干旱是全球最常见的极端气候事件,是人类当前面临的最严峻的挑战之一[1]。目前,干旱半干旱地区约占全球陆地总面积的41%,全球变暖、森林砍伐和城市化将导致未来全球干旱发生频率持续增加[2]。干旱限制了全球植物的地理分布,严重影响其生长和发育[3]。因此,揭示植物干旱胁迫响应机制,挖掘抗旱有关基因和代谢途径对农业生产实践意义重大。
在漫长的进化过程中,植物逐渐形成了从表型特征到内部生理生化反应、基因表达以及蛋白合成等变化来抵御和适应干旱逆境的机制[4]。表型及生理生化方面主要表现为在干旱胁迫下植物株高下降、叶片萎蔫、根系发达和气孔闭合等表型的变化以及光合系统、渗透调节物质、抗氧化酶活性和内源激素含量等生理的变化[5-6]。分子层面主要表现为鉴定和克隆了一些受干旱胁迫诱导表达的基因,并根据功能将这些基因分成2 类[7]:一类是功能蛋白基因,包括糖代谢调节基因、渗透物质合成酶基因、转运蛋白基因、抗氧化保护剂基因及分子伴侣基因等,它们在植物抗旱性中直接起保护作用[8];另一类是转录因子,参与调控干旱胁迫的信号转导和调控基因表达[9],主要有DREB[10]、MYB[11]、bZIP[12]、WRKY[13]和bHLH[14]。植物响应干旱胁迫是极其复杂的过程,植物抗旱性是由多基因控制的综合性状(数量性状),仅研究植物干旱胁迫下的表型变化、生理生化反应及个别基因的功能验证不足以全面揭示植物的抗旱机制[15]。目前,转录组测序技术已广泛应用于植物响应干旱胁迫的研究,如烟草[16]、葡萄[17]、高粱[18]、马缨杜鹃[19]和苹果[20]等。通过以上研究发现,许多与光合作用、信号转导、类黄酮合成和抗氧化酶等有关基因均参与植物对干旱胁迫的分子响应[21]。
火龙果(Hylocereus spp.)系仙人掌科(Cactaceae)量天尺属(Hylocereus)果用栽培植物,耐旱性强,是挖掘植物抗旱基因的理想材料[22]。前人关于火龙果响应干旱胁迫的研究多涉及生理生化和表型结构分析,也有利用芯片技术检测干旱相关表达序列标签[23];其他报道还包括火龙果抗旱相关基因的克隆及其在各种非生物胁迫下的表达水平[24]、利用超表达技术验证火龙果抗旱相关基因[25]、火龙果分子标记开发[26]及重要的表观遗传现象之甲基化[27]等。组学技术是从系统生物学层面研究植物干旱胁迫协同应答网络机制的重要手段,然而迄今为止对火龙果转录组水平的研究报道还很少。目前,仅有齐钊[28]以火龙果大红品种为试材,对自然干旱下火龙果根和茎进行了转录组测序,筛选了一些差异表达基因(differential expressed genes,DEGs)。众所周知,植物对干旱胁迫的响应存在品种差异性[6],同时,自然干旱是缓慢且渐进的过程,涉及变化的因素很多,不易控制,相比较而言,聚乙二醇(polyethylene glycol,PEG)作为不可吸收、不可代谢且无毒的渗透剂,其胁迫程度可定量,因此更适用于植物干旱胁迫响应的分子机制研究。笔者以贵州喀斯特山地火龙果高抗旱种质紫红龙(Hylocereus spp.‘Zihonglong’)为试材,比较分析了其在正常供水和PEG模拟的干旱胁迫(-4.9 MPa)条件下的丙二醛(malondialdehyde,MDA)含量、过氧化氢酶(catalase,CAT)和过氧化物酶(peroxidase,POD)活性变化及转录组差异,以期对前人研究结果进行补充,进一步挖掘与植物干旱胁迫响应相关的候选基因和途径,深入认识其对干旱胁迫的响应机制。该研究结果对抗旱种质培育具有重要理论意义。
1 材料和方法
1.1 试验材料
试验材料采用火龙果品种紫红龙(Hylocereus spp.‘Zihonglong’),为确保供试植株遗传背景一致,所有材料均为火龙果幼苗无性系,火龙果幼苗无性系创制参考Nie等[24]的方法。
1.2 试验设计
供试植株在MS培养基+0.1 mg·L-1 NAA+30 g·L-1蔗糖+7 g·L-1琼脂的培养基上生长28 d后,将大小一致(高度7~8 cm)的幼苗转移至Hoagland 培养液中进行预培养[29],每3 d 更换1 次营养液,气泵提供氧气。为了模拟干旱胁迫,将预培养14 d 后的火龙果幼苗转移到含有PEG-6000(-0.49 MPa)的霍格兰(Hoagland’s)营养液中,连续培养7 d,以仅含Hoagland营养液的处理为对照,分别于0、6 h、12 h、18 h、1 d、3 d、5 d、7 d 采集胁迫组和对照的火龙果肉质茎,立即液氮冷冻,-80 ℃贮存,用于生理指标测定,采集6 h和3 d火龙果肉质茎(干旱胁迫6 h和3 d的火龙果幼苗分别命名为OS6H和OS3D,相应的对照分别命名为NS6H和NS3D)进行转录组测序,3次生物学重复,每天光照14 h,光通量密度300 μmol·m-2·s-1,昼夜恒温为(25±1)℃,相对湿度(60±5)%。
1.3 MDA含量、CAT和POD活性测定
MDA含量采用硫代巴比妥酸法测定[30],532 nm吸光值计算;POD 活性参照Wang 等[31]的方法测定,470 nm 吸光值计算;CAT活性参照Sima等[32]的方法测定,240 nm 吸光值计算。采用Microsoft Excel 2007 软件记录和整理数据,DPS 7.05 进行单因素方差分析和Duncan新复极差法进行多重比较,差异显著性定义为p <0.05 或p <0.01,数据表示为3 次生物学重复的平均值±标准差(SE)。
1.4 cDNA文库的构建及转录组测序
采用RNAsimple 总RNA 提取试剂盒(Cat. NO.DP419)分别从12 份火龙果肉质茎样品中提取总RNA,具体步骤参见说明书。采用NanoPhotometerTM 超微量分光光度计(Implen GmbH,Munich,Germany)检测RNA 浓度、纯度及是否存在污染,采用Agilent 2100 生物分析仪(Agilent Technologies,Palo Alto,Calif。)分析RNA完整性。样品检测合格后,将胁迫组(OS6H 和OS3D)和对照(NS6H 和NS3D)的合格RNA等量混合到相应的文库中,用带有Oligo(dT)的磁珠富集mRNA,加入Fragmentation缓冲液裂解mRNA成短片段。以mRNA为模板,用6-base随机引物反转录合成第一链cDNA,然后加入缓冲液、dNTPs 和DNA 聚合酶Ⅰ合成二链cDNA。经过纯化的双链cDNA 经损伤修复、末端修复以及A尾连接测序接头,AMPure XP beads筛选片段后进行PCR 富集,得到最终的cDNA 文库。使用Qubit 2.0 对构建的12 个cDNA 文库进行初步定量,Agilent 2100 生物分析仪检测文库的Insert size,实时荧光定量PCR(quantitative real-time PCR,qRT-PC)方法确保各个文库有效物质的量>2 nmol。将12 个cDNA 文库按照预计下机数据量Pooling,在Illumina 测序平台测序,对获得的Raw data 进行数据过滤,去除接头Linker 序列和低质量Reads 获得高质量的Clean data。该Clean data 经过序列组装,获得火龙果品种紫红龙的Unigene库。
1.5 基因的注释和表达水平分析
使用BLAST 软件将Unigene 序列与基因本体论数据库(gene ontology,GO)、东京基因与基因组百科全书(kyoto encyclopedia of genes and genomes,KEGG)、蛋白质真核同源数据库(eukaryotic orthologous groups,KOG)、蛋白质序列数据库(swiss-prot protein database,Swiss-Prot)、蛋白质家族域数据库(protein families,Pfam)和非冗余蛋白质数据库(NCBI non-redundant protein database,NR)进行比对。使用KOBAS2.0 获得Unigene 序列在KEGG 中的KEGG orthology结果并预测Unigene序列的氨基酸序列。使用HMMER 软件与Pfam 数据库比对获得Unigene 序列的注释信息。使用FPKM(fragments per kilobase of transcript per million mapped reads)公式估算基因的表达水平。
1.6 差异表达基因的筛选
差异倍数(fold change)表示干旱胁迫组与其相应对照组(即OS6H vs NS6H 和OS3D vs NS3D)基因表达量的比值。错误发现率(false discovery rate,FDR)是通过校正差异显著性p 值(p-value)而获得的。采用公认的Benjamini-Hochberg校正方法对原有假设检验得到的显著性p 值(p-value)进行校正,最终以Fold Change≥2 且FDR <0.01 为上调标准,以Fold Change≤0.5 且FDR<0.01 为下调标准来筛选火龙果干旱胁迫下差异表达基因。进一步对这些差异表达基因进行GO功能富集分析和KEGG富集分析,确定差异表达基因显著富集的GO 条目和KEGG代谢通路。
1.7 实时荧光定量PCR分析
为了验证转录组测序的准确可靠性,随机选取12个差异表达基因,分析其qRT-PCR结果是否与转录组结果一致。利用Primer 6.0设计引物,以Actin7为内参基因[25],qRT-PCR 引物序列见表1,数据处理参考Livak等[33]方法,使用2-ΔΔCT计算基因的相对表达量。
表1 用于差异表达基因qRT-PCR 验证的引物
Table 1 Primers for qRT-PCR validation of DEGs

?
