澳洲坚果(Macadamia spp.)是世界上重要的坚果作物之一,原产于澳大利亚东海岸亚热带雨林[1-3]。我国大陆地区于20 世纪70 年代末开始澳洲坚果引种试种,至2020 年末,中国澳洲坚果种植面积已占全世界的63%。种质资源的收集与保护及优良品种的选育是澳洲坚果产业持续发展的关键。目前国内澳洲坚果育种工作以国外引种和国内实生选种为主,随着育种工作的推进,新品种登记及审定数量不断增加。在生产中,同物异名或同名异物的现象已开始出现,危害育种者和生产者的权益。在澳洲坚果种质资源收集过程中,如何对其快速鉴定以避免重复收集及实现有效利用,是澳洲坚果种质资源收集及保护工作的重要内容。
利用分子标记构建DNA 指纹图谱广泛用于苹果[4-5]、柑橘[6]、杏[7]、核桃[8-9]等果树的品种保护及种质鉴定。澳洲坚果相关研究中,蔡元保等[10]建立SCoT标记反应体系并对12 份澳洲坚果种质扩增,通过3对引物组合将供试材料完全区分,构建了12份材料的指纹图谱。SSR 标记具有共显性,重复性好等优点,被国际植物新品种保护联盟(UPOV)推荐为构建DNA指纹数据库的分子标记之一[11],亦是国内植物品种鉴定DNA 分子标记法推荐的方法之一[12]。将SSR标记应用于澳洲坚果DAN指纹图谱研究,在国内外尚未见报道。
笔者在本研究中基于已公布的基因组序列设计、筛选一套高多态性SSR 引物,通过毛细管电泳建立澳洲坚果部分种质的DNA指纹图谱,构建其分子身份证,以期为澳洲坚果新品种保护及种质资源快速分子鉴定提供依据。
1 材料和方法
1.1 材料
试验材料均取自农业部景洪澳洲坚果种质资源圃。
1.2 方法
1.2.1 DNA提取 每份材料从3个单株采集新鲜幼嫩叶片混合,采用CTAB法提取叶片基因组DNA。
1.2.2 SSR 引物 在NCBI 网站上搜索并下载澳洲坚果基因组序列(http://www.ncbi.nlm.nih.gov/genome/?term=CM023674.1),利用Kmer-SSR 搜索重复碱基位点,然后编程处理序列,使用Primer3 批量设计引物。
1.2.3 PCR扩增 PCR反应体系:10×Buffer 2 μL,2.5 mmol·L-1 dNTP 0.4 μL,正反引物各0.3 μL,DNA模板2 μL(20 ng·μL-1),5 U Taq 0.2 μL,ddH2O 14.8 μL。
PCR 扩增程序:94 ℃预变性5 min;94 ℃变性30 s,65 ℃到50 ℃降落复性30 s,72 ℃延伸40 s,共35个循环;最终72 ℃延伸3 min。
1.2.4 电泳检测 PCR 产物在2%琼脂糖凝胶电泳中检测,选取其中条带明亮单一、扩增效果好的进行后续PAGE 检测;将初步挑选出来的PCR 产物加入上样缓冲液,94 ℃变性10 min,于6%变性聚丙烯酰胺凝胶电泳1.5 h(恒定功率90 W),银染检测。从中挑选具有多态性且与预期扩增产物片段大小一致的引物;筛选后的SSR引物标记6-FAM荧光染料合成荧光引物,经PCR 扩增,取PCR 产物0.3 μL、分子质量内标0.5 μL和去离子甲酰胺9.5 μL混合加入PCR板,95 ℃变性5 min,4 ℃冷却后离心,使用3730XL测序仪进行毛细管电泳。
1.2.5 数据处理 利用Genemarker V2.2.0 软件对测序仪得到的原始数据进行分析,通过比较泳道内分子质量内标与样品峰值的位置,得到片段大小。统计各样品等位基因位点,纯合位点记录为X,杂合位点记录为X/Y,杂合位点中的小片段数据在前,大片段数据在后,等位变异缺失位点记录为0。参照杨文娟[13]和郭艳春等[14]的方法,将每对引物获得的全部等位基因进行数字+英文字母形式的编码,按分子质量从小到大的顺序用数字1~9 依次表示,超出9 的部分用英文字母A~Z 依次表示,构建供试材料的指纹图谱代码。通过在线条码生成器(http://barcode.cnaidc.com/html/BCGcode128b.php)和二维码生成器(https://cli.im/),分别生成条形码和二维码形式的DNA分子身份证。
