根际微生物是指生活在植物根系和土壤的交界面并与植物相互作用的微生物,其组成由微生物、植物根系和环境的相互作用决定[1-2],不同植物、土壤类型可形成特定的微生物群落,通过分泌植物生长激素促进根系生长并提高根系对营养物质和水分的吸收能力[3-5],富集的有益微生物可产生抑菌的抗菌化合物或激活植物免疫系统达到抑菌效果[6-8],富集的病原微生物将引起连作障碍并与植物竞争养分等[9-11],进而影响植物的生长发育[12-13]。因此,研究根际微生物多样性和组成对植物的生长发育、抗逆性和病害生物防治有重要意义[14]。
19世纪中叶,科技人员将欧洲种嫁接到美洲种上以防治葡萄根瘤蚜,筛选新的砧木基因型及研究砧穗互作机制成为现代种植的研究方向[15]。关于砧木研究多集中于根系吸收水分和矿质元素能力、对土壤病害和逆境胁迫等的抗性不同,进而影响植株光合能力、树势、抗逆性和果实品质[16-19]等方面,而对砧木根际微生物研究较少。Marasco 等[15]利用不同砧木161.49、420A 等嫁接葡萄品种巴贝拉,发现砧木显著影响根际微生物群落组成和多样性;王静等[20]研究樱桃砧木根际微生物时发现,适宜的樱桃砧木有利于樱桃树土壤生态环境;而徐龙晓等[21]研究认为,相比砧木差异,土壤质地对苹果根际微生物碳源利用类型影响更大。由于大部分土壤微生物不能培养,传统的微生物分析方法无法准确揭示微生物多样性,随着高通量测序技术的发展,利用基因测序技术极大地方便了根际微生物群落结构的检测。笔者在本研究中利用16S rRNA 高通量测序技术分析常用葡萄砧木1103P、5BB 和Beta 嫁接和自根苗阳光玫瑰根际微生物,揭示微生物群落结构和多样性,以期为阳光玫瑰筛选适宜砧木、改善土壤管理提供参考。
1 材料和方法
1.1 供试材料
土壤样品采自山东省招远市大户庄园葡萄园,暖温带大陆性季风气候,年平均气温11.5 ℃,年降水量607 mm,年日照数2503 h。砧木品种有1103P、5BB和Beta,自根苗为对照。以4个葡萄园地块3年生阳光玫瑰为试材,每个地块666.7 m2,南北行向,篱架栽培,单干单臂树形+V 形叶幕,所有葡萄地块统一常规管理。葡萄转色期测定4个地块土壤理化性质和微生物,无显著差异,葡萄采收期将取样器消毒灭菌,每个葡萄地块选取生长良好的阳光玫瑰植株5 株,按照五点取样法采集根际土壤,混合均匀,设置3 次生物学重复。采样时去除地表杂质,以树干为中心,挖取离地表约30 cm行内土壤,抖落挖出的土壤根系上附着的大块土壤,收集根系上附着的粉状或粒状土壤,经低温保存送回实验室,过2 mm无菌筛网去除杂质,分为两组,一组保存于无菌管中放置在-80 ℃冰箱用于微生物检测,另一组自然风干用于土壤理化性质测定。
1.2 土壤理化性质测定
土壤有机质采用重铬酸钾容量法-稀释热法测定;土壤pH值用水土质量比1∶1,pH计测定;碱解氮含量采用碱解扩散法进行分析;有效磷含量采用NaHCO3浸提并利用硫酸钼锑抗比色法进行测定;通过醋酸铵浸提土壤速效钾,并利用火焰光度法测定其含量;采用1 mol·L-1 NH4OAc 溶液浸提交换性钙镁,并运用原子吸收分光光度计测定浸提液中的钙镁含量。
1.3 土壤DNA提取与PCR扩增测序
用E.Z.N.A.® Soil DNA Kit(Omega Bio- tek,Norcross, GA, U.S.A.)试剂盒提取土壤总DNA,利用1%的琼脂糖凝胶进行电泳检测。土壤细菌扩增引物为341F(5’-CCTAYGGGRBGCASCAG-3’)和806R(5’-GGACTACNNGGGTATCTAAT-3’),土壤真菌扩增引物为ITS1F(5’-CTTGGTCATTTAGAGGAAGTAA-3’)和ITS2R(5’-GCTGCGTTCTTCATCGATGC-3’),PCR 反应程序:95 ℃预变性5 min;95 ℃变性30 s,55 ℃退火30 s,72 ℃延伸30 s,共25个循环;最后72 ℃延伸5 min。PCR 扩增产物经AxyPrepDNA 凝胶回收试剂盒(AXYGEN,美国)纯化回收后用于高通量测序。
1.4 序列处理分析
Illumina PE250 测序技术委托上海凌恩生物科技有限公司,按照公司质控规定进行:根据barcode得到样品的有效序列;过滤read尾部20以下质量值的碱基,设置10 bp窗口,若窗口内平均质量值低于20,自窗口截去后端碱基,过滤质控后50 bp以下read;根据PE reads的overlap关系,拼接成对reads成一条序列,最小overlap长度10 bp;拼接序列的overlap区允许最大错配比率0.2,筛除不符合的序列;区分样品时根据序列首尾两端barcode 和引物,调整序列方向,barcode 允许错配数为0,最大引物错配数为2;用Usearch软件和gold数据库,采用denovo和reference结合的方式去除嵌合体,区分样本后进行OTU聚类分析和物种分类学分析,采用RDP classifier 贝叶斯算法对97%相似水平的OTU代表序列进行分类学分析(置信度阈值为0.7),比对数据库为Silva(细菌)和Unite(真菌),统计各样本的群落组成。
