAAATCGTCAGATACTGCGACC F:GTGGAGGTTCTGTCACATGAAG R:CTTGCTGGTGCTAAAGTCATCA退火温度Tm/℃59.6 59.9 58.9 60.0 58.9 58.9 360
2 结果与分析
2.1 红夏橙与枳基因组间特征性SNP位点的筛选
对枳/枳嫁接组合的转录组数据进行组装后,共得到216 656 个枳的基因和253 978 个枳的转录本(表2),且其中有5381 个基因在4 个数据库中都有注释信息(表3)。得到可信的枳参考序列后,使用枳/枳嫁接组合中的枳的转录组数据、红夏橙/枳嫁接组合中红夏橙接穗转录组数据以及在NCBI上已报道的红夏橙转录组数据和红夏橙基因组数据,通过比对分析来确定红夏橙和枳间的特征性SNP 位点,最终得到共254 580个特征性SNP位点。
表2 枳转录本组装结果
Table 2 Assembly results of trifoliate transcript
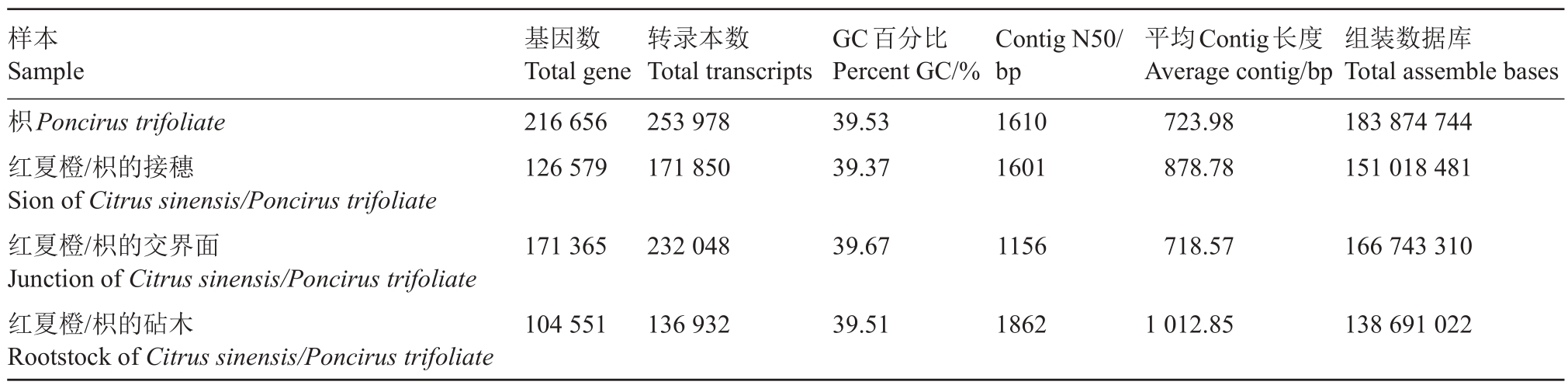
样本Sample枳Poncirus trifoliate红夏橙/枳的接穗Sion of Citrus sinensis/Poncirus trifoliate红夏橙/枳的交界面Junction of Citrus sinensis/Poncirus trifoliate红夏橙/枳的砧木Rootstock of Citrus sinensis/Poncirus trifoliate基因数Total gene 216 656 126 579转录本数Total transcripts 253 978 171 850 GC百分比Percent GC/%39.53 39.37 Contig N50/bp 1610 1601平均Contig长度Average contig/bp 723.98 878.78组装数据库Total assemble bases 183 874 744 151 018 481 171 365232 04839.671156718.57166 743 310 104 551136 93239.5118621 012.85138 691 022
表3 注释结果
Table 3 The result of annotation

总数Total 77 635 NR 数据库NR database 15 338 COG 数据库COG database 26 727 Swiss-Prot数据库Swiss-Prot database 56 565 TrEMBL数据库TrEMBL database 77 464 4个都有All have 5381