2 结果与分析
2.1 引物筛选
根据重复单元2 个碱基以上,重复次数8 次以上,在染色体上分布均匀的原则,从基因组中搜索设计的SSR引物中共挑选240对用于后续筛选。从供试样品中选取表型差异较大的4 份材料(J72、J99、HAES695、O.C)对240 对引物进行PCR 扩增,PCR产物经2%琼脂糖电泳检测后,初步筛选出155对扩增效果较好SSR 引物。上述155 对SSR 引物经6%变性聚丙烯酰胺凝胶电泳检测,评价扩增片段的多态性、稳定性,挑选了22 对多态性高、扩增稳定、带型较清晰的SSR 引物(表1)。将最终得到的22 对SSR 引物合成FAM 标记的荧光引物用于毛细管电泳检测。
表1 22 对SSR 引物信息
Table 1 Information of 22 pairs of SSR primers

引物Primer P4 P11 P15 P16 P28 P33 P35 P74 P84 P89 P95 P98 P108 P118 P127 P132 P141 P163 P202 P211 P238 P240重复基团Repeat types(AGGTCG)8(CGAAAACA)16(GCC)8(GCGT)10(CT)13(AG)16(AG)21(AAG)12(AT)12(TA)13(TCT)10(AAC)14(GA)12(AT)14(GTT)11(TA)12(TAT)11(TC)9(AT)14(AG)16(TAT)13(AAATAAA)10正向序列Forward primer GTGGTGGTCAAGGACTTGGT TGGGTCTTATGGAAACTGGAGA TCGTAAGTAGCCGCGTGATC GCAGGTCCCCATCCTTCATG GCAGCTCTTGTACAATGAGCC TGGATCTATGCACTTCCCCT TGGGTTTTCCTTGACTCACTTG AGAGGGTAAACGTGAGAAACCC AACCTGATGTCTGGCCCAAT AGTTGAATGAAGAGGCCGGG CCCCCTCAAGTGAAGAAAAGT TCTGACCCTTAGATCACTGTTGT TGTTGTTGATCTCCAATCGAAGC ACTCAAAACACAGATCAACGGC TGCCATTTAAATAGGATAAGGCTG ATTAGACACAAAACAAAAATACATGCT ACCCCATACATATCAGTAAGAGGG CTGTCACCCTCTTCCGGAAC GCTCTTCCACCTCATCTGGG AGTAGCAGCCGAGATCACTG AGCTCCCTCAATGGCCTAGA CGGACTCGATTGGGCCTATC反向序列Reverse primer TGTCAGAATGCCCATCTCTCA GGGCAAACTTAAGAGTAGATCAGC GCGGTGGTGAGATGGATCTT ACACCAATCTATCTGGCTTTGGA AGCTCACCTTCCAATCTAGTATCC TCTCTAATTAGGCGGTTGGGAC GCTACCCAGCCAGAAACAGA CGTCCTCTTCCTCTTCTTCTGA TGATACTGCATATCAAGAAGCCA TTGCTTTTCTTCGACACGCA TGTCCCTAACTCCCTATAAAAGGC TGACCATAGGAGAAGTAGTGAGGA TCCCCCTCCCTTCTTTTTGA ACTTGGCTGCGAAAGAGTTG TGGAATCAAGATTCAAGATTTCGGG CCAGTCCGACCTGGTTGG TGGCCCTTGTTTTCTTGGGA TCATCTGTTCAGAGGCTTGCT TGCTACGCTCTGATACGTCT TCTCTTCCCTTGCACATTCTCA GGAAACATCACAAGGTTGAAGGT TACCATTTCACACGCTCGGT多态性信息量PIC 0.740 1 0.875 9 0.459 3 0.820 5 0.735 6 0.834 0 0.770 7 0.821 3 0.788 8 0.800 2 0.638 2 0.635 6 0.843 8 0.834 9 0.757 9 0.767 6 0.823 6 0.744 2 0.600 4 0.800 1 0.722 3 0.790 3