2 结果与分析
2.1 干旱胁迫对火龙果MDA含量、CAT和POD活性的影响
在整个处理期间,胁迫组MDA 含量始终高于对照组,胁迫处理6 h 出现第1 个峰值,干旱胁迫处理1 d后,MDA含量先是略有下降,然后又开始升高(图1-A),其中,干旱胁迫6 h 和3 d 火龙果的MDA含量分别比对照提高了约1.32 和1.49 倍(p <0.01);在干旱胁迫处理期间,POD 活性呈先升后降再升高的趋势,胁迫处理12 h出现第1个峰值,随后下降并于1 d 后持续升高,其中,干旱胁迫6 h 和3 d火龙果的POD 活性分别比对照提高了约1.56 和1.61倍(p <0.01)(图1-B);CAT活性随着干旱胁迫处理时间的延长呈波动趋势,胁迫处理12 h 达到第1 个峰值(约为对照的2.25 倍,p <0.01),之后呈下降趋势,在第5 天出现第二个峰值且高于第一个峰值(约为对照的2.02 倍,p <0.01),然后再次下降(图1-C)。综合以上指标,初步确定处于上升期或峰值点的6 h和3 d作为转录组样品的取样点。

图1 火龙果在正常供水和干旱胁迫下的MDA 含量、POD 和CAT 活性变化
Fig.1 Changes in the MDA content,POD and CAT activities in pitaya plants under control and stressed conditions at different time points
图内数据为平均值±标准误;星号表示与对照组相比差异显著(*p <0.05)。
Data are presented as the mean±SE.Asterisks indicate significant difference(*p <0.05)in comparison with the control.
2.2 差异表达基因分析
转录组测序结果显示,2 个比较组中OS6H vs NS6H(6 h 时干旱胁迫与对照的比值)和OS3D vs NS3D(3 d 时干旱胁迫与对照的比值)共鉴定出432个DEGs;干旱胁迫6 h(OS6H vs NS6H)特有DEGs共288 个(88 个上调,200 个下调),占总差异表达基因的66.7%;干旱胁迫3 d(OS3D vs NS3D)特有DEGs 共126个(79个上调,47个下调),占总差异表达基因的29.2%;2 个比较组中共同表达的DEGs 有18个(12个共同上调,4个共同下调,2个表达模式相反),占总差异表达基因的4.2%(图2)。
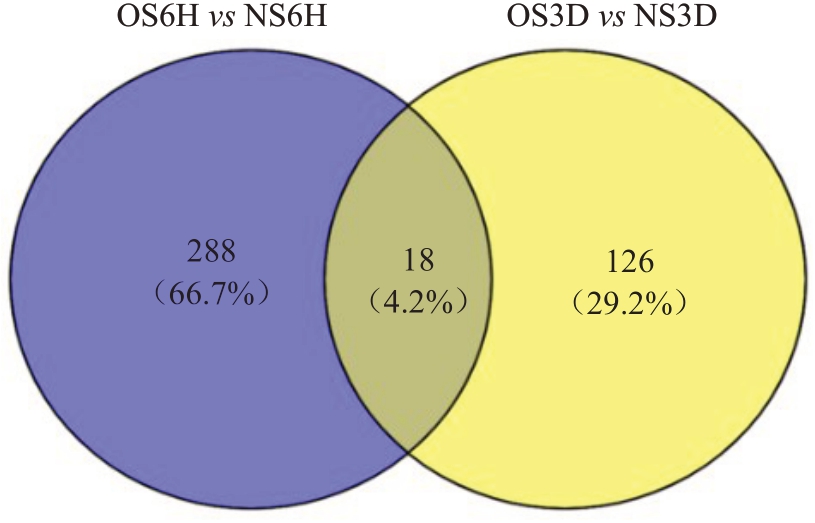
图2 不同比较组差异表达基因韦恩图
Fig.2 Venn diagrams of DEGs among different comparisons
2.3 差异表达基因功能富集分析
GO 富集分析表明,所鉴定的差异表达基因涉及15个生物学过程、10个细胞组分和8个分子功能(图3)。差异表达基因富集到生物学过程中的主要条目有代谢过程和细胞学过程;差异表达基因富集到分子功能的主要条目有催化活性和结合;差异表达基因富集到细胞组分的主要条目有膜、膜组分、细胞和细胞组分。OS6H vs NS6H 比较组比OS3D vs NS3D富集到的差异表达基因数目更多。
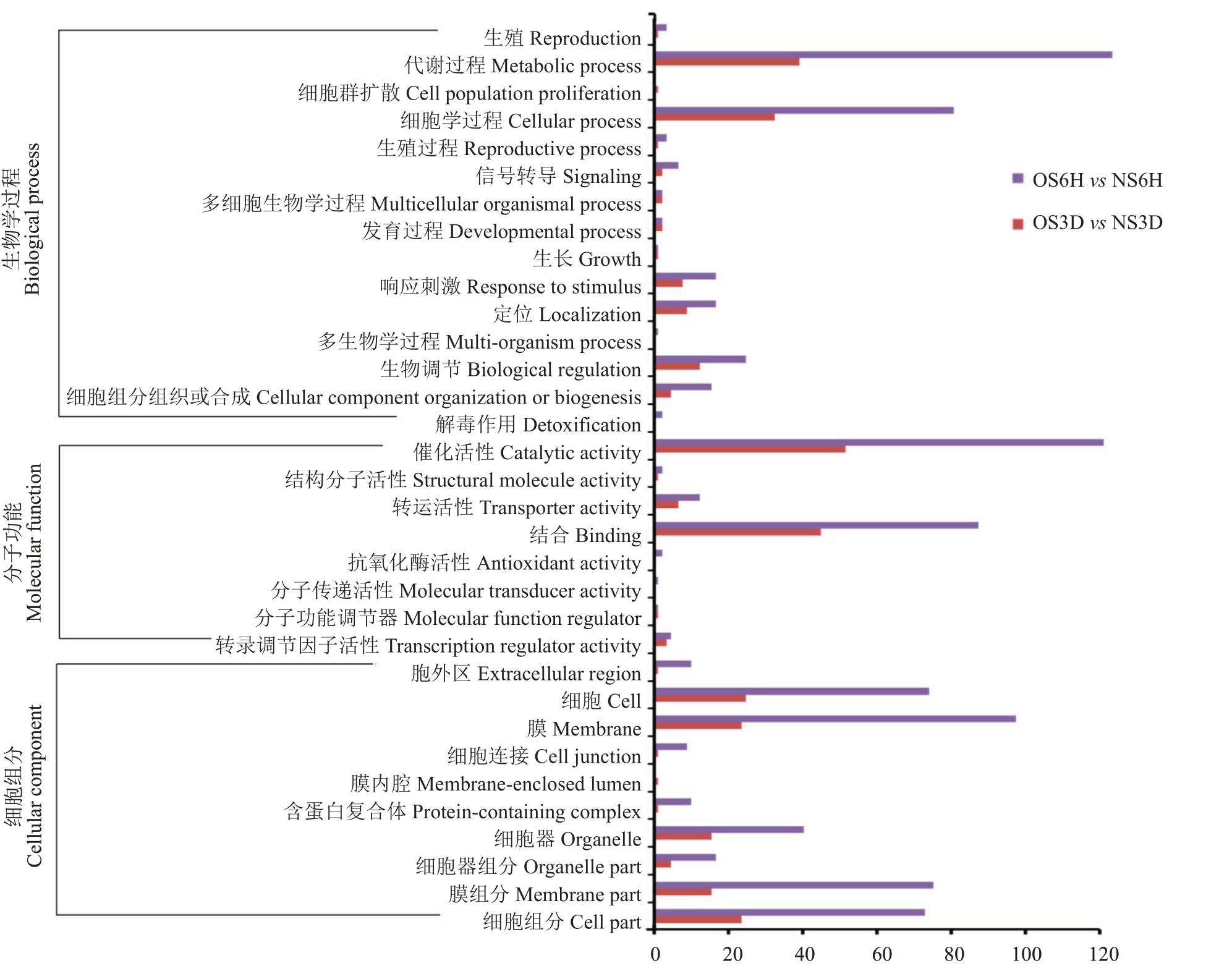
图3 差异表达基因的GO 分析
Fig.3 Gene ontology(GO)analysis of DEGs
KEGG通路富集分析发现,OS6H vs NS6H比较组最显著富集的通路依次是淀粉和蔗糖代谢、光合作用-天线蛋白、苯丙烷生物合成、氰基氨基酸代谢、二芳基庚烷和姜醇的生物合成等(图4-A);OS3D vs NS3D 比较组最显著富集的通路依次是丙氨酸、天冬氨酸和谷氨酸代谢、淀粉和蔗糖代谢、氰基氨基酸代谢、苯丙烷生物合成、β-丙氨酸代谢等(图4-B)。其中,淀粉和蔗糖代谢、苯丙烷生物合成、氰基氨基酸代谢、二芳基庚烷和姜醇的生物合成、类黄酮生物合成、β-丙氨酸代谢和植物激素信号转导这8 条途径为2 个比较组所共有。在GO 和KEGG 分析的基础上,主要关注了碳水化合物代谢、氨基酸代谢、次生代谢、脂质代谢、翻译和运输及信号转导相关的差异表达基因。
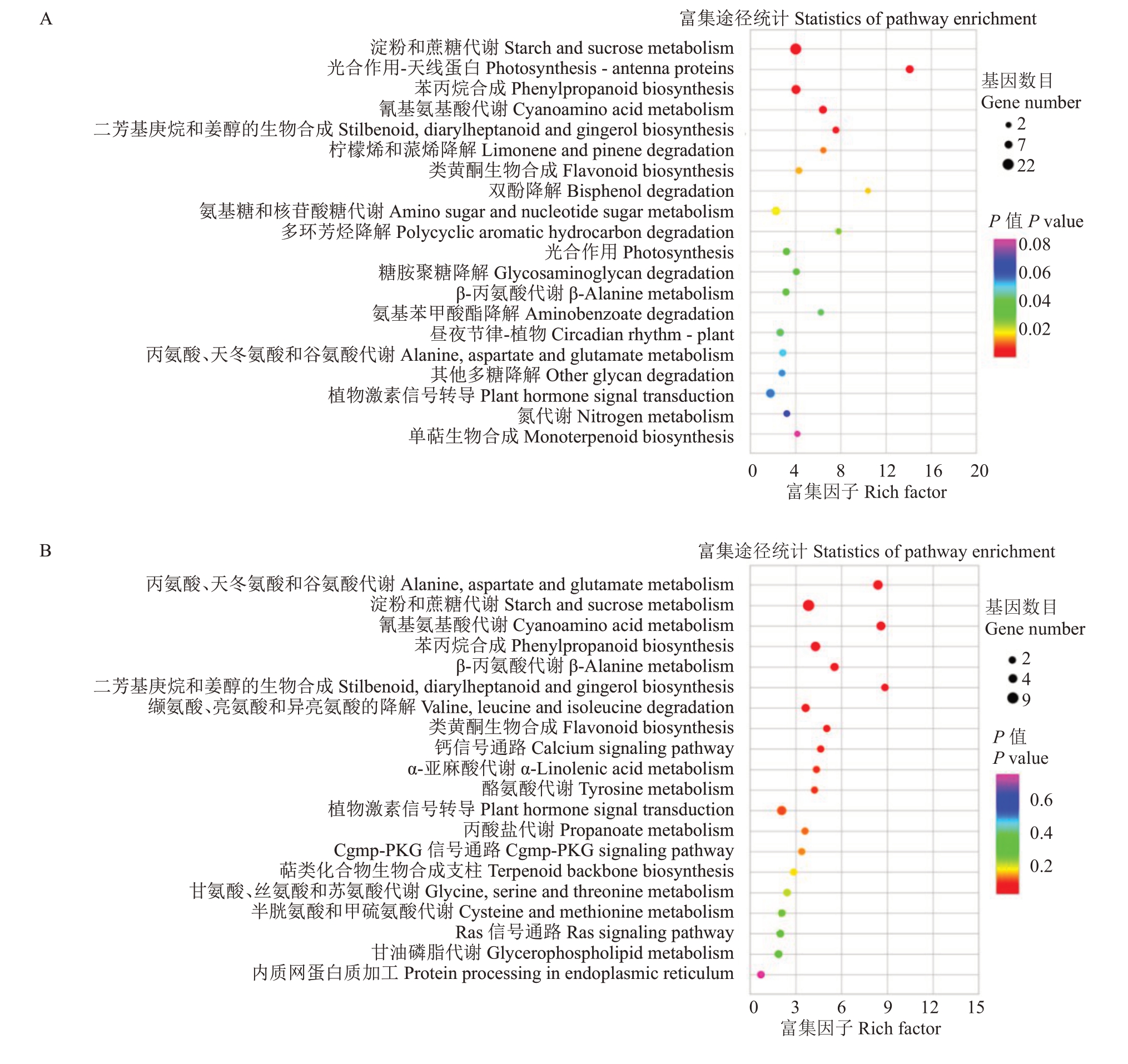
图4 干旱胁迫条件下火龙果茎DEGs 的KEGG 分析
Fig.4 KEGG analysis of DEGs in pitaya stems under drought stress conditions
A.干旱胁迫6 h;B.干旱胁迫3 d。
A.Drought stress conditions at 6 h.B.Drought stress conditions at 3 d.