2.2 基于特征性SNP 筛选柑橘砧穗间可传递mRNA
红夏橙和枳的特征性SNP 位点作为红夏橙/枳砧穗间mRNA是否传递的判断依据,共筛选出30个可从红夏橙转移到枳候选mRNA(表4),以及24 个可从枳转移到红夏橙的候选mRNA(表5)。
表4 基于物种特征性SNP 筛选的从红夏橙转移到枳的候选mRNA
Table 4 The candidate mRNAs screened by comparative transcriptome analysis can be Transmitted from sweet orange to trifoliate orange
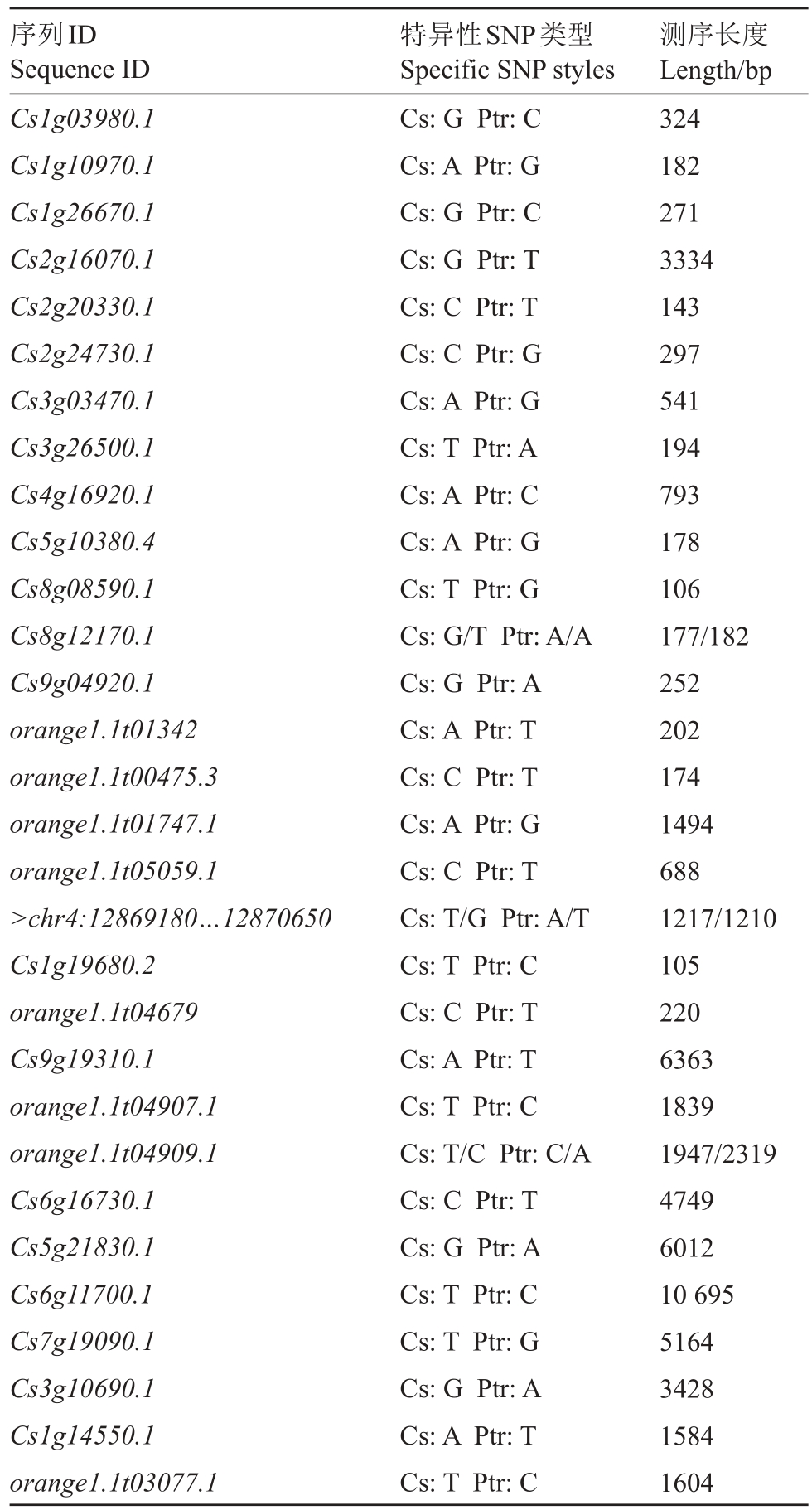
序列ID Sequence ID Cs1g03980.1 Cs1g10970.1 Cs1g26670.1 Cs2g16070.1 Cs2g20330.1 Cs2g24730.1 Cs3g03470.1 Cs3g26500.1 Cs4g16920.1 Cs5g10380.4 Cs8g08590.1 Cs8g12170.1 Cs9g04920.1 orange1.1t01342 orange1.1t00475.3 orange1.1t01747.1 orange1.1t05059.1>chr4:12869180…12870650 Cs1g19680.2 orange1.1t04679 Cs9g19310.1 orange1.1t04907.1 orange1.1t04909.1 Cs6g16730.1 Cs5g21830.1 Cs6g11700.1 Cs7g19090.1 Cs3g10690.1 Cs1g14550.1 orange1.1t03077.1特异性SNP类型Specific SNP styles Cs:G Ptr:C Cs:A Ptr:G Cs:G Ptr:C Cs:G Ptr:T Cs:C Ptr:T Cs:C Ptr:G Cs:A Ptr:G Cs:T Ptr:A Cs:A Ptr:C Cs:A Ptr:G Cs:T Ptr:G Cs:G/T Ptr:A/A Cs:G Ptr:A Cs:A Ptr:T Cs:C Ptr:T Cs:A Ptr:G Cs:C Ptr:T Cs:T/G Ptr:A/T Cs:T Ptr:C Cs:C Ptr:T Cs:A Ptr:T Cs:T Ptr:C Cs:T/C Ptr:C/A Cs:C Ptr:T Cs:G Ptr:A Cs:T Ptr:C Cs:T Ptr:G Cs:G Ptr:A