2.2 DNA指纹图谱构建
利用22 对多态性引物对83 份澳洲坚果种质进行毛细管电泳检测。选取引物P84、P118、P127、P132、P141、P163、P202、P211、P238 共9 对构建该群体的DNA指纹图谱。将每对引物扩增得到的多态性片段根据大小进行数字+英文字母形式的编码(表2),如引物P202 共有13 个多态性位点,从小到大依次编码为286(1)、286/288(2)、286/290(3)、286/292(4)、288(5)、288/290(6)、288/292(7)、288/294(8)、290(9)、290/292(A)、292(B)、292/294(C)、300(D)。9 对引物按照P211、P127、P238、P132、P118、P163、P202、P141、P84的顺序排列组合,将每份种质对应的引物位点编码统计组合,形成该种质的SSR 指纹图谱代码(表3)。如景哈57 的SSR 指纹图谱代码为H7A49A52I(图1)。对照表2,第1个字母“H”表示种质景哈57 在引物P211 中的扩增片段位于该引物多态性片段梯度中的第17位,第2个数字“7”表示种质景哈57在引物P127中的扩增片段位于该引物多态性片段梯度中的第7位,第3个字母“A”表示种质景哈57在引物P238中的扩增片段位于该引物多态性片段梯度中的第10位,其余代码依次类推。将每份种质的SSR指纹图谱代码导入在线条形码生成器,生成条形码DNA分子身份证,将每份种质的主要信息和SSR指纹图谱代码导入二维码生成程序,生成二维码DNA分子身份证,图2为部分种质的示例。
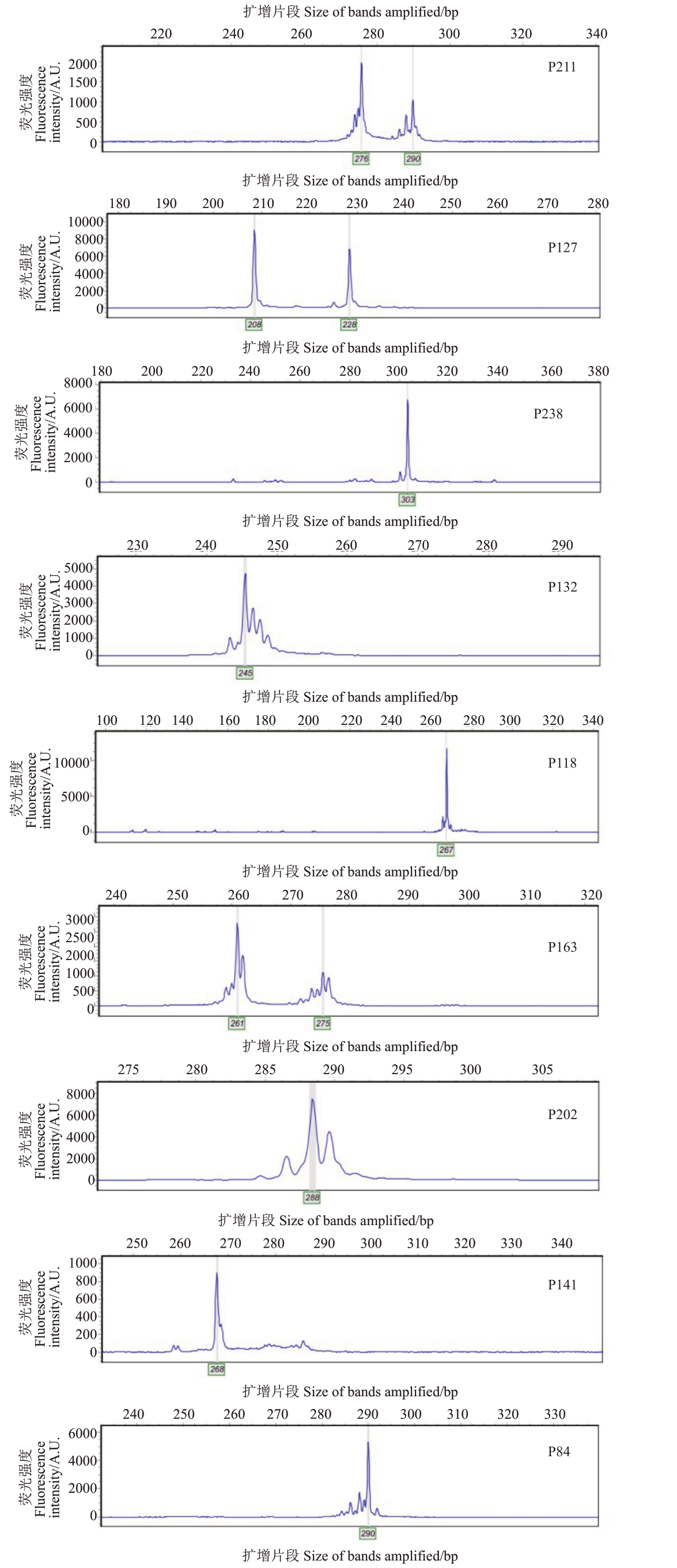
图1 景哈57 在9 个SSR 位点的毛细管电泳检测带型
Fig.1 The Band features of Jingha 57 detected by capillary electrophoresis at 9 SSR loci
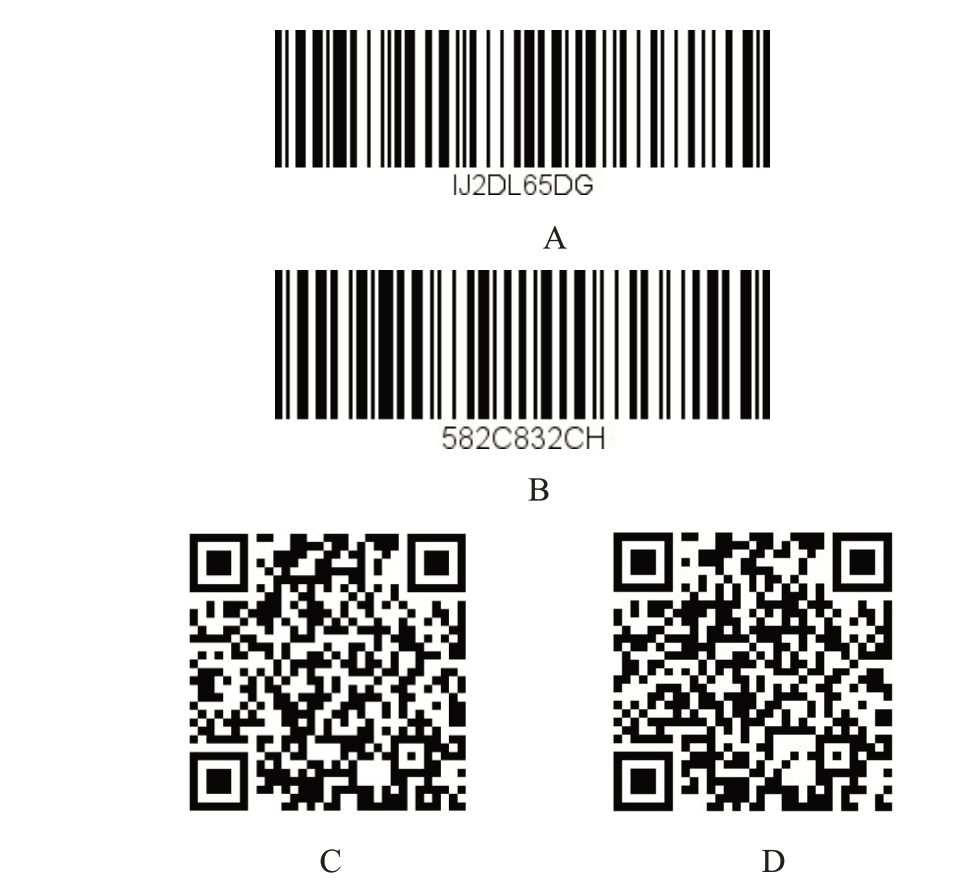
图2 澳洲坚果种质条形码和二维码DNA 分子身份证示例
Fig.2 Bar code and quick response code of the macadamia germplasm
A.O.C 条形码DNA 分子身份证;B.桂热一号条形码DNA 分子身份证;C.O.C 二维码DNA 分子身份证;D.桂热一号二维码DNA分子身份证。
A.Bar code DNA molecular identification of O.C; B.Bar code DNA molecular identification of Guire1;C.Quick response code DNA molecular identification of O.C;D.Quick response code DNA molecular identification of Guire1.
表2 9 对SSR 引物毛细管电泳结果及带型编码
Table 2 Amplification results and code typing of 9 pairs of SSR primers detected with capillary electrophoresis