2.4 碳水化合物代谢相关差异表达基因
有17个DEGs与碳水化合物代谢相关,其中,11个参与淀粉和蔗糖代谢,3 个参与糖酵解,3 个参与丙酮酸代谢(表2)。在干旱胁迫下,火龙果淀粉和蔗糖代谢的关键酶基因如1,4-a-葡聚糖分支酶3基因(SBE3)、糖基转移酶基因(PHO1)、4-α-葡聚糖转移酶基因(DPEP)和非活性-β-淀粉酶基因(BAM9)的积累在干旱胁迫6 h 显著增加,α-1,4 葡聚糖磷酸化酶亚基基因(TPPD)和海藻糖酶基因(Os10g0521000)在干旱胁迫3 d 显著增加。2 个多聚半乳糖醛酸抑制蛋白基因(PGIP)在2组比较中均显著下调,而果胶酯酶基因(PECS-2.1)仅在干旱胁迫6 h显著下调。此外,参与糖酵解的ATP依赖型6磷酸果糖激酶基因(PFK6)、乙醛脱氢酶基因(ALDH3F1)以及参与丙酮酸代谢的苹果酸合成酶基因(MLS)、磷酸烯醇式丙酮酸羧化酶基因(PPC1)在干旱胁迫6 h 的积累均显著增加,乙醇脱氢酶3 基因(ADH)在2组比较中均显著上调。
表2 干旱胁迫下火龙果碳水化合物代谢相关差异表达基因
Table 2 DEGs involved in carbohydrate metabolism in pitaya under drought stress
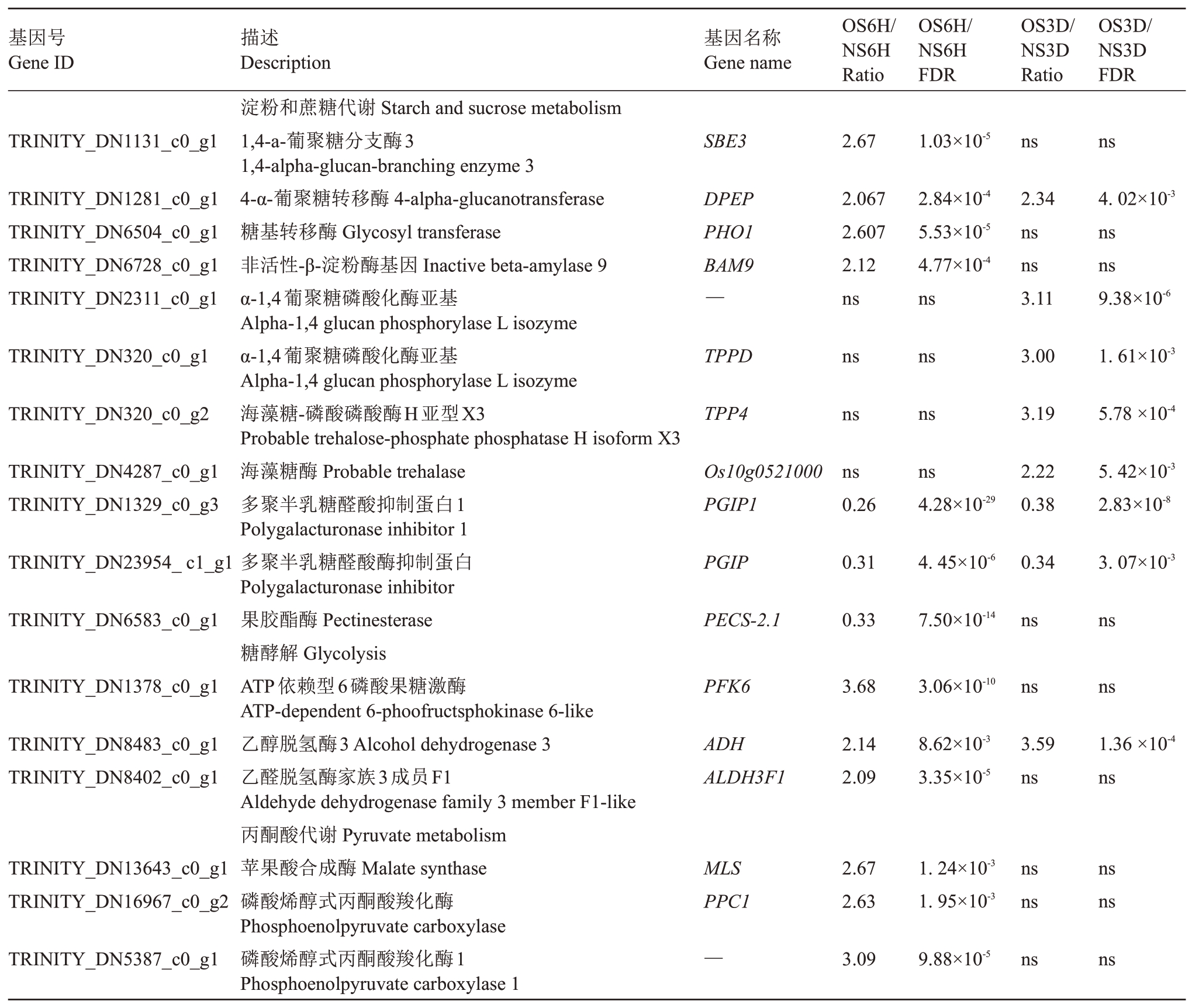
注:“—”表示未找到或不存在。“ns”表示差异不显著。下同。
Note:“—”Not found or not exist.“ns”There is no significant difference.The same below.
?
2.5 氨基酸代谢相关差异表达基因
干旱胁迫诱导的上调DEGs中,有8个参与了氨基酸代谢(表3)。丙氨酸-乙醛酸转氨酶2 同源物3基因(At3g08860)和同型半胱氨酸S甲基转移酶3基因(HMT3)在2 组比较中均显著上调。参与谷氨酸代谢的谷氨酸合酶基因(GLU1)和参与谷胱甘肽代谢的谷胱甘肽s-转移酶T1 亚型X1 基因(GSTT1)仅在OS6H vs NS6H 的比较组中显著上调,参与谷氨酸代谢的伴侣蛋白基因(ATJ11)仅在OS3D vs NS3D的比较组中显著上调,谷氨酸-乙醛酸氨基转移酶2基因(GGAT2)仅在OS3D vs NS3D 的比较组中显著下调,参与谷氨酸代谢的类谷氨酸脱羧酶基因(GAD)在OS6H vs NS6H 的比较组中显著下调,在OS3D vs NS3D的比较组中显著上调。
表3 干旱胁迫下火龙果氨基酸代谢相关差异表达基因
Table 3 DEGs involved in amino acid metabolism in pitaya under drought stress

?