Cs:A Ptr:T Cs:T Ptr:C测序长度Length/bp 324 182 271 3334 143 297 541 194 793 178 106 177/182 252 202 174 1494 688 1217/1210 105 220 6363 1839 1947/2319 4749 6012 10 695 5164 3428 1584 1604
表5 基于物种特征性SNP 分析筛选的从枳转移到红夏橙的候选mRNA
Table 5 The candidate mRNAs screened by comparative transcriptome analysis can be Transmitted from trifoliate orange to sweet orange

序列ID Sequence ID Pt7g010320.1 Pt2g028010.4 Pt2g009090.1 Pt5g022680.1 Pt3g021050.1 Pt5g020570.1 Pt5g006030.2 Pt1g014530.1 Pt3g004160.1 Pt3g005620.2 Pt3g005430.1 Pt3g007590.1 Pt3g021870.2 Pt3g020940.1 PtUn023860.3测序长度Length/bp 2711/3457/3391/3360/3292 6080 11 700 799/1016 1594 4751 6529/5947 1432 1125 3173 2898 1516 5462 8736 1271/1365/1304/1303 Pt6g018000.1 Pt6g003120.1 Pt6g003360.1 Pt4g012460.1 Pt8g003160.1 Pt9g017320.1 Cs9g04560.1 Pt1g015050.1特异性SNP类型Specific SNP styles Cs:C/T/C/C/A Ptr:A/A/T/G/C Cs:G Ptr:A Cs:A Ptr:G Cs:C/G Ptr:T/T Cs:A Ptr:C Cs:T Ptr:C Cs:G/A Ptr:A/G Cs:C Ptr:G Cs:T Ptr:G Cs:G Ptr:A Cs:C Ptr:G Cs:A Ptr:T Cs:C Ptr:T Cs:A Ptr:G Cs:G/C/A/A Ptr:C/A/T/C Cs:A Ptr:G Cs:T Ptr:G Cs:C Ptr:G Cs:T/C Ptr:C/T Cs:T Ptr:G Cs:A Ptr:G Cs:C Ptr:G Cs:C/T/T/T Ptr:T/A/C/G 7481 8265 2630 12 884/10 565 5537 9608 11 692 1205/1215/1193/1083