引物名称Primer name P211 P127 P238 P132 P118 P163 P202 P141 P84毛细管电泳多态性片段及编码Code of capillary electrophoresis polymorphism fragments 266/278(1)268(2)268/270(3)268/276(4)268/278(5)268/280(6)268/290(7)270/276(8)270/278(9)270/280(A)270/290(B)274/278(C)274/288(D)276/278(E)276/280(F)276/288(G)276/290(H)278(I)278/290(J)280(K)280/290(L)284(M)286(N)290(O)205(1)205/208(2)208(3)208/215(4)208/222(5)208/225(6)208/228(7)208/232(8)215(9)218/222(A)215/225(B 215/228(C)215/232(D)218/225(E)222/225(F)222/232(G)225(H)225/228(I)225/232(J)228(K)228/232(L)232(M)254/257(N)233(1)248(2)248/281(3)281(4)281/284(5)284(6)297(7)297/303(8)297/306(9)303(A)303/309(B)309(C)311(D)319/331(E)326(F)334(G)344(H)243/248(1)243/250(2)243/254(3)245(4)245/248(5)245/250(6)245/252(7)245/254(8)245/258(9)245/264(A 248(B)248/252(C)248/254(D)248/258(E)248/264(F)250(G)250/252(H)250/254(I)250/264(J)250/266(K)252(L)252/254(M)252/258(N)254/264(O)256(P)264(Q)270(R)120(1)120/227(2)120/267(3)120/269(4)120/273(5)153/227(6)227(7)227/275(8)267(9)267/269(A)267/271(B 267/300(C)269(D)271(E)271/273(F)271/275(G)271/277(H)273(I)273/275(J)273/277(K)275(L)275/277(M 275/279(N)277(O)277/279(P)0/0(Q)257(1)257/261(2)257/263(3)257/271(4)257/275(5)257/278(6)261(7)261/263(8)261/271(9)261/275(A)263(B 263/275(C)263/278(D)271/275(E)285/298(F)285/300(G)292(H)294(I)298/304(J)302/306(K)286(1)286/288(2)286/290(3)286/292(4)288(5)288/290(6)288/292(7)288/294(8)290(9)290/292(A)292(B)292/294(C)300(D)266/280(1)268(2)268/287(3)268/290(4)268/293(5)270/280(6)278(7)278/284(8)278/287(9)278/310(A)278/332(B)280(C)280/284(D)280/287(E)280/290(F)280/293(G)280/295(H)284/287(I)284/295(J)287(K)287/290(L 287/293(M)287/295(N)287/298(O)287/308(P)290(Q)290/293(R)290/294(S)290/303(T)0(U)271(1)273(2)275/277(3)275/290(4)279(5)279/282(6)279/284(7)279/288(8)282(9)282/286(A)282/288(B)284(C)284/286(D)284/288(E)284/290(F)286/288(G)288(H)290(I)0(J)