1.5 数据分析
应用SPSS 19.0对土壤样品理化性质、微生物多样性指数统计和微生物门属丰度进行显著性分析(p<0.05);应用上海凌恩生物科技有限公司云平台R语言包进行主坐标分析作图(Principal Coordinates Analysis,PCoA)和组间差异检验(Adonis,ANOSIM)。由于样品数少(12个)而物种数目(细菌1056属,真菌412属)很大,比较各样品微生物组成差异时,在细菌和真菌属水平上做PCoA分析,并进行组间差异检验,以检验组间的差异是否显著大于组内差异。
2 结果与分析
2.1 土壤理化性质
不同砧木和自根苗阳光玫瑰根际土壤理化性质检测结果如表1 所示,4 个样品pH 值存在显著差异(p<0.05),其中1103P 最低,5BB 最高;5BB 和自根苗土壤碱解氮(N)含量显著高于1103P 和Beta;1103P有机质含量显著低于5BB和自根苗的根际土壤样品;Beta 有效磷(P)含量显著低于其他3 个样品;自根苗根际土壤速效钾(K)含量显著高于砧木;自根苗交换性钙(Ca)含量和砧木无显著差异,5BB显著低于Beta;交换性镁(Mg)含量4个样品无显著差异。
表1 土壤样品理化性质
Table 1 Physical and chemical properties of soil samples
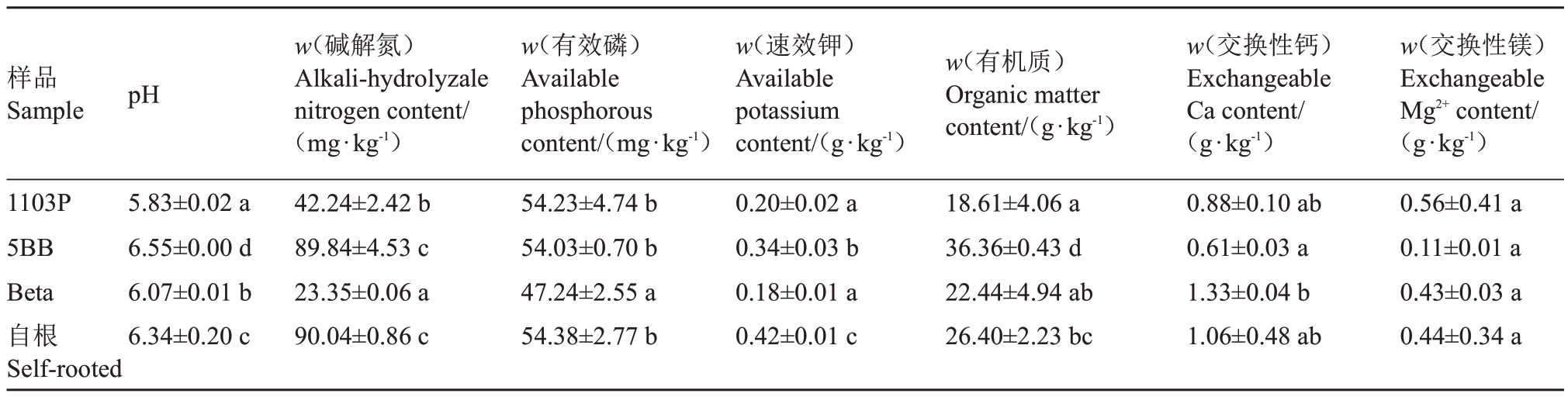
注:不同小写字母表示在p<0.05 差异显著。下同。
Note:Different small letters indicate significant difference at p<0.05.The same below.
样品Sample pH w(有机质)Organic matter content/(g·kg-1)1103P 5BB Beta自根Self-rooted 5.83±0.02 a 6.55±0.00 d 6.07±0.01 b 6.34±0.20 c w(碱解氮)Alkali-hydrolyzale nitrogen content/(mg·kg-1)42.24±2.42 b 89.84±4.53 c 23.35±0.06 a 90.04±0.86 c w(有效磷)Available phosphorous content/(mg·kg-1)54.23±4.74 b 54.03±0.70 b 47.24±2.55 a 54.38±2.77 b w(速效钾)Available potassium content/(g·kg-1)0.20±0.02 a 0.34±0.03 b 0.18±0.01 a 0.42±0.01 c 18.61±4.06 a 36.36±0.43 d 22.44±4.94 ab 26.40±2.23 bc w(交换性钙)Exchangeable Ca content/(g·kg-1)0.88±0.10 ab 0.61±0.03 a 1.33±0.04 b 1.06±0.48 ab w(交换性镁)Exchangeable Mg2+content/(g·kg-1)0.56±0.41 a 0.11±0.01 a 0.43±0.03 a 0.44±0.34 a
2.2 根际土壤微生物多样性
4 个根际土壤样品中细菌和真菌的α 多样性指数见表2:细菌OTU显著高于真菌,随着测序深度的增加,土壤细菌测序覆盖率为96.35%~97.17%。按照97%的相似水平将序列划分成不同的OTU,用chao1 算法估计样本中所含OTU 数目即为Chao 指数,OTU 和Chao 反映了土壤微生物群落丰度,细菌OTU 数和群落丰度指数Chao 均为1103P 最大;Shannon 指数和Simpson 指数估算样本中微生物多样性,Shannon指数越大,Simpson指数越小,群落多样性指数越高,细菌Shannon指数1103P显著高于其他砧木,Simpson 指数1103P 略低于其他砧木,细菌群落多样性排序为1103P>自根>5BB>Beta。
表2 阳光玫瑰根际土壤中细菌和真菌α 多样性指数统计
Table 2 α diversity index statistics of fungi and bacteria in the rhizosphere soil of Shine Muscat