2.3 利用SNP-RT-PCR 验证柑橘砧穗间可传递mRNA
通过SNP-RT-PCR 的方法,对候选的可传递mRNA 进行验证。为确保结果的可信度,选择同一种嫁接组合的3 个单株样品作为模板对特征性SNP 位点进行3 次反复验证。对24 个由枳转移到红夏橙候选基因扩增产物双向测序的结果表明,有2 组杂合SNP 位点处在特征性SNP 位点的位置且SNP 类型与特征性SNP 类型一致,证明这2 个mRNA( 对 应 基 因 编 号 Pt2g028010.4 和Pt3g020940.1)可从枳转移到红夏橙。同样对30 个由红夏橙转移至枳的候选基因扩增产物双向测序的结果表明,有1 组杂合SNP 位点处在特征性SNP位点的位置且SNP类型与特征性SNP类型一致,证明该mRNA(对应基因编号Cs2g16070.1)从红夏橙转移到枳中(表6)。基于已确定的特异性SNP 位点进行PCR 扩增后,扩增产物双向测序的结果表明(图2)。
表6 测序所得的SNP 位点类型及位置
Table 6 The SNP styles and site by sequencing
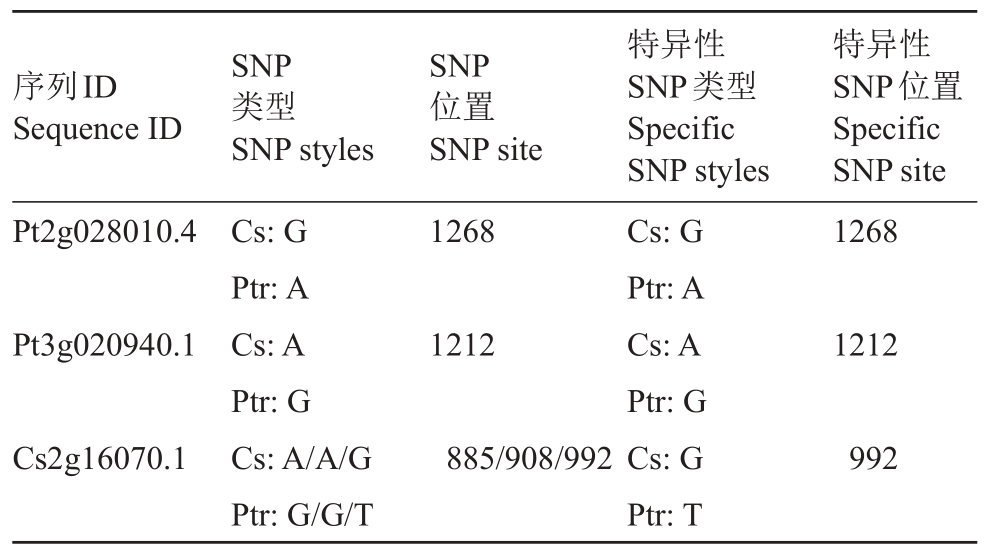
序列ID Sequence ID Pt2g028010.4 SNP位置SNP site 1268特异性SNP位置Specific SNP site 1268 Pt3g020940.112121212 Cs2g16070.1 SNP类型SNP styles Cs:G Ptr:A Cs:A Ptr:G Cs:A/A/G Ptr:G/G/T 885/908/992特异性SNP类型Specific SNP styles Cs:G Ptr:A Cs:A Ptr:G Cs:G Ptr:T 992
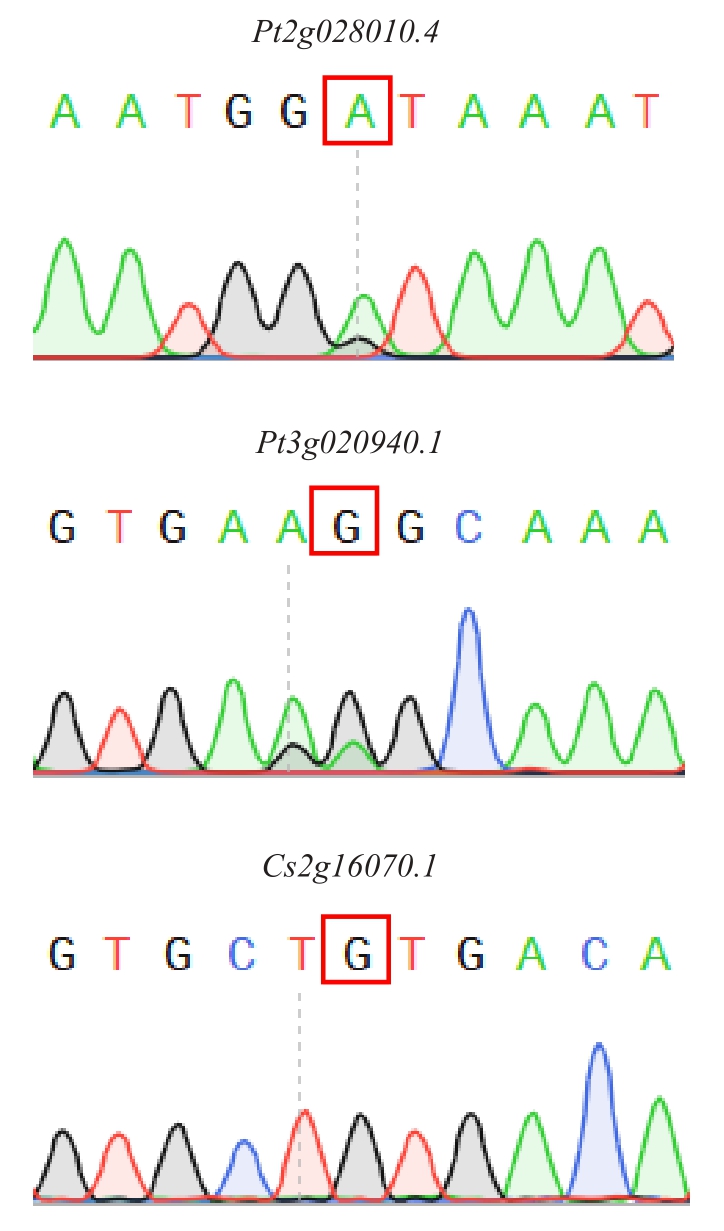
图2 Pt2g028010.4、Pt3g020940.1、Cs2g16070.1 在特异性SNP 位点处出现重叠峰
Fig.2 The overlapping peak of the specific SNP in Pt2g028010.4,Pt3g020940.1 and Cs2g16070.1
3 讨 论