表3 83 份澳洲坚果种质资源的SSR 指纹图谱代码
Table 3 SSR fingerprinting of 83 macadamia germplasm resources
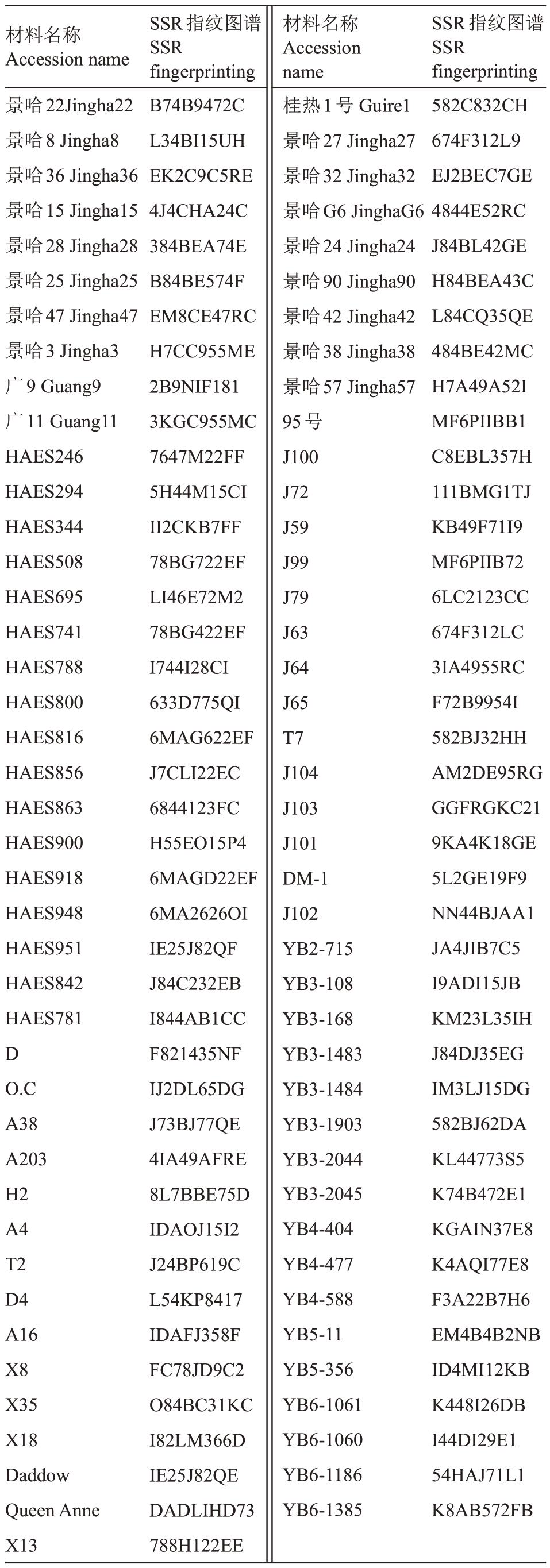
材料名称Accession name景哈22Jingha22景哈8 Jingha8景哈36 Jingha36景哈15 Jingha15景哈28 Jingha28景哈25 Jingha25景哈47 Jingha47景哈3 Jingha3广9 Guang9广11 Guang11 HAES246 HAES294 HAES344 HAES508 HAES695 HAES741 HAES788 HAES800 HAES816 HAES856 HAES863 HAES900 HAES918 HAES948 HAES951 HAES842 HAES781 D O.C A38 A203 H2 A4 T2 D4 A16 X8 X35 X18 Daddow Queen Anne X13 SSR指纹图谱SSR fingerprinting B74B9472C L34BI15UH EK2C9C5RE 4J4CHA24C 384BEA74E B84BE574F EM8CE47RC H7CC955ME 2B9NIF181 3KGC955MC 7647M22FF 5H44M15CI II2CKB7FF 78BG722EF LI46E72M2 78BG422EF I744I28CI 633D775QI 6MAG622EF J7CLI22EC 6844123FC H55EO15P4 6MAGD22EF 6MA2626OI IE25J82QF J84C232EB I844AB1CC F821435NF IJ2DL65DG J73BJ77QE 4IA49AFRE 8L7BBE75D IDAOJ15I2 J24BP619C L54KP8417 IDAFJ358F FC78JD9C2 O84BC31KC I82LM366D IE25J82QE DADLIHD73 788H122EE材料名称Accession name桂热1号Guire1景哈27 Jingha27景哈32 Jingha32景哈G6 JinghaG6景哈24 Jingha24景哈90 Jingha90景哈42 Jingha42景哈38 Jingha38景哈57 Jingha57 95号J100 J72 J59 J99 J79 J63 J64 J65 T7 J104 J103 J101 DM-1 J102 YB2-715 YB3-108 YB3-168 YB3-1483 YB3-1484 YB3-1903 YB3-2044 YB3-2045 YB4-404 YB4-477 YB4-588 YB5-11 YB5-356 YB6-1061 YB6-1060 YB6-1186 YB6-1385 SSR指纹图谱SSR fingerprinting 582C832CH 674F312L9 EJ2BEC7GE 4844E52RC J84BL42GE H84BEA43C L84CQ35QE 484BE42MC H7A49A52I MF6PIIBB1 C8EBL357H 111BMG1TJ KB49F71I9 MF6PIIB72 6LC2123CC 674F312LC 3IA4955RC F72B9954I 582BJ32HH AM2DE95RG GGFRGKC21 9KA4K18GE 5L2GE19F9 NN44BJAA1 JA4JIB7C5 I9ADI15JB KM23L35IH J84DJ35EG IM3LJ15DG 582BJ62DA KL44773S5 K74B472E1 KGAIN37E8 K4AQI77E8 F3A22B7H6 EM4B4B2NB ID4MI12KB K448I26DB I44DI29E1 54HAJ71L1 K8AB572FB