微生物类群Microbial community细菌Bacteria真菌Fungi样品Samples 1103P 5BB Beta自根Self-rooted 1103P 5BB Beta自根Self-rooted 97%相似水平Similarity level of 97%OTU 5 316.00±104.65 b 4 724.50±181.73 a 4 748.50±61.52 ab 5 112.00±350.72 ab 682.00±73.54 a 761.50±12.02 a 662.00±24.04 a 704.50±33.23 a Chao 6 332.26±161.74 b 5 828.09±23.25 a 5 877.14±26.75 a 6 253.27±257.85 ab 734.51±51.12 a 850.77±8.84 b 704.67±22.94 a 788.31±39.05 ab Shannon 7.55±0.06 b 7.35±0.05 a 7.21±0.08 a 7.37±0.08 ab 4.71±0.22 ab 4.52±0.09 ab 4.74±0.04 b 4.48±0.26 a Simpson 0.001 6±0.000 4 a 0.002 2±0.000 1 a 0.003 8±0.000 4 b 0.001 8±0.000 1 a 0.021 9±0.004 6 a 0.030 9±0.004 4 a 0.021 9±0.003 1 a 0.036 6±0.021 1 a Coverage/%96.35 96.82 96.38 97.17 99.83 99.73 99.87 99.76
4 个根际土壤样品真菌测序覆盖率达到99.73%~99.87%,OTU数和丰度指数Chao高低顺序均为5BB>自根>1103P>Beta,不同砧木根际土壤真菌Shannon 指数Beta 显著高于自根,砧木间无显著差异,4 个样品的Simpson 指数无显著差异,真菌群落多样性为Beta>1103P>5BB>自根。
2.3 微生物分类学组成分析
2.3.1 细菌分类学组成分析 4个根际土壤样品细菌OTU 归类到44 门133 纲345 目536 科1056 属,图1-A为样品细菌在门分类学水平上的主要物种分布,其中,门水平分类选取各砧木相对丰度大于1%的物种,色块长度代表物种相对丰度所占比例。由图1可知,细菌优势门类群主要为变形菌门(Proteobacteria,24.15% ~33.57% )、酸 杆 菌 门(Acidobacteriota,14.83% ~22.82%)、放 线 菌 门(Actinobacteriota,7.24%~10.99%),而厚壁菌门(Firmicutes,0.91%~2.55%)和硝化螺旋菌门(Nitrospirota,1.01%~2.85%)相对丰度比例较低。其中,变形菌门、酸杆菌门和放线菌门相对丰度比例最高的分别为5BB、1103P和自根苗。

图1 细菌的水平分布柱状图和主坐标分析
Fig.1 Horizontal distribution histogram of bacteria and principal coordinates analysis
与自根苗相比,砧木增加了酸杆菌门、髌骨菌门(Patescibacteria)和疣微菌门(Verrucomicrobiota)的相对丰度比例,降低了放线菌门和芽单胞菌门(Gemmatimonadota)丰度比例,其中1103P和Beta的酸杆菌门分别比自根增加53.87%和40.54%,不同砧木之间细菌门组成和相对丰度也不一样,5BB 的拟杆菌门(Bacteroidota,12.27%)相对丰度比例显著高于其他组(3.98%~5.23%)(图1-A)。
图1-B展示4个根际土壤样品中细菌在属分类学水平上相对丰度比例排名前15的物种。除暂未命名的细菌外,相对丰度较高的细菌为RB41(0.65%~5.29%)、Candidatus Udaeobacter(1.28%~3.12%)、苔藓杆菌属(Bryobacter,2.03%~2.15%)、硝化螺菌属(Nitrospira,1.01%~2.85%)和芽单胞菌属(Gemmatimonas,0.80%~1.99%)。其中,1103P的优势细菌属为Candidatus Udaeobacter、RB41和硝化螺菌属,5BB的优势细菌属为苔藓杆菌属、邱贾伊杆菌属(Chujaibacter)和鞘氨醇单胞菌属(Sphingomonas),Beta的优势细菌属为立体杆菌属(Steroidobacter)、CandidatusUdaeobacter 和RB41,自根的优势细菌属为硝化螺菌属、苔藓杆菌属(Bryobacter)和假丝酵母属。