2.6 次生代谢相关差异表达基因
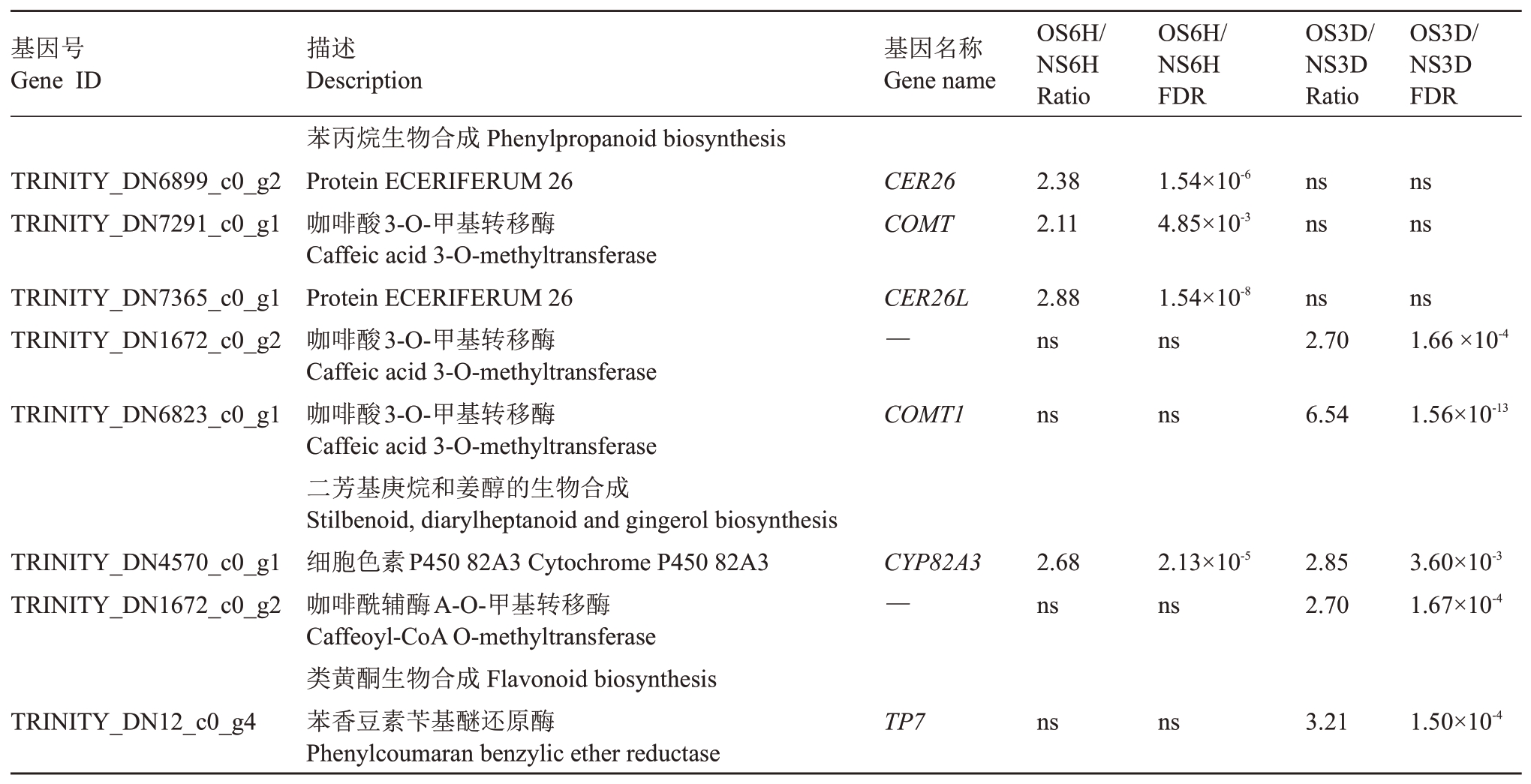
干旱胁迫也促进了次生代谢相关基因表达(表4)。细胞色素P450 基因(CYP82A3)和咖啡酸3-O-甲基转移酶基因(COMT)在2 个比较组中均显著上调,参与苯丙烷生物合成的2 个Protein ECERIFERUM 26 基因(CER26)仅在OS6H vs NS6H 的比较组中显著上调,咖啡酰辅酶A-O-甲基转移酶基因(caffeoyl-CoA-O-methyltransferase)和参与类黄酮生物合成的苯香豆素苄基醚还原酶基因(TP7)仅在OS3D Vs NS3D比较组中显著上调。
表4 干旱胁迫下火龙果次生代谢相关差异表达基因
Table 4 DEGs involved in secondary metabolism in pitaya under drought stress
?
2.7 脂质代谢、翻译和运输相关差异表达基因
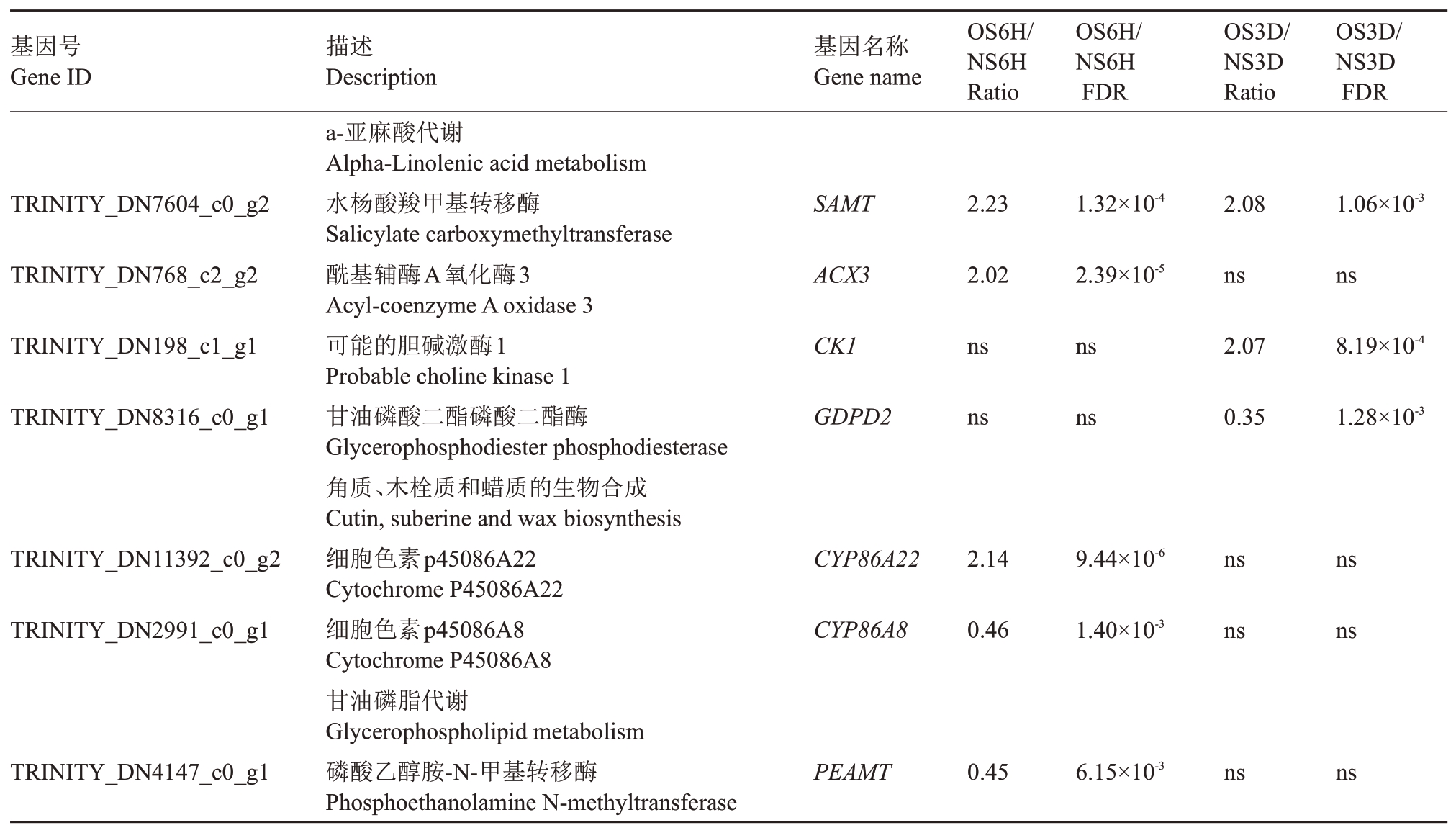
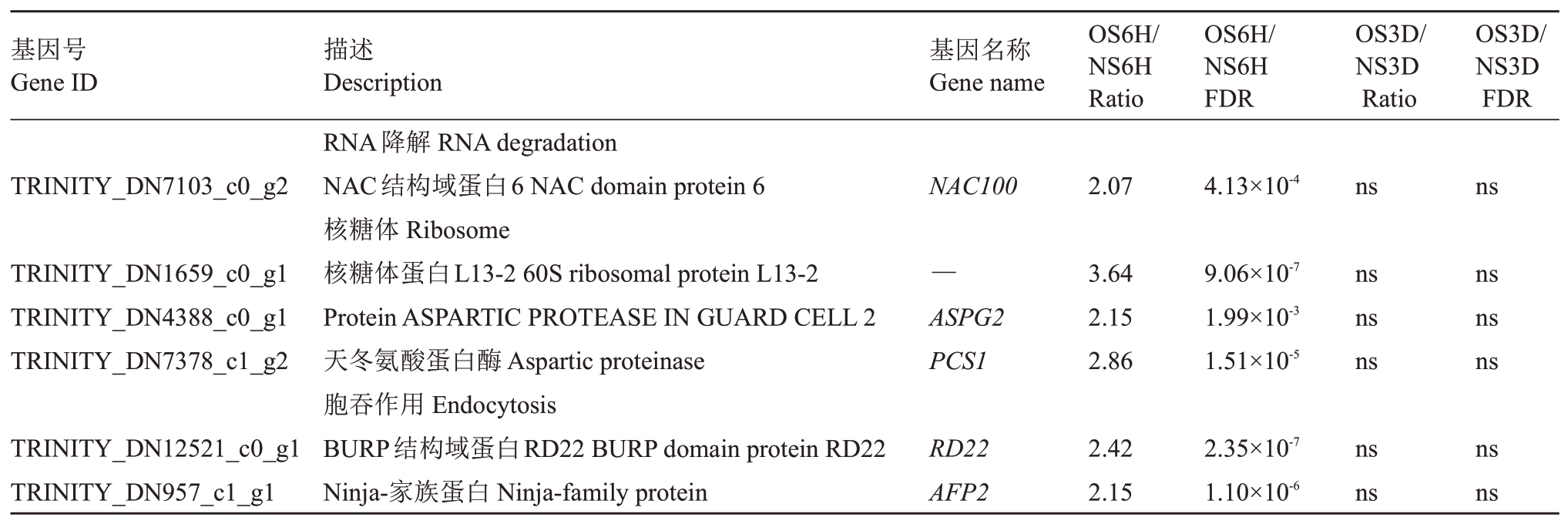
干旱胁迫也引起а-亚麻酸和甘油磷脂代谢相关基因的变化(表5),其中,参与а-亚麻酸代谢的3 个上调基因为水杨酸羧甲基转移酶基因(SAMT)、酰基辅酶A 氧化酶3 基因(ACX3)和可能的胆碱激酶基因(CK1),下调基因为甘油磷酸二酯磷酸二酯酶基因(GDPD2);参与甘油磷脂代谢的磷酸乙醇胺-N-甲基转移酶基因(PEAMT)仅在仅在OS6H vs NS6H的比较组下调表达。参与翻译和运输的差异表达基因仅在OS6H vs NS6H 的比较组中显著上调表达(表6),主要有NAC 结构域蛋白6 基因(NAC100)、60s 核糖体蛋白基因(60S ribosomal protein,L13-2)、天冬氨酸蛋白酶基因(ASPG2 和PCS1)和Ninja-家族蛋白基因(AF2)等。
表5 干旱胁迫下火龙果脂质代谢相关差异表达基因
Table 5 DEGs involved in lipid metabolism in pitaya under drought stress
?