3 讨 论
3.1 SSR标记的选择
随着分子标记技术不断发展,多种DNA 标记被应用于植物DNA 指纹图谱的构建。相较于其他分子标记类型,SSR 标记有其独特的优势,目前仍然比任何其他传统的DNA 指纹方法更重要[15-16]。文雁成等[17]研究表明,与SRAP 标记相比,SSR 标记以其扩增谱带少、易于识别和统计而更适合用于引物组合法构建品种指纹图谱。李志远等[18]认为SNP 标记检测成本较高,鉴别相同数量品种所需要的SNP 标记数多于SSR 标记。早期SSR 标记开发存在成本高、周期长的问题,鉴于高通量测序费用的降低及越来越多物种基因组的发布,从已知序列识别重复位点开发SSR 标记变得简单、快速和经济。
SSR标记的检测技术主要为聚丙烯酰胺凝胶电泳后银染及毛细管电泳的荧光检测。银染技术存在人工读取胶板误差较大问题。荧光标记的毛细管电泳检测无论是检测效率还是数据读取的自动化和准确性,都优于银染检测,特别是针对较大群体材料的检测[19-20]。与基于普通SSR标记的DNA指纹鉴定方法相比,基于SSR荧光标记的DNA指纹鉴定效率更高、结果更准确[21]。本次研究选择用聚丙烯酰胺凝胶电泳对引物进行初步筛选,最后将多态性引物荧光标记用于供试材料的分析。兼顾了引物筛选中的费用成本及检测效率,获得了澳洲坚果高效的SSR引物。
3.2 指纹图谱的构建
DNA 指纹图谱是鉴别品种、品系的有力工具,非常适合于品种资源的鉴定工作[22]。在我国不同澳洲坚果种植区,存在新登记品种的主要特征相似、对同一优株不同命名的现象,易造成育种工作的重复及品种权纠纷。有些“假品种”与既有品种高度相似,苗期不易鉴别,后期给种植户带来一定的经济损失。DNA 指纹图谱可在早期从分子水平上为品种鉴别保护提供技术支持。随着澳洲坚果种质资源保存数量的增加,利用分子标记技术构建澳洲坚果种质资源DNA指纹图谱,可有效避免种质资源的重复性收集和保存。
指纹库构建时采用什么样的数据记录方式不仅影响数据库构建的难易程度,还影响数据库使用的方便程度[23]。早期构建数据库时根据谱带的有无进行1/0 形式编号的方法已很少使用,目前,对等位基因进行排列赋值的方法受到越来越多研究者的青睐。笔者选择扩增稳定、重复性好的特异引物按固定顺序排列组合,建立部分澳洲坚果种质的指纹数据库。并对各引物扩增的等位基因由小到大进行排列,以数字、英文字母的方式开始赋值,将每份材料的DNA数据转换为字符串,再将字符串转化为条形码及二维码形式,使其更具直观性,易被扫描识别,特别是二维码可添加文本图片信息,在资源圃种质资源的管理中应用更广。笔者在本研究中部分引物的等位基因位点可能随着建库规模的扩大而增加,造成数据库的变动。但笔者认为,鉴于目前澳洲坚果种质资源收集保存较少,该简易直观的建库方法可满足现阶段的工作需求。
4 结 论
基于澳洲坚果基因组设计、筛选的SSR 标记,可有效用于澳洲坚果种质资源的鉴定。
[1] MAST A R,WILLIS C L,JONES E H,DOWNS K M,WESTON P H.A smaller macadamia from a more vagile tribe:inference of phylogenetic relationships,divergence times,and diaspore evolution in macadamia and relatives (tribe Macadamieae;Proteaceae)[J].American Journal of Botany,2008,95(7):843-870.