根际细菌OTU 组间差异检验R 值为0.955(p<0.05),表明组间差异大于组内差异。4 个根际土壤样品细菌属水平PCoA 分析结果表明,在两个坐标轴上Beta和自根苗根际细菌属均不能较好地区分,说明Beta 和自根根际土壤细菌群落属水平组成相似性高,1103P 和5BB 根际细菌属能较好地区分自根苗,说明1103P 和5BB 改变了阳光玫瑰根际土壤细菌群落组成(图1-C)。
2.3.2 真菌分类学组成分析 4个根际土壤样品真菌OTU 归类到8 门33 纲82 目188 科412 属,门分类学水平上的真菌物种分布如图2-A 所示,真菌优势菌群为子囊菌门(Ascomycota,55.57%~64.86%)、毛霉菌门(Mucoromycota,5.84%~22.24%)和担子菌门(Basidiomycota,12.72%~19.22%),相对丰度比例平均占89.72%。其中,1103P 的子囊菌门和担子菌门(Basidiomycota)相对丰度比例均低于其他3 组样品,毛霉菌门显著高于其他3组样品。
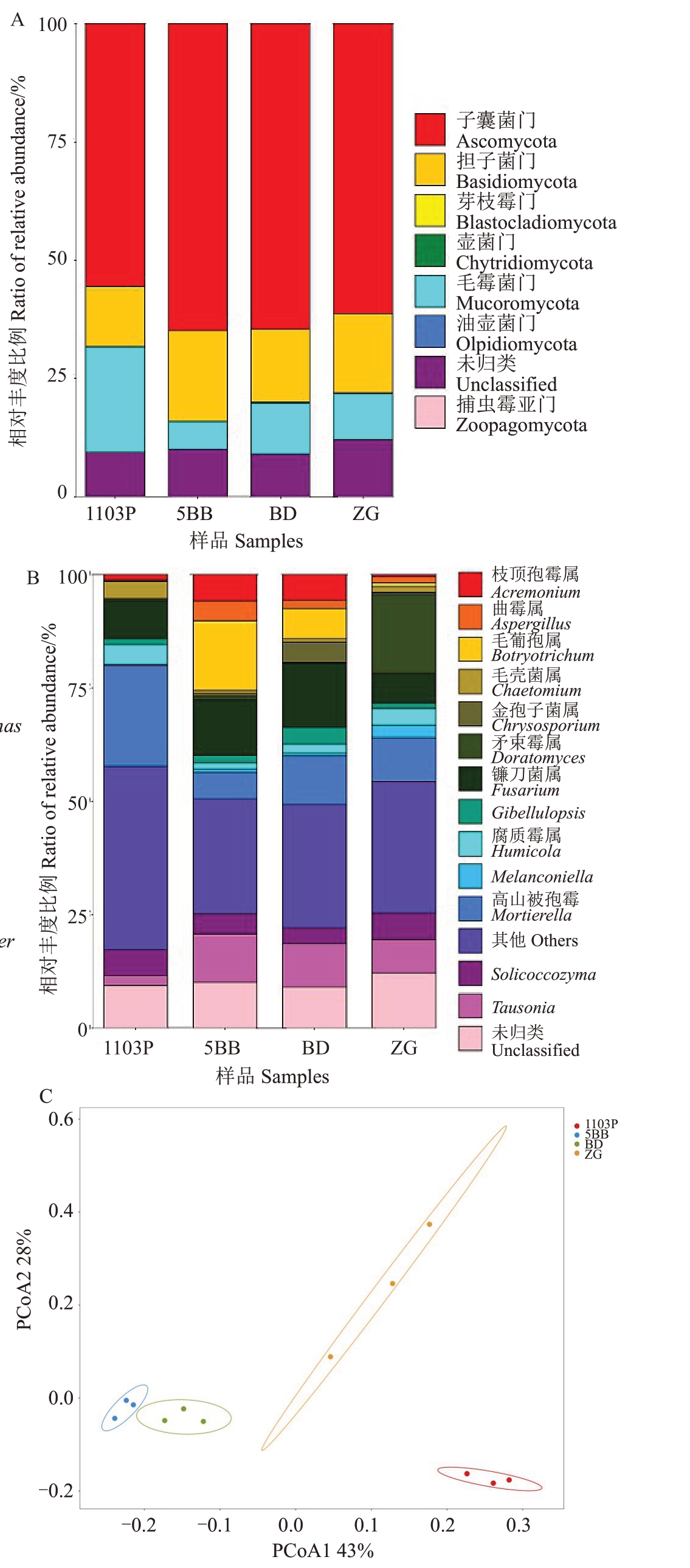
图2 真菌的水平分布柱状图和主坐标分析
Fig.2 Horizontal distribution histogram of fungi and principal coordinates analysis
A.真菌门;B.真菌属;C.主坐标分析。
A.Phylum of fungi; B.Genus of fungi; C.Principal coordinates analysis.
图2-B为土壤样品真菌在属分类学水平上相对丰度比例排名前15 的物种。优势真菌为高山被孢霉(Mortierella,5.84%~22.19%)、镰刀菌属(Fusarium,6.57% ~14.22% )和 Tausonia 属(2.29% ~10.58%)。不同处理之间优势真菌属组成不同,1103P的优势真菌属为高山被孢霉、镰刀菌属和Solicoccozyma;5BB的优势真菌属为毛葡孢属(Botryotrichum)、镰刀菌属和Tausonia 属;Beta 的优势真菌属为镰刀菌属、高山被孢霉和Tausonia属;自根的优势真菌属为矛束霉属(Doratomyces)、高山被孢霉和Tausonia属。
根际真菌OTU 组间差异检验R 值为0.857(p<0.05),表明组间差异大于组内差异。4 个根际土壤样品真菌OTU主坐标分析如图2-C所示,自根苗和1103P 根际真菌群落在第一主成分正半轴,5BB 和Beta 根际真菌群落在负半轴,4 个样品根际真菌均能较好区分,即不同砧木根际土壤真菌群落分布差异较大。
2.4 土壤细菌群落与土壤理化性质的冗余分析
通过对4个根际土壤样品细菌群落结构与土壤理化性质的冗余分析,发现有机质含量、交换性镁含量和pH 值对根际土壤优势细菌的影响较大。变形菌门和拟杆菌门的丰度变化与有机质含量、pH值及碱解氮、有效磷和速效钾含量呈正相关,与交换性镁、交换性钙含量呈负相关;酸杆菌门和放线菌门丰度变化趋势与有机质含量、pH值和碱解氮含量呈负相关,与交换性镁和交换性钙含量呈正相关(图3)。
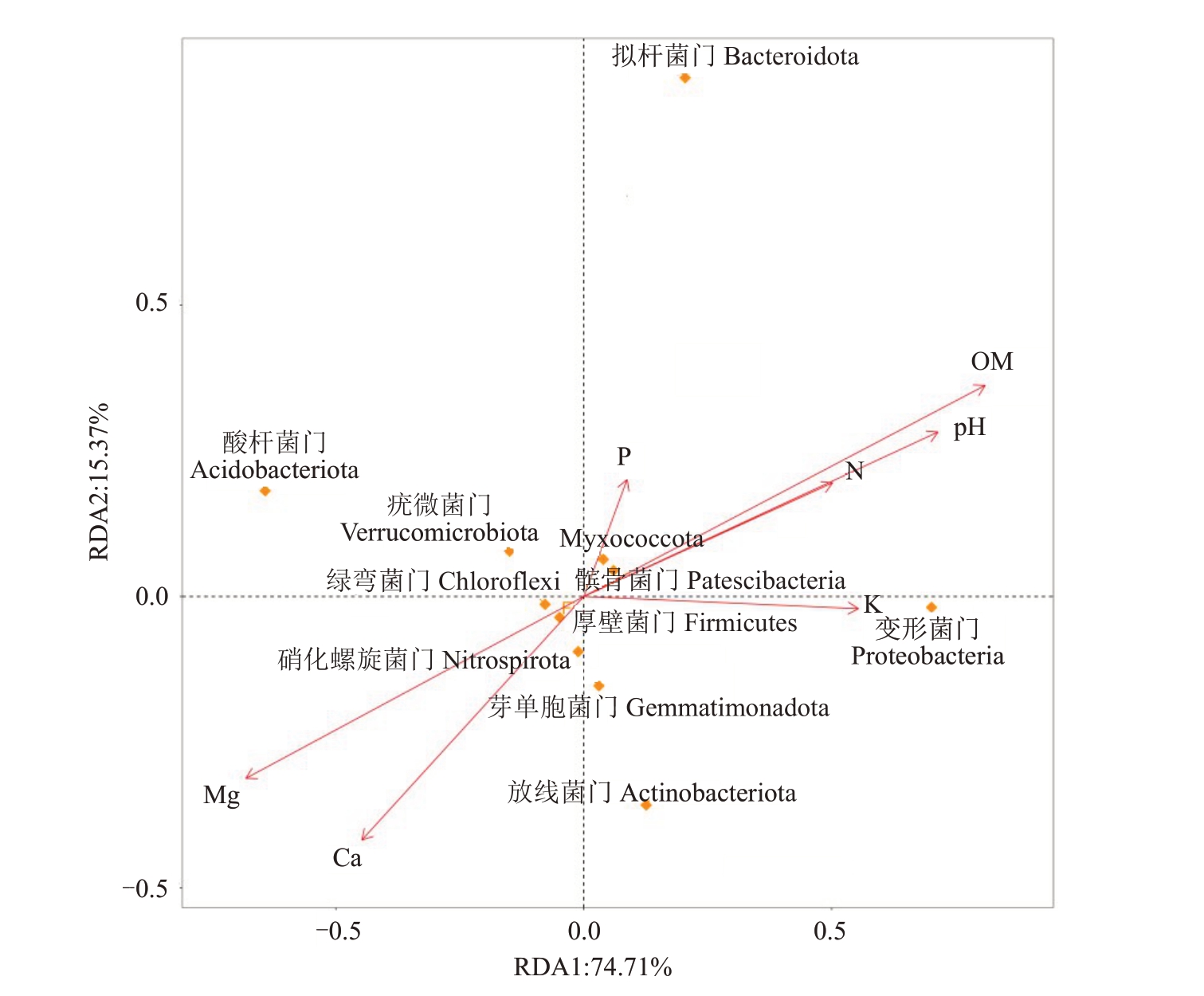
图3 土壤细菌群落与土壤理化性质的冗余分析
Fig.3 Redundancy analysis between bacterial communities and soil chemical properties
3 讨 论
Clegg等[22]和邵微等[23]研究均表明,微生物群落多样性的增加有利于提高土壤系统的稳定性和应对生态环境恶化时的缓冲能力,微生物功能多样性与细菌α 多样性呈正相关。本研究中,高通量测序结果表明1103P 细菌α 多样性最高,能够显著增加根际细菌的多样性和丰度,孙茜[24]研究表明,1103P、5BB、SO4、3309C这4种葡萄砧木的抗旱、耐盐和耐碱能力存在显著差异,1103P 均为最好,可能是1103P根际细菌多样性的增加提高了根系对环境胁迫的缓冲能力。笔者在本研究中发现细菌多样性显著高于真菌,与前人对森林生态系统、落叶果树和酿酒葡萄的根际微生物研究结果相一致[23,25-26]。本文真菌属水平的PCoA分析结果表明,4个样品真菌组成差异较大,说明砧木对真菌组成影响较大。