表6 干旱胁迫下火龙果转运和运输相关差异表达基因
Table 6 DEGs involved in translation and transport in pitaya under drought stress
?
2.8 信号转导相关差异表达基因
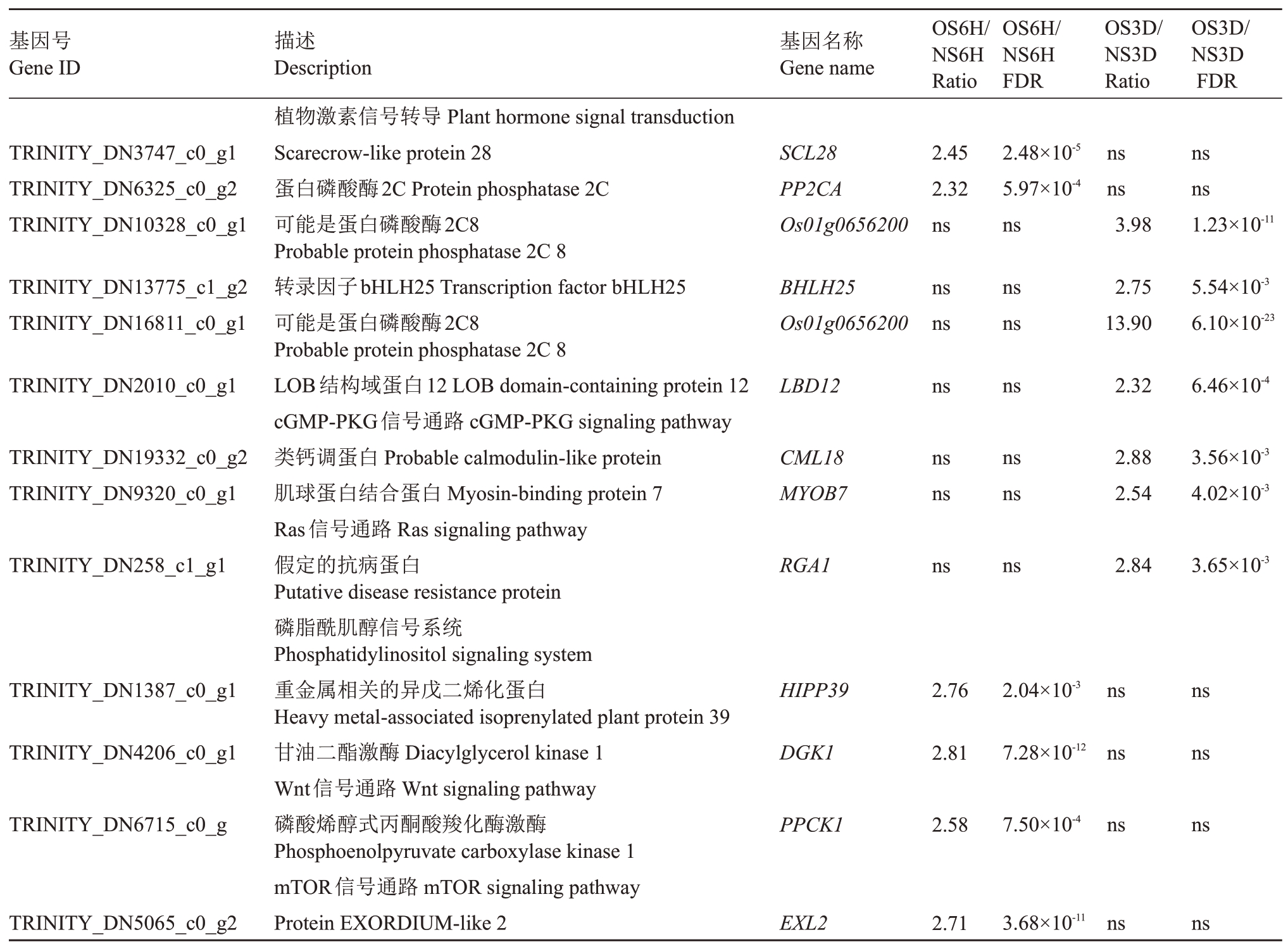
干旱胁迫下,一系列差异响应信号转导的相关基因在火龙果幼苗中被鉴定出来(表7)。干旱胁迫后6 h和3 d共同激活了植物激素信号转导途径,其中,Scarecrow-like protein 28(SCL28)基因和蛋白磷酸酶基因(PP2CA)仅在OS6H vs NS6H 的比较组中显著上调,可能的蛋白磷酸酶2C8 基因(Os01g0656200)、转录因子基因bHLH25 和LOB 结构域蛋白12 基因(LBD12)仅在OS3D vs NS3D 的比较组中显著上调。cGMP-PKG 信号通路相关基因[可能的类钙调蛋白基因(CML18)和肌球蛋白结合蛋白基因(MYOB7)]和Ras 信号通路基因[假定的抗病蛋白基因(RGA1)]仅在OS3D vs NS3D的比较组中显著上调。磷脂酰肌醇信号系统相关基因[重金属相关的异戊二烯化蛋白基因(HIPP39)和甘油二酯激酶基因(DGK1)],Wnt信号通路基因(磷酸烯醇式丙酮酸羧化酶激酶(PPCK1)和mTOR 信号通路基因(EXL2)仅在OS6H vs NS6H的比较组中显著上调。
表7 干旱胁迫下火龙果信号转导相关差异表达基因
Table 7 DEGs involved in signal transduction in pitaya under drought stress
?
2.9 实时荧光定量PCR验证
为验证转录组测序结果,本研究随机挑取了12个DEGs 进行实时qRT-PCR 检测,结果显示这12 个基因的变化趋势与转录组结果基本一致,表明转录组测序结果准确可信(图5)。
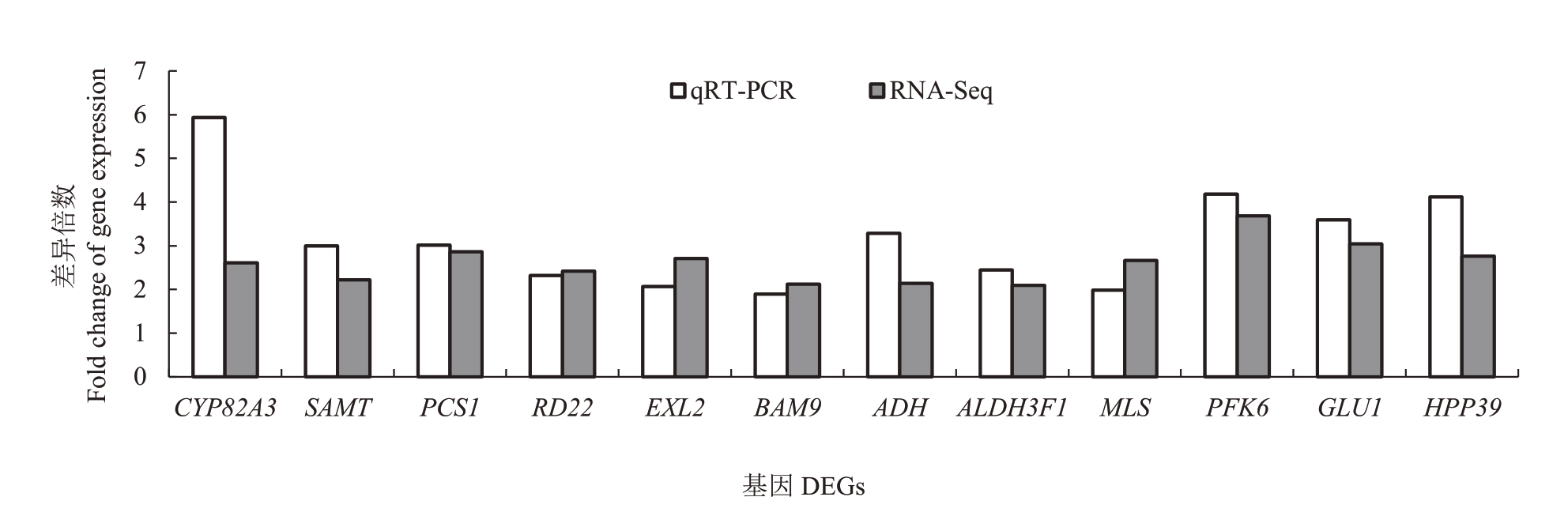
图5 通过qRT-PCR 对12 个DEGs 进行验证
Fig.5 Validation of 12 selected DEGs via qRT-PCR
3 讨 论
3.1 碳水化合物代谢
碳是能量循环和生存所不可或缺的,即使在不利条件下,碳也会被植物不断代谢[34]。淀粉是植物中最重要的碳储备,许多酶参与了淀粉的生物合成和降解[35]。在干旱胁迫下,火龙果淀粉和蔗糖代谢的关键酶如1,4-α-葡聚糖分支酶3(SBE3)、糖基转移酶(PHO1)、4-α-葡聚糖转移酶(DPEP)、非活性β-淀粉酶(BAM9)和α-1,4 葡聚糖磷酸化酶亚基(TPPD)的积累显著增加,促进了淀粉的转化和降解。海藻糖参与植物对逆境胁迫的响应及适应过程,在维持植物体内渗透压、稳定膜系统和参与信号转导等方面发挥着重要的生理调节作用[36]。与前人研究一致,本研究中海藻糖酶相关基因(TPP4 和Os10g0521000)在干旱胁迫3 d后显著上调表达。
糖酵解是所有生物体内碳水化合物代谢的重要代谢途径,本研究有3 个参与糖酵解的酶基因在干旱胁迫下均上调表达,其中,磷酸果糖激酶(PFK6)是糖酵解的限速酶,而乙醇脱氢酶(ADH)在响应非生物胁迫中的转录调控作用已被广泛揭示。Shi等[37]研究发现,AtADH1响应盐、干旱、寒冷和病原体感染等生物和非生物胁迫,AtADH1 的过表达可提升多个胁迫相关基因的转录水平及促进可溶性糖的积累和胼胝质的沉积,AtADH1转基因植物对盐、干旱、寒冷和病原菌感染的抗逆性均有所提高。Khan等[38]报道乙醛脱氢酶在大豆干旱后复苏过程中扮演关键角色,本研究中醛脱氢酶家族3 成员基因(ALDH3F1)也显著上调表达。据报道应激相关的醛脱氢酶对于清除ROS 和催化有毒醛氧化产生无毒的羧酸具有重要作用,此外还参与多种途径并调节多种信号转导[39]。逆境胁迫能够强烈诱导磷酸烯醇丙酮酸羧化酶(PPC)和苹果酸合成酶(MLS)的表达,参与植物对环境的抗逆反应[40]。PPC催化磷酸烯醇式丙酮酸β-羧化生成草酰乙酸[41],MLS 催化乙醛酸与乙酰辅酶A 结合生成苹果酸,苹果酸脱氢重新形成草酰乙酸进入三羧酸循环,它们可能增加了干旱条件下火龙果能量供应,以用于启动其他适应干旱胁迫代谢途径。
3.2 氨基酸代谢
氨基酸代谢途径在水分亏缺时被激活,并在信号转导和渗透调节中发挥重要作用[42]。β-丙氨酸是植物中重要的渗透保护剂,At3g08860 参与β-丙氨酸代谢,与前人研究一致,本研究中At3g08860在渗透胁迫下特异性上调[43]。有研究证实,干旱降低了番薯(Ipomoea batatas)的谷氨酰胺合成酶基因(GS)和glutamine oxoglutarateammo transeferase 基因(GOGAT)的转录水平[44]。也有报道称干旱胁迫下杉木幼苗(Cunninghamia lanceolata)的GS 和GOGAT活性增强[45]。此外,谷氨酸合酶1(GLU1)参与拟南芥缺铁反应和远距离运输[46],本研究的GLU1的上调表达说明其可能在火龙果缓解干旱胁迫的不利影响中发挥着重要作用。谷氨酸脱羧酶(GAD)是催化L-谷氨酸脱羧生成γ-氨基丁酸(GABA)的酶,可通过Ca2+信号转导激活,促进植物在干旱胁迫下GABA 的积累,调节气孔关闭,防止水分流失[47]。本研究中GAD 在干旱胁迫6 h 时显著下调,在干旱胁迫3 d 显著上调,说明植物响应干旱胁迫的复杂性。谷胱甘肽-s-转移酶(GST)广泛存在于植物中,主要参与外源化学物质的代谢解毒和氧化应激[48],转GST基因植株对甘露醇诱导的渗透胁迫的抗性也增强[49],GST 的解毒和抗氧化活性可能是火龙果适应干旱胁迫的重要因素。
3.3 次生代谢
干旱胁迫显著影响了火龙果次生代谢物的合成,本研究中参与苯丙烷生物合成的3个咖啡酸-O-甲基转移酶基因(COMT)和2 个Protein ECERIFERUM 26 基因(CER26)在火龙果干旱胁迫条件下均差异表达。COMT在调节植物生长发育和胁迫反应中起着重要作用,是植物维管细胞壁中控制S亚基合成的木质素单体特异性酶,参与木质素和褪黑素的生物合成[50]。有研究证实COMT1 的积累量在干旱胁迫下普遍减少,另有研究表明TaCOMT 基因在拟南芥中过量表达可促进褪黑素的生成,进而增强转基因植株的抗旱能力[51],本研究中观察到的COMT1 上调表达表明该基因可能参加了褪黑素生物合成途径[52]。VvCCoAOMT是多功能的甲基转移酶,烟草中PoCCoAOMT 的结构性过表达显著增强了烟草对干旱的耐受性。CCoAOMT1 通过调节木质素合成、H2O2积累和ABA信号通路在应对干旱胁迫中发挥积极作用[53]。Protein ECERIFERUM 26(CER26)基因参与茎角质层蜡质的积累[54],本研究中2个CER26基因均上调表达。苯香豆素苯醚还原酶(TP7)被认为是新木质素生物合成的关键酶[55]。有研究表明苯香豆素苯醚还原酶可能参与了木质素类和异黄酮发挥生物活性的关键步骤[56],也有研究报道苯香豆素苯醚还原酶可以降低植物中苯丙二聚体的生成,保护植物免受逆境胁迫产生的氧化损伤[57]。