[2] NOCK C J,BATEN A,BARKLA B J,FURTADO A,HENRY R J,KING G J.Genome and transcriptome sequencing characterises the gene space of Macadamia integrifolia (Proteaceae)[J].BMC Genomics,2016,17(1):937.
[3] O’CONNOR K,HAYES B,HARDNER C,NOCK C,BATEN A,ALAM M,HENRY R J,TOPP B.Genome-wide association studies for yield component traits in a macadamia breeding population[J].BMC Genomics,2020,21(1):199.
[4] 王立新,张小军,史星雲,高华,赵政阳.苹果栽培品种SSR 指纹图谱的构建[J].果树学报,2012,29(6):971-977.WANG Lixin,ZHANG Xiaojun,SHI Xingyun,GAO Hua,ZHAO Zhengyang.Establishment of SSR fingerprinting database on major apple (Malus×domestica) cultivars[J].Journal of Fruit Science,2012,29(6):971-977.
[5] 高源,刘凤之,王昆,王大江,龚欣,刘立军.苹果部分种质资源分子身份证的构建[J].中国农业科学,2015,48(19):3887-3898.GAO Yuan,LIU Fengzhi,WANG Kun,WANG Dajiang,GONG Xin,LIU Lijun.Establishment of molecular ID for some apple germplasm resources[J].Scientia Agricultura Sinica,2015,48(19):3887-3898.
[6] 李益,马先锋,唐浩,李娜,江东,龙桂友,李大志,牛英,韩瑞玺,邓子牛.柑橘品种鉴定的SSR 标记开发和指纹图谱库构建[J].中国农业科学,2018,51(15):2969-2979.LI Yi,MA Xianfeng,TANG Hao,LI Na,JIANG Dong,LONG Guiyou,LI Dazhi,NIU Ying,HAN Ruixi,DENG Ziniu.SSR markers screening for identification of citrus cultivar and construction of DNA fingerprinting library[J].Scientia Agricultura Sinica,2018,51(15):2969-2979.
[7] 刘娟,廖康,曼苏尔·那斯尔,孙琪,刘欢,贾杨.新疆杏品种(系)遗传多样性分析及DNA 指纹图谱库构建[J].中国农业科学,2015,48(4):748-758.LIU Juan,LIAO Kang,Mansuer·Nasir,SUN Qi,LIU Huan,JIA Yang.Analysis of genetic diversity and construction of DNA fingerprint database of Xinjiang apricot varieties(lines)[J].Scientia Agricultura Sinica,2015,48(4):748-758.
[8] 周于波,朱鹏,龚伟,王景燕,闫思宇,吴开志.四川核桃良种SSR 指纹图谱构建及遗传多样性分析[J].西北植物学报,2018,38(7):1254-1261.ZHOU Yubo,ZHU Peng,GONG Wei,WANG Jingyan,YAN Siyu,WU Kaizhi.SSR fingerprint construction and genetic diversity analysis of elite Juglans regia cultivars in Sichuan[J].Acta Botanica Boreali-Occidentalia Sinica,2018,38(7):1254-1261.
[9] 何旭东,郑纪伟,田雪瑶,教忠意,窦全琴.薄壳山核桃品种亲缘关系分析与指纹图谱构建[J].林业科学研究,2021,34(4):95-102.HE Xudong,ZHENG Jiwei,TIAN Xueyao,JIAO Zhongyi,DOU Quanqin.Genetic relationship analysis and fingerprint construction of Carya illinoensis carieties[J].Forest Research,2021,34(4):95-102.
[10] 蔡元保,杨祥燕,陈显国,曾黎明,郭凌飞,林玉虹,崔明勇.澳洲坚果SCoT 反应体系的建立及应用[J].热带亚热带植物学报,2013,21(3):253-258.CAI Yuanbao,YANG Xiangyan,CHEN Xianguo,ZENG Liming,GUO Lingfei,LIN Yuhong,CUI Mingyong.Establishment and application of SCoT amplification system for macadamia[J].Journal of Tropical and Subtropical Botany,2013,21(3):253-258.
[11] International Union for the Protection of New Varieties of Plants.Guidelines for DNA-profiling:Molecular markers selection and database construction[M].Geneva:UPOV,2006.
[12] 中华人民共和国农业部.植物品种鉴定DNA 分子标记法总则:NY/T 2594—2016[S].北京:中国农业出版社,2016.Ministry of Agriculture and Rural of the People’s Republic of China.General guideline for identification of plant varieties using DNA markers:NY/T 2594—2016[S].Beijing:Chinese Agriculture Press,2016.