根际微生物群落被认为是植物的第二基因组[27],是根际微生态的重要组成部分,植物根系通过分泌代谢产物、改变根际环境等,逐渐形成特定的微生物群落[28-31]。已有研究表明酸杆菌门含有可编码纤维素酶和半纤维素酶的基因,可以通过降解木质素和纤维素,提高土壤养分含量[32-33]。前人分别通过分析云南丘北县、香格里拉和河北怀来酿酒葡萄园根际土壤微生物情况,发现在门水平上,不同产区不同品种优势细菌相似,均为变形菌门、酸杆菌门、放线菌门和芽单胞菌门等,但不同产区不同品种优势菌相对丰度不同[26,34-35],这些报道与本研究的结果相吻合。杨敏等[26]和宋雪洁[35]报道表明放线菌门相对丰度高于酸杆菌门,而本研究表明,酸杆菌门相对丰度比例平均为18.48%,显著高于放线菌门(8.26%),且相对自根苗,砧木均增加根际土壤的酸杆菌门丰度,所以,这些葡萄砧木可通过改善菌群结构进而有效提高土壤的养分含量。在细菌属水平上,本研究表明1103P 根际土壤中硝化螺菌属相对丰度相对较高,薛银刚等[36]研究认为部分该类细菌参与土壤氮磷循环,有固氮解磷作用,因此,1103P可能有助于改善果园土壤环境。特别地,Brewer等[37]研究认为Candidatus Udaeobacter 是一类需氧异养微生物,有许多氨基酸和维生素营养缺陷,通过牺牲代谢的多功能性以提高效率和相对丰度,本研究中证实4个样品的Candidatus Udaeobacter相对丰度均较高,特别是1103P根际土壤中此类细菌相对丰度最高,可能是1103P 土壤微生物多样性较高引起的。真菌分类学组成比较单一,子囊菌门、毛霉菌门和担子菌门相对丰度比例较高,与Hibbett等[38]研究结果一致,同自根苗相比,1103P显著增加了毛霉菌门的相对丰度比例,毛霉菌门腐生,广泛分布于酒曲、植物残体、腐败有机物、动物粪便和土壤中,可产生蛋白酶、淀粉酶、谷氨酰胺酶等复杂酶系,分解大分子有机物成小分子,可能利于植物根吸收。综上所述,阳光玫瑰不同砧木和自根根际土壤样品门水平上优势菌群较为一致,不同样品优势菌群相对丰度不同,1103P微生物结构可能更有利于根系对土壤营养的吸收。
除了根系分泌物等生物因素,土壤理化情况等非生物因素对根际微生物群落组成也发挥了重要作用。宋雪洁[35]研究表明随着葡萄园有机肥含量增加,土壤细菌多样性增加,放线菌门、浮菌门和拟杆菌门丰度增加,变形菌门、芽单胞菌门和厚壁菌门丰度显著下降。杨敏等[26]研究香格里拉不同葡萄园土壤微生物和土壤理化性质间关系,认为土壤电导率、碱解氮、有机质和速效钾含量是根际细菌群落组成的关键因素,pH 和速效磷含量影响不显著。此外,前人多项研究表明酸杆菌门相对丰度与pH 值呈负相关[39-41],本研究结果与之相一致,再次证实了酸杆菌门可能更适合酸性环境生长,而影响土壤微生物群落组成和分布的环境因素不完全一致,可能原因为根际微生物结构的影响因素较多,除了土壤理化性质,根系分泌物[42-43]、重金属、有机污染物[44-45]等也是重要影响因素。因此,在固定产区、管理方式和根系等情况下,可通过改善土壤营养情况调节微生物群落组成和分布。
4 结 论
砧木可以改变根际土壤微生物多样性和组成,其中,1103P和5BB对根际微生物影响较大,土壤理化性质中有机质、交换性镁含量和pH值对根际土壤优势细菌的影响较大。
[1] TRIVEDI P,LEACH J E,TRINGE S G,SA T,SINGH B K.Plant-microbiome interactions: From community assembly to plant health[J].Nature Reviews Microbiology,2020,18(11):607-621.
[2] WALTERS W A,JIN Z,NICHOLAS Y,WALLACE J G,JESSICA S,ZHANG W,ANTONIO G P,JASON P,OMRY K,SHI Q.Large-scale replicated field study of maize rhizosphere identifies heritable microbes[J].Proceedings of the National Academy of Sciences of the United States of America,2018,115(28):7368-7373.
[3] YUAN J,ZHAO J,WEN T,ZHAO M,LI R,GOOSSENS P,HUANG Q,BAI Y,VIVANCO J M,KOWALCHUK G A,BERENDSEN R L,SHEN Q.Root exudates drive the soilborne legacy of aboveground pathogeninfection[J].Microbiome,2018,6:156.