为了探究mRNA 分子能否在砧木和接穗之间传递,此前的研究采用的主要方法有:(1)不少可传递mRNA 具有修饰碱基5-甲基胞嘧啶(m5C)的特点,因此根据该特点可筛选砧穗间可传递mRNA[21];(2)前人研究发现南瓜、苹果、梨等韧皮部汁液中存在可长距离运输的RNP(ribonucleoprotein complexes)核糖核蛋白复合体,由此鉴定到PP16-1、GAIP、GAI、SCL14P、STMP、PbWoxT1、ERFP、和MybP等可传递的mRNA[24]。但采用这种方法也有局限性,即只能鉴定到与该复合体特异结合的mRNA,从而忽视了以其他形式运输的mRNA;(3)前人对杜梨的侧枝环剥后发现其韧皮部中一些基因的信号消失了,从而判断其mRNA可能由其他部位转移而来[17]。此方法比较直观,但具有一定的随机性且无法准确的判断mRNA 消失的具体原因。由于采用的红夏橙和枳的韧皮部汁液都较难提取,故很难采用检测韧皮部运输物质的方法来筛选砧穗间传递的mRNA分子。所以笔者采用异源嫁接红夏橙和枳的方法,以物种特征性SNP 作为判断依据对红夏橙与枳间可传递的mRNA进行鉴定。
笔者在本研究中基于红夏橙和枳各自的特征性SNP分析及SNP-RT-PCR验证,最终鉴定了3个可靠的能在柑橘砧穗间传递的mRNA 分子。与前人相关研究相比,这个数目是比较少的,推测有以下可能原因:(1)柑橘为多年生木本作物,木质部发达、韧皮部汁液少,相比于韧皮部汁液丰富的草本植物,其体内可传递的mRNA 相对较少;(2)分析比对不同物种间的特征性SNP 是本研究筛选可传递mRNA 的前提,但研究发现并不是所有SNP 都能稳定遗传,能够从父母本稳定遗传下来的SNP 仅占基因组的30%左右[25],那些不能稳定遗传的SNP 位点可能受各种因素如季节、环境胁迫、生长周期的影响,这可能导致大多数可传递的mRNA 被遗漏;(3)相比前人在葡萄砧穗互作研究中将2个样本同时出现杂合SNP 位点定义为可传递的mRNA[19],笔者设置了更为严格的筛选条件,即只有在3 个植株个体中均出现杂合的同一SNP位点,才视其为可在砧穗间传递的mRNA,尽管这样得到的候选可传递mRNA 数量较少,但更为可靠;同时在筛选SNP位点时保证3个生物学重复的枳转录本均为相同的纯合子且与枳基因型一致,红夏橙转录本均为另一纯合子且与红夏橙基因组基因型一致,即可排除因为单株植物之间的差异。
对这3 个可传递mRNA 的功能预测发现:从红夏橙转移到枳中的mRNA 其基因Cs2g16070.1 编码1个邻甲基转移酶,在MP(methoxypyrazines)的合成过程中发挥重要作用[26];从枳转移到红夏橙中的mRNA 其基因Pt2g028010.4 编码1 个泛素蛋白连接酶UPL5(ubiquitin protein ligase),有研究表明拟南芥的UPL5 可以直接与植物细胞质中转录因子WRKY53 相互作用从而参与叶片衰老的调控[27];另一个从枳转移到红夏橙中的mRNA 其基因Pt3g020940.1 编码1 个尿苷二磷酸糖基转移酶,在植物中该酶通过将碳水化合物从活化的单糖供体转移到醇、酸、胺或硫醇上来催化低分子糖苷的产生[28]。