[13] 杨文娟.芝麻应用核心种质DNA 分子身份证和种质资源数据库共享平台构建[D].北京:中国农业科学院,2018.YANG Wenjuan.Establishment of DNA molecular identification of Sesamum indicum applied core germplasm and information database of germplasm resource[D].Beijing:Chinese Academy of Agricultural Sciences,2018.
[14] 郭艳春,张力岚,陈思远,祁建民,方平平,陶爱芬,张列梅,张立武.黄麻应用核心种质的DNA 分子身份证构建[J].作物学报,2021,47(1):80-93.GUO Yanchun,ZHANG Lilan,CHEN Siyuan,QI Jianmin,FANG Pingping,TAO Aifen,ZHANG Liemei,ZHANG Liwu.Establishment of DNA molecular fingerprint of applied core germplasm in jute (Corchorus spp.)[J].Acta Agronomica Sinica,2021,47(1):80-93.
[15] SELKOE K A,TOONEN R J.Microsatellites for ecologists:a practical guide to using and evaluating microsatellite markers[J].Ecology Letters,2006,9(5):615-629.
[16] GUICHOUX E,LAGACHE L,WAGNER S,CHAUMEIL P,LÉGER P,LEPAIS O,LEPOITTEVIN C,MALAUSA T,REVARDEL E,SALIN F,PETIT R J.Current trends in microsatellite genotyping[J].Molecular Ecology Resources,2011,11(4):591-611.
[17] 文雁成,王汉中,沈金雄,刘贵华.SRAP 和SSR 标记构建的甘蓝型油菜品种指纹图谱比较[J].中国油料作物学报,2006,28(3):233-239.WEN Yancheng,WANG Hanzhong,SHEN Jinxiong,LIU Guihua.Comparision of cultivar fingerprints constructed with SRAP and SSR markers in Brassica napus L.[J].Chinese Journal of Oil Crop Sciences,2006,28(3):233-239.
[18] 李志远,于海龙,方智远,杨丽梅,刘玉梅,庄木,吕红豪,张扬勇.甘蓝SNP 标记开发及主要品种的DNA 指纹图谱构建[J].中国农业科学,2018,51(14):2771-2788.LI Zhiyuan,YU Hailong,FANG Zhiyuan,YANG Limei,LIU Yumei,ZHUANG Mu,LÜ Honghao,ZHANG Yangyong.Development of SNP markers in cabbage and construction of DNA fingerprinting of main varieties[J].Scientia Agricultura Sinica,2018,51(14):2771-2788.
[19] 易红梅,王凤格,赵久然,王璐,郭景伦,原亚萍.玉米品种SSR 标记毛细管电泳荧光检测法与变性PAGE 银染检测法的比较研究[J].华北农学报,2006,21(5):64-67.YI Hongmei,WANG Fengge,ZHAO Jiuran,WANG Lu,GUO Jinglun,YUAN Yaping.Comparison of two maize SSR detection methods:Capillary electrophoresis with fluorescence detection method and denaturing PAGE silver- staining detection method[J].Acta Agriculturae Boreali-Sinica,2006,21(5):64-67.
[20] 郝晨阳,王兰芬,贾继增,董玉琛,张学勇.SSR 荧光标记和银染技术的比较分析[J].作物学报,2005,31(2):144-149.HAO Chenyang,WANG Lanfen,JIA Jizeng,DONG Yuchen,ZHANG Xueyong.Comparison of fluorescence and silver-staining detection systems of microsatellite markers[J].Acta Agronomica Sinica,2005,31(2):144-149.
[21] 郑永胜,张晗,王东建,孙加梅,王雪梅,段丽丽,李华,王玮,李汝玉.基于荧光检测技术的小麦品种SSR 鉴定体系的建立[J].中国农业科学,2014,47(19):3725-3735.ZHENG Yongsheng,ZHANG Han,WANG Dongjian,SUN Jiamei,WANG Xuemei,DUAN Lili,LI Hua,WANG Wei,LI Ruyu.Development of a wheat variety identification system based on fluorescently labeled SSR markers[J].Scientia Agricultura Sinica,2014,47(19):3725-3735.
[22] 王忠华.DNA 指纹图谱技术及其在作物品种资源中的应用[J].分子植物育种,2006,4(3):425-430.WANG Zhonghua.DNA fingerprinting technology and its application in crop germplasm resources[J].Molecular Plant Breeding,2006,4(3):425-430.
[23] 王凤格,杨扬,易红梅,赵久然,任洁,王璐,葛建镕,江彬,张宪晨,田红丽,侯振华.中国玉米审定品种标准SSR 指纹库的构建[J].中国农业科学,2017,50(1):1-14.WANG Fengge,YANG Yang,YI Hongmei,ZHAO Jiuran,REN Jie,WANG Lu,GE Jianrong,JIANG Bin,ZHANG Xianchen,TIAN Hongli,HOU Zhenhua.Construction of an SSR-based standard fingerprint database for corn variety authorized in China[J].Scientia Agricultura Sinica,2017,50(1):1-14.