3.4 脂质代谢
在干旱胁迫下火龙果脂质代谢相关基因差异表达。水杨酸是植物中重要的激素之一,水杨酸在植物抵抗非生物胁迫及植物防御反应中的作用已被广泛证实[58]。水杨酸甲酯(MeSA)生物合成相关基因水杨酸羧甲基转移酶基因(SAMT)的上调表达表明,水杨酸可能在火龙果抗旱中发挥了重要作用。Han等[59]报道细胞色素P450(CYP86A22)是重要的ω-脂酰基辅酶a 羟化酶,参与矮牵牛柱头ω-羟基脂肪酸的生产和三酰基甘油-/二酰基甘油基酯内酯的生物合成。磷酸乙醇胺-N-甲基转移酶(PEAMT)是甜菜碱生物合成途径的关键酶,据报道甜菜碱在长期胁迫的幼嫩组织中合成,并向成熟组织转移[60]。干旱胁迫下,火龙果幼嫩的肉质茎顶部首先失水受到伤害,可能是本研究中PEAMT基因下调的原因。
3.5 翻译和运输
NAC(NAM-ATAF-CUC)转录调控因子参与植物非生物胁迫调控。Greve等[61]首先报道了NAC的结构域,并在矮牵牛NAM 基因、拟南芥ATAF1/2 和CUC2 基因编码的蛋白的N端发现均包含一段高度保守的NAC 结构域,可结合DNA 和其他蛋白。本研究中,NAC结构域蛋白6基因(NAC100)在火龙果干旱胁迫中上调表达,其功能有待进一步证实。天冬氨酸蛋白酶是4 大类蛋白水解酶之一,广泛存在于多种生物体中,这类蛋白酶在植物的整个生长发育过程中,尤其是胁迫反应、衰老、程序性死亡以及蛋白质的加工和降解发挥着重要功能,本研究检测到2 个上调的天冬氨酸蛋白酶基因(ASPG2 和PCS1)。据报道保卫细胞天冬氨酸蛋白酶1 基因(ASPG1)在马铃薯和拟南芥中与干旱胁迫有关,其只在保卫细胞中表达。拟南芥在干旱胁迫条件下,ABA 诱导ASPG1 基因表达,积累的ASPG1 会关闭气孔减少水分流失,同时激活抗过氧化物酶系统保护植物免受氧化损伤[62]。最近很多试验证明拟南芥中的天冬氨酸蛋白酶(PCS1)参与了细胞的程序性死亡,PCS1 的功能缺失使得植物细胞死亡,不能形成有活性的花粉,胚胎也不能正常发育[63]。BURP结构域基因是庞大的植物特异性家族,所有Gh-BURPs 特别是RD22-like 亚家族的成员,都能被不同的胁迫诱导,据报道GmRD22 在转基因拟南芥和转基因水稻中均与细胞壁过氧化物酶相互作用,增加盐胁迫下的木质素产量[64]。火龙果中RD22 基因上调表达说明其可能参与了调节细胞壁过氧化物酶,从而在干旱胁迫条件下增强细胞壁的完整性。
3.6 信号转导
蛋白磷酸酶2C(PP2Cs)是植物对非生物胁迫反应的重要调控因子。在拟南芥中,PP2CA作为脱落酸(ABA)信号的中枢调控因子,负调控植物的生长、发育和对多种逆境的响应[65]。本研究在火龙果中共鉴定出3 个PP2CA 转录因子,它们是火龙果应对干旱胁迫的重要调控因子。钙离子是植物响应逆境胁迫中重要的第二信使,Yang等[66]报道,转甜瓜类钙调蛋白CmCML13 拟南芥通过减少植株的Na+浓度使其抗旱性增强,本研究中可能的类钙调蛋白(CML18)转录因子上调表达表明其可能在火龙果应对非生物胁迫方面发挥着重要作用[67]。碱性螺旋-环-螺旋蛋白(basic helix-loop-helix protein,bHLH)是真核生物中普遍存在的转录因子,通过与靶基因的特定基序相互作用来调控基因表达,bHLH 转录因子广泛参与植物的生长和代谢,在植物对胁迫的响应中发挥着重要作用[14]。干旱胁迫下,拟南芥AtMYC2(bHLH)在脱落酸信号转导中起转录激活作用,Scarecrow-like protein(SCL)是由外源激素诱导的GRAS 家族公认的转录因子,响应干旱和渗透胁迫[68]。干旱胁迫下,火龙果转录因子bHLH25和SCL28均上调表达说明它们参与了火龙果对干旱应答的信号通路。
火龙果磷脂酰肌醇信号通路的2 个基因[重金属相关的异戊二烯化蛋白(HIPP39)和甘油二酯激酶(DGK1)]在干旱胁迫6 h 后上调表达,据报道,重金属相关的异戊二烯化蛋白HIPPs在维管植物发育中起重要作用,也参与重金属稳态和解毒机制,响应寒冷和干旱胁迫以及与植物病原体相互作用。据报道OsHIPP41基因在水稻(Oryza sativa)对寒冷和干旱胁迫的响应中上调表达,其产物定位于细胞质和细胞核中[69]。二酰基甘油激酶(DGK)使二酰基甘油磷酸化生成磷脂酸,二酰基甘油和磷脂酸都是细胞中的脂质介质。研究表明,DGK1 基因在水稻根系生长发育中起着重要作用[70]。拟南芥exodium-like1(EXL1)基因是适应碳和能量限制生长条件所必需的,Schröder等[71]推测,EXL1基因抑制油菜素类固醇依赖的生长,并控制碳在细胞中的分配。磷酸烯醇式丙酮酸羧化酶(PEPC)在光合作用和中间代谢物的生物合成中具有重要作用,其活性是由磷酸烯醇式丙酮酸羧化酶激酶(PPCK)催化的磷酸化控制的,Liu等[72]报道,干旱胁迫下,PPCK活性和转录水平在c4-PEPC 转基因水稻中得到提升,火龙果PPCK1 和Protein EXORDIUM-like 2(EXL2)基因的上调表达说明其参与了火龙果对干旱胁迫的响应和调节。
4 结 论
干旱胁迫启动了火龙果一系列信号转导途径(如类钙调蛋白CML18、二酰基甘油激酶DGK1、蛋白磷酸酶PP2CA 及BHLH25、NAC100、SCL28 转录因子等),调控下游基因表达,通过碳水化合物(淀粉、海藻糖及苹果酸等)的降解和转化、氨基酸(如丙氨酸、谷氨酸、酪氨酸及谷胱甘肽等)代谢及次生代谢(如类黄酮、苯丙烷及二芳基庚烷和姜醇的生物合成)等增强了火龙果的渗透调节和解毒能力,火龙果的高抗旱性归因于其高渗透调节、解毒和抗氧化能力,从而避免了显著的氧化损伤。
[1] 张冰,巩远发,徐影,张帅.CMIP5 全球气候模式对中国地区干旱变化模拟能力评估[J].干旱气象,2014,32(5):694-700.ZHANG Bing,GONG Yuanfa,XU Ying,ZHANG Shuai.Evaluation on the simulation of the drought change in China based on global climate models from CMIP5[J].Journal of Arid Meteorology,2014,32(5):694-700.