但这3个基因在柑橘砧穗间传递的意义是什么,具体发挥什么功能,还有待进一步研究。前人研究发现,在韧皮部长距离运输的一些mRNA分子中含有PTB 基序(UUCUCUCUCUU)[29],PTB 基序能够高度亲和且特异地结合PTB蛋白形成RNP(ribonucleoprotein)复合体,进而在韧皮部中远距离传递[30-31]。但本研究得到的这3 个可在柑橘砧穗间传递的mRNA分子没有PTB基序,说明该结构可能不是mRNA传递的必需条件。
4 结 论
笔者在本研究中通过分析比对柑橘红夏橙接穗(Citrus sinensis)和枳砧木(Poncirus trifoliate)各自特征性单核苷酸多态性(single nucleotide polymorphisms,SNPs),筛选出了可在柑橘砧穗间传递的mRNA分子,进一步采用SNP-PCR对这些mRNA进行了验证得到3 个可靠的可传递mRNA,鉴定了柑橘砧穗间传递的mRNA,为深入探究这些mRNA的功能、推进砧穗互作分子机制研究提供了候选基因。
[1]GOLDSCHMIDT E E. Plant grafting:New mechanisms,evolutionary implications[J].Frontiers in Plant Science,2014,5:727.
[2]STOVER E,INCH S,RICHARDSON M L,HALL D G. Conventional citrus of some scion/rootstock combinations show field tolerance under high huanglongbing disease pressure[J].Scientia Horticulturae,2016,51(2):127-132.
[3]LAINO P,RUSSO M P,GUARDO M,REFORGIATO-RECUPERO G,VALÈ G,CATTIVELLI L,MOLITERNI V M.Rootstockscion interaction affecting citrus response to CTV infection:A proteomic view[J].Plant Physiology,2016,156(4):444-467.