[4] KANG S M,KHAN A L,WAQAS M,YOU Y H,KIM J H,KIM J G,HAMAYUN M,LEE I J.Plant growth-promoting rhizobacteria reduce adverse effects of salinity and osmotic stress by regulating phytohormones and antioxidants in Cucumis sativus[J].Journal of Plant Interactions,2014,9(1):673-682.
[5] KIM Y C,LEVEAU J H J,GARDENER B B M,PIERSON E A,PIERSON L S,RYU C M.The multifactorial basis for plant health promotion by plant-associated bacteria[J].Applied and Environmental Microbiology,2011,77(5):1548-1555.
[6] KWAK M J,KONG H G,CHOI K,KWON S K,SONG J Y,LEE J,LEE P A,CHOI S Y,SEO M,LEE H J,JUNG E J,PARK H,ROY N,KIM H,LEE M M,RUBIN E M,LEE S W,KIM J F.Rhizoshere microbiome structure alters to enable wilt resistance in tomato[J].Nature Biotechnology,2018,36(11):1100-1109.
[7] WELLER D M,RAAIJMAKERS J M,GARDENER B B M,THOMASHOW L S.Microbial populations responsiblefor specific soil suppressiveness to plant pathogens[J].Annual Review of Phytopathology,2002,40:309-348.
[8] ROSENZWEIG N,TIEDJE J M,QUENSEN J F,MENG Q X,HAO J J J.Microbial communities associated with potato common scab-suppressive soil determined by pyrosequencing analyses[J].Plant Disease,2012,96(5):718-725.
[9] LI X G,DING C F,HUA K,ZHANG T L,ZHANG Y N,ZHAO L,YANG Y R,LIU J G,WANG X X.Soil sickness of peanuts is attributable to modifications in soil microbes induced by peanut root exudates rather than to direct allelopathy[J].Soil Biology and Biochemistry,2014,78:149-159.
[10] MACFARLANE S A.Molecular determinants of the transmission of plant viruses by nematodes[J].Molecular Plant Pathology,2003,4(3):211-215.
[11] XU X L,OUYANG H,RICHTER A,WANEK W,CAO G M,KUZYAKOV Y.Spatio-temporal variations determine plant-microbe competition for inorganic nitrogen in an alpine meadow[J].Journal of Ecology,2011,99(2):563-571.
[12] HEIJDEN M G A,BARDGETT R D,STRAALEN N M.The unseen majority: Soil microbes asdrivers of plant diversity and productivity in terrestrial ecosystems[J].Ecology Letters,2008,11(3):296-310.