[2] 管晓丹,马洁茹,黄建平,黄瑞新,张镭,马柱国.海洋对干旱半干旱区气候变化的影响[J].中国科学:地球科学,2019,49(6):895-912.GUAN Xiaodan,MA Jieru,HUANG Jianping,HUANG Ruixin,ZHANG Lei,MA Zhuguo. Impact of oceans on climate change in drylands[J].Scientia Sinica(Terrae),2019,49(6):895-912.
[3] CAO Y,LUO Q X,TIAN Y,MENG F. Physiological and proteomic analyses of the drought stress response in Amygdalus mira(Koehne)Yü et Lu roots[J].BMC Plant Biology,2017,17(1):1-16.
[4] CLAEYS H,INZÉ D. The agony of choice:how plants balance growth and survival under water-limiting conditions[J]. Plant Physiology,2013,162(4):1768-1779.
[5] ZHAO P J,LIU P,SHAO J F,LI C Q,WANG B,GUO X,YAN B,XIA Y J,PENG M. Analysis of different strategies adapted by two cassava cultivars in response to drought stress:ensuring survival or continuing growth[J]. Journal of Experimental Botany,2014,66(5):1477-1488.
[6] ZHANG C M,SHI S L. Physiological and proteomic responses of contrasting Alfalfa (Medicago sativa L.) varieties to PEG-induced osmotic stress[J].Frontiers in Plant Science,2018,9:242.
[7] MA X W,WANG P,ZHOU S H,SUN Y,LIU N,LI X,HOU Y.De novo transcriptome sequencing and comprehensive analysis of the drought-responsive genes in the desert plant Cynanchum komarovii[J].BMC Genomics,2015,16(1):1-17.
[8] JIANG T B,FOUNTAIN J,DAVIS G,KEMERAIT R,SCULLY B,GUO L B. Root morphology and gene expression analysis in response to drought stress in maize(Zea mays)[J]. Plant Molecular Biology Reporter,2012,30(2):360-369.
[9] SAIBO N J M,LOURENÇO T,OLIVEIRA M M.Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses[J]. Annals of Botany,2009,103(4):609-623.
[10] PAUL S,GAYEN D,DATTA S K,DATTA K. Dissecting root proteome of transgenic rice cultivars unravels metabolic alterations and accumulation of novel stress responsive proteins under drought stress[J].Plant Science,2015,234:133-143.
[11] LI J L,HAN G L,SUN C F,SUI N.Research advances of MYB transcription factors in plant stress resistance and breeding[J].Plant Signaling&Behavior,2019,14(8):1613131.
[12] YING S,ZHANG D F,FU J,SHI Y S,SONG Y C,WANG T Y,YU L. Cloning and characterization of a maize bZIP transcription factor,ZmbZIP72,confers drought and salt tolerance in transgenic Arabidopsis[J].Planta,2012,235(2):253-266.
[13] AGARWAL P,REDDY M P,CHIKARA J. WRKY:its structure,evolutionary relationship,DNA-binding selectivity,role in stress tolerance and development of plants[J]. Molecular Biology Reports,2011,38(6):3883-3896.
[14] SUN X,WANG Y,SUI N. Transcriptional regulation of bHLH during plant response to stress[J]. Biochemical and Biophysical Research Communications,2018,503(2):397-401.
[15] 李瑞雪,孙任洁,汪泰初,陈丹丹,李荣芳,李龙,赵卫国.植物抗旱性鉴定评价方法及抗旱机制研究进展[J].生物技术通报,2017,33(7):40-48.LI Ruixue,SUN Renjie,WANG Taichu,CHEN Dandan,LI Rongfang,LI Long,ZHAO Weiguo. Research progress on identification and evaluation methods,and mechanism of drought resistance in plants[J].Biotechnology Bulletin,2017,33(7):40-48.
[16] YANG H J,ZHAO L,ZHAO S M,WANG J,SHI H.Biochemical and transcriptomic analyses of drought stress responses of LY1306 tobacco strain[J].Scientific Reports,2017,7(1):17442.
[17] XU Y,HUANG B R. Comparative transcriptomic analysis reveals common molecular factors responsive to heat and drought stress in Agrostis stolonifera[J]. Scientific Reports,2018,8(1):15181.
[18] DEVNARAIN N,CRAMPTON B G,OLIVIER N,VAN DER W,ESTHUYZEN C. BECKER J V W,O'KENNEDY M M.Transcriptomic analysis of a Sorghum bicolor Landrace identifies a role for beta-alanine betaine biosynthesis in drought tolerance[J].South African Journal of Botany,2019,127:244-255.
[19] CAI Y F,WANG J H,ZHANG L,SONG J,PENG L C,ZHANG S B. Physiological and transcriptomic analysis highlight key metabolic pathways in relation to drought tolerance in Rhododendron delavayi[J]. Physiology and Molecular Biology of Plants,2019,25(4):991-1008.
[20] GENG D L,LU L Y,YAN M J,SHEN X X,JIANG L J.Physiological and transcriptomic analyses of roots from Malus siever-sii under drought stress[J]. Journal of Integrative Agriculture,2019,18(6):1280-1294.
[21] MA X S,XIA H,LIU Y H,WEI H,ZHENG X,SONG C,CHEN L,LIU H,LUO L.Transcriptomic and metabolomic studies disclose key metabolism pathways contributing to well-maintained photosynthesis under the drought and the consequent drought-tolerance in rice[J].Frontiers in Plant Science,2016,7:1886.
[22] NIE Q,QIAO G,PENG L,WEN X. Transcriptional activation of long terminal repeat retrotransposon sequences in the genome of pitaya under abiotic stress[J]. Plant Physiology and Biochemistry,2019,135:460-468.
[23] FAN Q J,YAN F X,QIAO G,ZHANG B X,WEN X P.Identification of differentially-expressed genes potentially implicated in drought response in pitaya (Hylocereus undatus) by suppression subtractive hybridization and cDNA microarray analysis[J].Gene,2014,533(1):322-331.
[24] NIE Q,GAO G L,FAN Q J,QIAO G,WEN X P,LIU T,PENG Z J,CAI Y Q. Isolation and characterization of a catalase gene“HuCAT3”from pitaya (Hylocereus undatus) and its expression under abiotic stress[J].Gene,2015,563(1):63-71.
[25] 刘小翠.火龙果HubHLH1-like 基因的克隆及功能分析[D].贵阳:贵州大学,2017.LIU Xiaocui. Cloning and function analysis of HubHLH1-like from pitaya(Hylocereus undatus)[D].Guiyang:Guizhou University,2017.
[26] 陶金,乔光,文晓鹏,刘涛,彭志军.火龙果IRAP 分子标记反应体系的建立与优化[J].华中农业大学学报,2014,33(4):33-38.TAO Jin,QIAO Guang,WEN Xiaopeng,LIU Tao,PENG Zhiju.Establishment and optimization of IRAP marker methodology in dragon fruit[J]. Journal of Huazhong Agricultural University,2014,33(4):33-38.
[27] 刘鹏飞,乔光,文晓鹏.火龙果组培苗DNA 甲基化变化及应答赤霉素效应[J].华中农业大学学报,2016,35(5):18-26.LIU Pengfei,QIAO Guang,WEN Xiaopeng. DNA methylation variation of in vitro pitaya shoots and its response to exogenous GA application[J].Journal of Huazhong Agricultural University,2016,35(5):18-26.
[28] 齐钊. 火龙果干旱胁迫下的生理生化及基因差异表达研究[D].海口:海南大学,2018.QI Zhao. Research of physiological and biochemical changes and differentially expressed genes of pitaya under drought stress[D].Haikou:Hainan University,2018.
[29] HOAGLAND D R,ARNON D. The water-culture method for growing plants without soil[M]. California:University of California Agricultural Experiment Station,1938.
[30] CASTREJÓN S E,YATSIMIRSKY A K.Cyclodextrin enhanced fluorimetric determination of malonaldehyde by the thiobarbituric acid method[J].Talanta,1997,44(6):951-957.
[31] WANG L,WANG Y F,WANG X Y,LI Y,PENG F,WANG L.Regulation of POD activity by pelargonidin during vegetative growth in radish(Raphanus sativus L.) [J]. Scientia Horticulturae,2014,174:105-111.
[32] SIMA Y H,YAO J M,HOU Y S,WANG L,ZHAO L C.Variations of hydrogen peroxide and catalase expression in Bombyx eggs during diapause initiation and termination[J]. Archives of Insect Biochemistry and Physiology,2011,77(2):72-80.
[33] LIVAK K J,SCHMITTGEN T D.Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method[J].Methods,2001,25(4):402-408.
[34] HAO P C,ZHU J T,GU A Q,LÜ D,GE P,CHEN G,LI X,YAN Y. An integrative proteome analysis of different seedling organs in tolerant and sensitive wheat cultivars under drought stress and recovery[J].Proteomics,2015,15(9):1544-1563.
[35] BUCHNER P,BORISJUK L,WOBUS U.Glucan phosphorylases in Vicia faba L.:cloning,structural analysis and expression patterns of cytosolic and plastidic forms in relation to starch[J].Planta,1996,199(1):64-73.
[36] 王文静,张悦,余明芳,王棚涛.海藻糖调节植物响应非生物胁迫的研究进展[J].分子植物育种,2020,18(10):3433-3440.WANG Wenjing,ZHANG Yue,YU Mingfang,WANG Pengtao.Research progress of trehalose in regulating plant response to abiotic stress[J]. Molecular Plant Breeding,2020,18(10):3433-3440.