[4]SOUZA J,SILVA E M,FILHO M A, MORILLON R, BONATTO D,MICHELI F,DA SILVA G A.Different adaptation strategies of two citrus scion/rootstock combinations in response to drought stress[J].PLoS One,2017,12:e0177993.
[5]TEDESCO S,PINA A,FEVEREIRO P,KRAGLER F.A phenotypic search on graft compatibility in grapevine[J]. Agronomy,2020,10(5):706.
[6]NANDA A K,MELNYK C W. The role of plant hormones during grafting[J].Journal of Plant Research,2017,131(1):49-58.
[7]SHAHID M A,BALAL R M,KHAN N,SIMÓN-GRAO S,ALFOSEA-SIMÓN M,CÁMARA-ZAPATA J M,MATTSON N S,GARCIA-SANCHEZ F. Rootstocks influence the salt tolerance of Kinnow mandarin trees by altering the antioxidant defense system,osmolyte concentration,and toxic ion accumulation[J].Scientia Horticulturae,2019,250:1-11.
[8]张志焕,韩敏,张逸,王允,刘灿玉,曹逼力,徐坤.水分胁迫对不同抗旱性砧木嫁接番茄生长发育及水气交换参数的影响[J].中国农业科学,2017,50(2):391-398.ZHANG Zhihuan,HAN Min,ZHANG Yi,WANG Yun,LIU Canyu,CAO Bili,XU Kun. Effect of water stress on development and H2O and CO2 exchange in leaves of tomato grafted with different drought resistant rootstocks[J]. Scientia Agricultura Sinica,2017,50(2):391-398.
[9]周开兵,夏仁学.柑橘砧穗生理互作研究进展与展望[J].中国农学通报,2006,22(2):239-245.ZHOU Kaibing,XIA Renxue. The development and tendencies in the study on the physiological interaction between scions and rootstocks for citrus[J]. Chinese Agricultural Science Bulletin,2006,22(2):239-245.
[10]NOTAGUCHI M,KUROTANI K I,SATO Y,TABATA R,KAWAKATSU Y,OKAYASU K,HIGASHIYAMA T. Cell-cell adhesion in plant grafting is facilitated by β-1,4-glucanases[J].Science,2020,369(6504):698-702.
[11]HUANG T K,HAN C L,LIN S I,CHEN Y J,TSAI Y C,CHEN Y R,CHEN J W,LIN W Y,CHEN P M,LIU T Y,CHEN Y S,SUN C M,CHIOU T J. Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots[J]. The Plant Cell,2013,25(10):4044-4060.
[12]DEEKEN R,ACHE P,KAJAHN I,KLINKENBERG J,BRINGMANN G,HEDRICH R. Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments[J].The Plant Journal,2008,55(5):746-759.
[13]GAUPELS F,BUHTZ A,KNAUER T,DESHMUKH S,WALLER F,VAN BEL A J,KEHR J.Adaption of aphid stylectomy for analyses of proteins and mRNA in barley phloem sap[J].Journal of Experimental Botany,2008,59(12):3297-3306.
[14]OMID A,KEILIN T,GLASS A,LESHKOWITZ D,WOLF S.Characterization of phloem-sap transcription profile in melon plants[J]. Journal of Experimental Botany,2007,58(13):3645-3656.
[15]YANG H W,YU T S. Arabidopsis floral regulators FVE and AGL24 are phloem-mobile RNAs[J]. Botanical Studies,2010,51(1):17-26.
[16]MAHAJAN A,BHOGALE S,KANG I H,HANNAPEL D J,BANERJEE A K. The mRNA of a Knotted 1-like transcription factor of potato is phloem mobile[J]. Plant Molecular Biology,2012,79(6):595-608.
[17]DUAN X W,ZHANG W N,HUANG J,ZHAO L M,MA C,HAO L,YUAN H,HARADA T,LI T Z. KNOTTED1 mRNA undergoes long-distance transport and interacts with movement protein binding protein 2C in pear (Pyrus betulaefolia)[J]. Plant Cell,Tissue and Organ Culture (PCTOC),2015,121(1):109-119.