[13] HU L F,ROBERT C A M,CADOT S,ZHANG X,YE M,LI B B,MANZO D,CHERVET N,STEINGER T,VAN DER HEIJDEN M G A,SCHLAEPPI K,ERB M.Root exudate metabolites drive plant-soil feedbacks on growth anddefense by shaping the rhizosphere microbiota[J].Nature Communications,2018,9:2738.
[14] 刘京伟,李香真,姚敏杰.植物根际微生物群落构建的研究进展[J].微生物学报,2021,61(2):231-248.LIU Jingwei,LI Xiangzhen,YAO Minjie.Research progress on assembly of plant rhizosphere microbial community[J].Acta Microbiologica Sinica,2021,61(2):231-248.
[15] MARASCO R,ROLLI E,FUSI M,MICHOUD G,DAFFONCHIO D.Grapevine rootstocks shape underground bacterial microbiome and networking but not potential functionality[J].Microbiome,2018,6:1-17.
[16] 袁园园,门洪文,马盼,金仲鑫,王庆杰,翟衡,姚玉新.不同砧木嫁接对‘金手指’葡萄生长和一些生理特性的影响[J].西北农业学报,2015,24(8):110-115.YUAN Yuanyuan,MEN Hongwen,MA Pan,JIN Zhongxin,WANG Qingjie,ZHAI Heng,YAO Yuxin.Impacts if different rootstocks on growth and some physiological characters of‘Gold Finger’grapevine[J].Acta Agriculturae Boreali-Occidentalis Sinica,2015,24(8):110-115.
[17] CORSO M,VANNOZZI A,ZILIOTTO F,ZOUINE M,MAZA E,NICOLATO T,VITULO N,MEGGIO F,VALLE G,BOUZAYEN M,MVLLER M,MUNNÉ-BOSCH S,LUCCHIN M,BONGHI C.Grapevine rootstocks differentially affect the rate of ripening and modulate auxin-related genes in Cabernet Sauvignon berries[J].Frontiers in Plant Science,2016,7:1-14.
[18] 郝燕,马麒龙,张坤,白耀栋,杨瑞.河西走廊不同砧木对‘贵人香’葡萄生长与果实品质的影响[J].果树学报,2017,34(10):1286-1293.HAO Yan,MA Qilong,ZHANG Kun,BAI Yaodong,YANG Rui.Effects of different rootstocks on the growth and fruit quality of‘Italian Riesling’in Hexi Corridor[J].Journal of Fruit Science,2017,34(10):1286-1293.
[19] 李艳,杜远鹏,付艳东,翟衡.不同砧木嫁接的赤霞珠葡萄对淹水的生理响应[J].园艺学报,2013,40(11):2105-2114.LI Yan,DU Yuanpeng,FU Yandong,ZHAI Heng.Physiological responses of waterlogging on different rootstock combinations of Cabernet Sauvignon grape[J].Acta Horticulturae Sinica,2013,40(11):2105-2114.
[20] 王静,刘艳梅,王春梅,张明莉,王小娟,朱列霞.不同樱桃砧木根际微生物和养分的动态变化[J].西北农业学报,2015,24(1):123-129.WANG Jing,LIU Yanmei,WANG Chunmei,ZHANG Mingli,WANG Xiaojuan,ZHU Liexia.Dynamic changes of rhizosphere soil microbial biomass and nutrition of different cherry rootstocks[J].Acta Agriculturae Boreali-Occidentalis Sinica,2015,24(1):123-129.
[21] 徐龙晓,荀咪,宋建飞,田孝志,殷方鹏,黄伟男,张玮玮,杨洪强.土壤质地和砧木对苹果根际微生物功能多样性及其碳源利用的影响[J].园艺学报,2020,47(8):1530-1540.XU Longxiao,XUN Mi,SONG Jianfei,TIAN Xiaozhi,YIN Fangpeng,HUANG Weinan,ZHANG Weiwei,YANG Hongqiang.Effect of Soil textures and rootstock on rhizosphere microorganism and carbon source utilization of apple roots[J].Acta Horticulturae Sinica,2020,47(8):1530-1540.
[22] CLEGG C D,RITZ K,GRIFFITHS B S.%G + C profiling and cross hybridisation of microbial DNA revealsgreat variation in below-ground community structure in UK upland grasslands[J].Applied Soil Ecology,2000,14(2):25-134.
[23] 邵微,于会丽,张培基,徐国益,乔宪生,高登涛,王志强,田鹏,司鹏.不同落叶果树根际微生物群落代谢与组成的差异性研究[J].果树学报,2020,37(9):1371-1383.SHAO Wei,YU Huili,ZHANG Peiji,XU guoyi,QIAO Xiansheng,GAO Dengtao,WANG Zhiqiang,TIAN Peng,SI Peng.Differences in metabolism and composition of microbial communities in rhizosphere soils with different deciduous fruit trees[J].Journal of Fruit Science,2020,37(9):1371-1383.
[24] 孙茜.四种葡萄砧木抗旱、耐盐碱性的比较研究[D].银川:宁夏大学,2014.SUN Qian.Comparative studies on drought-tolerance,salt-tolerance and alkalinity tolerance of four Vitis rootstocks[D].Yinchuan:Ningxia University,2014.
[25] URBANOVΆ M,ŠNAJDR J,BALDRIAN P.Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees[J].Soil Biology and Biochemistry,2015,84:53-64.
[26] 杨敏,殷绒,张国涛,邵建辉,杜飞,邓维萍,朱书生.基于高通量测序技术的香格里拉葡萄酒产区根际微生物多样性研究[J].云南农业大学学报(自然科学),2020,35(3):392-400.YANG Min,YIN Rong,ZHANG Guotao,SHAO Jianhui,DU Fei,DENG Weiping,ZHU Shusheng.Study on the grape rhizosphere microbial diversity in ShangriLa wine region by highthroughput sequencing technology[J].Journal of Yunnan Agricultural University(Natural Science),2020,35(3):392-400.