[37] SHI H T,LIU W,YAO Y,WEI Y,CHAN Z.Alcohol dehydrogenase 1(ADH1)confers both abiotic and biotic stress resistance in Arabidopsis[J].Plant Science,2017,262:24-31.
[38] KHAN M N,KOMATSU S. Proteomic analysis of soybean root including hypocotyl during recovery from drought stress[J].Journal of Proteomics,2016,144:39-50.
[39] KIRCH H H,SCHLINGENSIEPEN S,KOTCHONI S,SUNKAR R,BARTELS D. Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana[J]. Plant Molecular Biology,2005,57(3):315-332.
[40] 赵晋锋,杜艳伟,王高鸿,李颜方,赵根有,王振华,王玉文,余爱丽.谷子PEPC 基因的鉴定及其对非生物逆境的响应特性[J].作物学报,2020,46(5):700-711.ZHAO Jinfeng,DU Yanwei,WANG Gaohong,LI Yanfang,ZHAO Genyou,WANG Zhenhua,WANG Yuwen,YU Aili.Identification of PEPC genes from foxtail millet and its response to abiotic stress[J]. Acta Agronomica Sinica,2020,46(5):700-711.
[41] SÁNCHEZ R,FLORES A,CEJUDO F J.Arabidopsis phosphoenolpyruvate carboxylase genes encode immunologically unrelated polypeptides and are differentially expressed in response to drought and salt stress[J].Planta,2006,223(5):901-909.
[42] SMEEKENS S,ROOK F.Sugar sensing and sugar-mediated signal transduction in plants[J].Plant Physiology,1997,115(1):7-13.
[43] PARTHASARATHY A,ADAMS L E,SAVKA F C,HUDSON A O. The Arabidopsis thaliana gene annotated by the locus tag At3g08860 encodes alanine aminotransferase[J]. Plant Direct,2019,3(9):e00171.
[44] XIA H Q,XU T,ZHANG J,SHEN K,LI Z,LIU J.Drought-induced responses of nitrogen metabolism in Ipomoea batatas[J].Plants,2020,9(10):1341.
[45] LI S B,ZHOU L L,ADDO-DANSO S D,DING G,SUN M,WU S,LIN S.Nitrogen supply enhances the physiological resistance of Chinese fir plantlets under polyethylene glycol(PEG)-induced drought stress[J].Scientific Reports,2020,10(1):1-8.
[46] CUI M,GU M J,LU Y R,ZHANG Y,CHEN C,LING HQ,WU H. Glutamate synthase 1 is involved in iron-deficiency response and long-distance transportation in Arabidopsis[J]. Journal of Integrative Plant Biology,2020,62(12):1925-1941.
[47] MEKONNEN D W,FLÜGGE U I,LUDEWIG F. Gamma-aminobutyric acid depletion affects stomata closure and drought tolerance ofArabidopsis thaliana[J].Plant Science,2016,245:25-34.
[48] MA L G,ZHANG Y H,MENG Q L,SHI F,LIU J,LI Y.Molecular cloning,identification of GSTs family in sunflower and their regulatory roles in biotic and abiotic stress[J]. World Journal of Microbiology&Biotechnology,2018,34(8):109.
[49] XU J,XING X J,TIAN Y S,PENG R H,XUE Y,ZHAO W,YAO Q H.Transgenic Arabidopsis plants expressing tomato glutathione S-transferase showed enhanced resistance to salt and drought stress[J].PLoS One,2015,10(9):e0136960.
[50] ZHANG K,CUI H T,CAO S H,YAN L,LI M,SUN Y.Overexpression of CrCOMT from Carex rigescens increases salt stress and modulates melatonin synthesis in Arabidopsis thaliana[J].Plant Cell Reports,2019,38(12):1501-1514.
[51] YANG W J,DU Y T,ZHOU Y B,CHEN J,XU Z S,MA Y Z,CHEN M,MIN D H.Overexpression of TaCOMT improves melatonin production and enhances drought tolerance in transgenic Arabidopsis[J]. International Journal of Molecular Sciences,2019,20(3):652.
[52] LI W,LU J X,LU K,YUAN J,HUANG J,DU H,LI J.Cloning and phylogenetic analysis of Brassica napus L. caffeic acid Omethyltransferase 1 gene family and its expression pattern under drought stress[J].PLoS One,2016,11(11):e0165975.
[53] ZHAO D Q,LUAN Y T,SHI W B,ZHANG X,MENG J,TAO J. A Paeonia ostii caffeoyl-CoA O-methyltransferase confers drought stress tolerance by promoting lignin synthesis and ROS scavenging[J].Plant Science,2021,303:110765.
[54] WANG X,OH M,SAKATA K,KOMATSU S. Gel-free/labelfree proteomic analysis of root tip of soybean over time under flooding and drought stresses[J]. Journal of Proteomics,2016,130:42-55.
[55] NUOENDAGUL A,KAMIMURA N,MORI T,NAKABAYASHI R,TSUJI Y,HISHIYAMA S,SAITO K,MASAI E,KAJITA S. Expression and functional analyses of a putative phenylcoumaran benzylic ether reductase in Arabidopsis thaliana[J]. Plant Cell Reports,2016,35(3):513-526.
[56] MIN T,KASAHARA H,BEDGAR D L,YOUN B,LAWRENCE P K,GANG D R,HALLS S C,PARK H,HILSENBECK J L,DAVIN L B,LEWIS N G,KANG C. Crystal structures of pinoresinol-lariciresinol and phenylcoumaran benzylic ether reductases and their relationship to isoflavone reductases[J].Journal of Biological Chemistry,2003,278(50):50714-50723.
[57] NICULAES C,MORREEL K,KIM H,LU F,MCKEE L S,IVENS B,HAUSTRAETE J,VANHOLME B,RYCKE R D,HERTZBERG M,FROMM J,BULONE V,POLLE A,RALPH J,BOERJAN W. Phenylcoumaran benzylic ether reductase prevents accumulation of compounds formed under oxidative conditions in poplar xylem[J].The Plant Cell,2014,26(9):3775-3791.
[58] DENG B,WANG W J,RUAN C Q,DENG L,YAO S,ZENG K. Involvement of CsWRKY70 in salicylic acid-induced Citrus fruit resistance against Penicillium digitatum[J].Horticulture Research,2020,7:157.
[59] HAN J X,CLEMENT J M,LI J,KING A,NG S,JAWORSKI J G.The cytochrome P450 CYP86A22 is a fatty acyl-CoA omegahydroxylase essential for estolide synthesis in the stigma of Pe-tunia hybrida[J].The Journal of Biological Chemistry,2010,285(6):3986-3996.
[60] ANNUNZIATA M G,CIARMIELLO L F,WOODROW P,DELL’AVERSANA E,CARILLO P. Spatial and temporal profile of Glycine betaine accumulation in plants under abiotic stresses[J].Frontiers in Plant Science,2019,10:230.
[61] GREVE K,LA COUR T,JENSEN M K,POULSEN F M,SKRIVER K. Interactions between plant RING-H2 and plantspecific NAC(NAM/ATAF1/2/CUC2)proteins:RING-H2 molecular specificity and cellular localization[J].The Biochemical Journal,2003,371(1):97-108.
[62] YAO X,XIONG W,YE T T,WU Y.Overexpression of the aspartic protease ASPG1 gene confers drought avoidance in Arabidopsis[J].Journal of Experimental Botany,2012,63(7):2579-2593.
[63] 葛伟娜,李超,张家琦,张岚,王振怡.植物天冬氨酸蛋白酶的研究进展[J].生物技术通报,2016,32(1):8-14.GE Weina,LI Chao,ZHANG Jiaqi,ZHANG Lan,WANG Zhenyi. Research advances on plant aspartic proteinase[J]. Biotechnology Bulletin,2016,32(1):8-14.
[64] WANG H M,ZHOU L,FU Y P,CHEUNG M Y,WONG F L,PHANG T H,SUN Z,LAM H M.Expression of an apoplast-localized BURP-domain protein from soybean(GmRD22)enhances tolerance towards abiotic stress[J]. Plant,Cell & Environment,2012,35(11):1932-1947.
[65] BAEK W,LIM C W,LUAN S,LEE S C. The RING finger E3 ligases PIR1 and PIR2 mediate PP2CA degradation to enhance abscisic acid response in Arabidopsis[J]. The Plant Journal,2019,100(3):473-486.
[66] YANG S L,XIONG X,ARIF S,GAO L,ZHAO L,SHAH I H,ZHANG Y. A calmodulin-like CmCML13 from Cucumis melo improved transgenic Arabidopsis salt tolerance through reduced shoot's Na+,and also improved drought resistance[J].Plant Physiology and Biochemistry,2020,155:271-283.
[67] MA Q P,ZHOU Q Q,CHEN C M,CUI Q,ZHAO Y,WANG K,ARKORFUL E,CHEN X,SUN K,LI X. Isolation and expression analysis of CsCML genes in response to abiotic stresses in the tea plant(Camellia sinensis)[J]. Scientific Reports,2019,9(1):8211.
[68] SOLÉ A,SÁNCHEZ C,VIELBA J M,VALLADARES S,ABARCA D,DíAZ-SALA C. Characterization and expression of a Pinus radiata putative ortholog to the Arabidopsis SHORTROOT gene[J].Tree Physiology,2008,28(11):1629-1639.
[69] DE A J B,TURCHETTO-ZOLET A C,DE O L F V,ZANETTINI M H B,MARGIS-PINHEIRO M. Heavy metal-associated isoprenylated plant protein(HIPP):characterization of a family of proteins exclusive to plants[J]. The FEBS Journal,2013,280(7):1604-1616.
[70] YUAN S,KIM S C,DENG X J,HONG Y,WANG X. Diacylglycerol kinase and associated lipid mediators modulate rice root architecture[J].The New Phytologist,2019,223(1):261-276.
[71] SCHRÖDER F,LISSO J,MÜSSIG C. Expression pattern and putative function of EXL1 and homologous genes in Arabidopsis[J].Plant Signaling&Behavior,2012,7(1):22-27.
[72] LIU X L,LI X,ZHANG C,DAI C,ZHOU J,REN C,ZHANG J. Phosphoenolpyruvate carboxylase regulation in C4-PEPC-expressing transgenic rice during early responses to drought stress[J].Physiologia Plantarum,2017,159(2):178-200.