[18]YANG Y Z,MAO L Y,JITTAYASOTHORN Y,KANG Y M,JIAO C,FEI Z J,ZHONG G Y. Messenger RNA exchange between scions and rootstocks in grafted grapevines[J].BMC Plant Biology,2015,15(1):251-264.
[19]NOTAGUCHI M,WOLF S,LUCAS W J. Phloem-mobile Aux/IAA transcripts target to the root tip and modify root architecture[J]. Journal of Integrative Plant Biology,2012,54(10):760-772.
[20]YANG L,PERRERA V,SAPLAOURA E,APELT F,BAHIN M,KRAMDI A,OLAS J,MUELLER- ROEBER B,SOKOLOWSKA E,ZHANG W N,LI R S,PITZALIS N,HEINLEIN M,ZHANG S D,GENOVESIO A,COLOT V,KRAGLER,F.m5C methylation guides systemic transport of messenger RNA over graft junctions in plants[J].Current Biology,2019,29(15):2465-2476.
[21]XU H Y,ZHANG W N,LI M F,HARADA T,HAN Z H,LI T Z. Gibberellic acid inensive mRNA transport in both directions between stock and scion in Malus[J]. Tree Genetics & Genomes,2010,6(6):1013-1019.
[22]KANEHIRA A,YAMADA K,IWAYA T,TSUWAMOTO R,KASAI A,NAKAZONO M,HARADA T. Apple phloem cell contains some mRNAs transported over long distances[J]. Tree Genetics&Genomes,2010,6(5):635-645.
[23]DUAN X W,ZHANG W N,HUANG J,HAO L,WANG S N,WANG A D,MENG D,ZHANG Q L,CHEN Q J,LI T Z. Pb-WoxT1 mRNA from pear (Pyrus betulaefolia) undergoes longdistance transport assisted by a polypyrimidine tract binding protein[J].The New Phytologist,2016,210(2):511-524.
[24]CHO S K,KANG I H,CARR T,HANNAPEL D J. Using the yeast three-hybrid system to identify proteins that interact with a phloem-mobile mRNA[J]. Frontiers in Plant Science,2012,3:189.
[25]陈发毅.利用转录组分析研究柑橘砧穗间mRNA 的移动[D].武汉:华中农业大学,2017.CHEN Fayi. Using transcriptome analysis to investigate the movement of mRNA between citrus rootstook and scion[D].Wu han:Huazhong Agricultural University,2017.
[26]GREGAN S M,JORDAN B. Methoxypyrazine accumulation and O-Methyltransferase gene expression in sauvignon blanc grapes:The role of leaf removal,light exposure,and berry development[J].Journal of Agricultural and Food Chemistry,2016,64(11):2200-2208.
[27]MIAO Y,ZENTGRAF U.A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53[J].The Plant Journal:For Cell and Molecular Biology,2010,63(2):179-188.
[28]SUN G X,PUTKARADZE N,BOHNACKER S,JONCZYK R,FIDA T,HOFFMANN T,BERNHARDT R,HäRTL K,SCHWAB W. Six uridine-diphosphate glycosyltransferases catalyze the glycosylation of bioactive C13-apocarotenols[J]. Plant Physiology,2020,184(4):1744-1761.
[29]HAM B K,BRANDOM J L,XOCONOSTLE-CÁZARES B,RINGGOLD V,LOUGH T J,LUCAS W J.A polypyrimidine tract binding protein pumpkin RBP50 forms the basis of phloem mobile ribonicleoprotein complex[J]. The Plant Cell,2009,21(1):197-215.
[30]LUCAS W J,BOUCHÉ-PILLON S,JACKSON D P,NGUYEN L,BAKER L,DING B,HAKE S. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata[J].Science,1995,270(5244):1980-1983.
[31]LI P F,HAM B K,LUCAS W J.CmRBP50 protein phosphorylation is essential for assembly of a stable phloem-mobile high-affinity ribonucleoprotein complex[J].Journal of Biological Chemistry,2011,286(26):23142-23149.