[27] BERENDSEN R L,PIETERSE C M J,BAKKER P A H M.The rhizosphere microbiomeand plant health[J].Trends in Plant Science,2012,17(8):478-486.
[28] 张锡洲,李廷轩,王永东.植物生长环境与根系分泌物的关系[J].土壤通报,2007,38(4):785-789.ZHANG Xizhou,LI Tingxuan,WANG Yongdong.Relationship between growth environment and root exudates of plants:A review[J].Chinese Journal of Soil Science,2007,38(4):785-789.
[29] BAETZ U,MARTINOIA E.Root exudates:The hidden part of plant defense[J].Trends in Plant Science,2014,19(2):90-98.
[30] FANG C,ZHUANG Y,XU T,LI Y,LI Y,LIN W.Changes in rice allelopathy and rhizosphere microflora by inhibitingrice phenylalanine ammonia- lyase gene expression[J].Journal of Chemical Ecology,2013,39(2):204-212.
[31] MICHALET S,ROHR J,WARSHAN D.BARDON C,ROGGY J C,DOMENACH A M,CZARNES S,POMMIER T,COMBOURIEU B,GUILLAUMAUD N,BELLVERT F,COMTE G,POLY F.Phytochemical analysis of mature tree root exudates in situ and their role in shaping soil microbial communities in relation to tree N-acquisition strategy[J].Plant Physiology Biochemistry,2013,72:169-177.
[32] PANKRATOV T A,IVANOVA A O,OEDYSH S N,LIESACK W.Bacterial populations and environmental factors controlling cellulose degradation in an acidic Sphagnum peat[J].Environmental Microbiology,2011,13(7):1800-1814.
[33] KANOKRATANA P,UENGWETWANIT T,RATTANACHOMSRI U,BUNTERNGSOOK B,NIMCHUA T,TANPHATSORNRUANG S,PLENGVIDHYA V,CHAMPREDA V,EURWILAICHITR L.Insights into the phylogeny and metabolic potential of a primary tropical peat swamp forest microbial community by metagenomic analysis[J].Microbial Ecology,2011,61(3):518-528.
[34] 刘芳.葡萄长期种植对根际土壤微生物群落的影响[D].昆明:云南大学,2020.LIU Fang.Effects of long term planting of grapevines on the rhizosphere soil microbiota[D].Kunming:Yunnan University,2020.
[35] 宋雪洁.葡萄园土壤细菌多样性研究[D].济南:齐鲁工业大学,2017.SONG Xuejie.The study on bacterial diversity of vineyard soil[D].Jinan:Qilu University of Technology,2017.
[36] 薛银刚,刘菲,周璐璐,金珊,姜逸,王颖聪,江晓栋,王倩,施昕澜,薛柯.基于高通量测序的工业园区地下水和土壤细菌群落结构比较研究[J].生态毒理学报,2017,12(6):107-115.XUE Yingang,LIU Fei,ZHOU Lulu,JIN Shan,JIANG Yi,WANG Yingcong,JIANG Xiaodong,WANG Qian,SHI Xinlan,XUE Ke.Comparison study of bacterial community structure between groundwater and soil in industrial park based on high throughput sequencing[J].Asian Journal of Ecotoxicology,2017,12(6):107-115.
[37] BREWER T E,HANDLEY K M,CARINI P,GILBERT J A,FIERER N.Genome reduction in an abundant and ubiquitous soil bacterium‘Candidatus Udaeobacter copiosus’[J].Nature Microbiology,2017,2:16198.
[38] HIBBETT D S,BINDER M,BISCHOFF J F,BLACKWELL M,CANNON P F,ERIKSSON O E,HUHNDORF S,JAMES T,KIRK P M,LÜCKING R,LUMBSCH H T,LUTZONI F,MATHENY B,MCLAUGHLIN D J,POWELL M J,REDHEAD S,SCHOCH C L,SPATAFORA J W,ZHANG N.A higher-level phylogenetic classification of the fungi[J].Mycological Research,2007,111(5):509-547.
[39] SHEN C,XIONG J,ZHANG H,FENG Y,LIN X,LI X,LIANG W,CHU H.Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain[J].Soil Biology and Biochemistry,2013,57:204-211.
[40] JONES R T,ROBESON M S,LAUBER C L,HAMADY M,KNIGHT R,FIERER N.A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses[J].The ISME Journal,2009,3(4):442-453.
[41] 高秀宏,李敏,卢萍,吕桂芬,牛艳芳.呼和浩特市大青山白桦根际土壤细菌群落结构研究[J].生态学报,2019,39(10):3586-3596.GAO Xiuhong,LI Min,LU Ping,LÜ Guifen,NIU Yanfang.Bacterial community in the rhizosphere soil of Betula platyphylla in the Daqing Mountains,Hohhot[J].Acta Ecologica Sinica,2019,39(10):3586-3596.
[42] BADRI D V,WEIR T L,LELIE D,VIVANCO J M.Rhizosphere chemical dialogues: Plant-microbeinteractions[J].Current Opinion in Biotechnology,2009,20(6):642-650.
[43] 李冬洁.植物根系分泌物与根际微生物的相互作用[J].广东蚕业,2018,52(4):21.LI Dongjie.Interaction between plant root exudates and rhizospheremicroorganisms[J].GuangdongSericulture,2018,52(4):21.
[44] GANS J,WOLINSKY M,DUNBAR J.Computational improvements reveal great bacterial diversity and high metal toxicity in soil[J].Science,2005,309(5739):1387-1390.
[45] TAN Y,CUI Y,LI H,KUANG A,LI X,WEI Y,JI X.Rhizospheric soil and root endogenous fungal diversity and composition in response to continuous Panax notoginseng cropping practices[J].Microbiological Research,2017,194:10-19.