NAC 转录因子是近些年发现的一类陆生植物特有的转录调控因子,数目众多,是一个庞大的转录因子家族。NAC蛋白N端为高度保守的150~160个氨基酸残基组成的结构域,而C 端则是高度多样化的转录调控区[1]。研究结果表明,NAC 因子在植物生长发育中具有重要的调控作用,包括花期[2]、根系发育[3]、器官衰老[4-6]、激素信号传导[7-8]、响应生物与非生物胁迫(盐/干旱/寒冷)[9-10]以及与病原菌的互作[11-12]等。目前报道显示,NAC 也参与植株果实成熟的各个方面。罗云波等[13]系统研究了NAC 转录因子NOR-like1 对番茄果实成熟的调控网络,表明NOR-like1 能直接作用于乙烯合成、叶绿素降解、类胡萝卜素积累以及果实软化途径中的基因,是番茄果实成熟的正调控因子。Zhu等[14]鉴定了一个新的番茄NAC 转录因子SlNAC4,其在萼片和果实成熟开始时表现出较高的积累性。沉默SlNAC4 基因后可引起番茄果实中多个成熟相关的基因显著下调,研究表明SlNAC4 通过影响乙烯合成调节果实的成熟。张琪静等[15]以金帅苹果果实为试材,筛选出与果实发育成熟阶段显著差异表达的NAC 转录因子13 个,并对差异表达显著的MdNAC78 和MdNAC80 2个基因进行了功能鉴定,结果显示MdNAC80能够结合MdACS1 和MdACS3a 启动子,而MdNAC78 不能,表明MdNAC80是苹果果实乙烯生成的正调控因子,MdNAC78 是苹果果实乙烯生成的负调控因子。安建平等[16]克隆了1 个NAC 转录因子MdNAC029,推测其可通过促进MdMYB1 基因的表达正向调控花青苷的积累。Liu等[17]以奉节脐橙(Citrus sinensis Osbeck)的晚熟突变体为试材,分离了1 个NAC 基因citNAC。基因表达分析表明,在果实成熟或衰老阶段,在果皮和果肉中表达高,其他发育阶段表达相对较低。通过系统发育分析,发现CitNAC基因功能与AtNAP 相似,推测CitNAC 与果实发育和衰老有关。武肖琦等[18]从桃树津柳早红分离克隆转录因子PpNAC072,定量表达分析表明PpNAC072在桃树各个部位均有表达,其中在果实的表达量最大,同时,该基因在果实发育过程中呈现增加-减少-增加的趋势,符合桃果实的生长规律,说明该基因参与桃的生长发育并影响果实的成熟。Zhou 等[19]鉴定了NAC转录因子BL(BLOOD)与调控桃红果性状的基因图谱连锁,研究表明,BL 与PpNAC1 形成异质二聚体调控转录因子PpMYB10.1 表达,调控红肉果实花青素合成,从而导致桃果肉呈血红色。
尽管葡萄基因组中存在有大量的NAC基因,但是在葡萄果实发育成熟过程中NAC 家族成员的潜在功能研究鲜有报道。笔者在前人研究[20]的基础上,对参与葡萄果实成熟过程的NAC转录因子进行初步筛选,为进一步深入研究NAC基因家族调控葡萄果实成熟的功能奠定基础。
1 材料和方法
1.1 植物材料
试验于2020 年在石家庄果树研究所葡萄示范园进行。试材为5 年生红地球、红巴拉多葡萄,南北行向栽植,行间自然生草,田间肥水与病虫害防治按照葡萄园常规措施进行管理。选择树势中庸的植株,各品种选择花期相近的果穗标记并进行花序整形,每结果枝留1 个果穗并整穗疏果,保证穗型一致。于5 月29 日进行第1 次采样,每隔10 d 采样1 次(依次记为:T1、T2、T3……),直至成熟。每次选取3 穗果实,均匀从每穗上中下剪取果粒,测定单果质量,然后随机抽取50~70 粒果实存放于自封袋内,做好标记,液氮速冻,-80 ℃超低温冰箱保存备用。
1.2 试验方法
1.2.1 果实理化性质分析 随机选取20粒果实,利用电子天平称量。果实中的可溶性糖含量利用液相色谱仪测定,使用外标法定性定量[21]。花色苷含量的测定参考杨夫臣等[22]的方法,并略有改动:称取液氮研磨的果皮粉末1 g,以1%盐酸-无水甲醇溶液(pH=2)作为浸提液,4 ℃遮光浸提1 h,4800 r·min-1离心10 min,上清液定容到50 mL。测定450~600 nm的吸收光谱,确定色素的最大吸收峰,并测定各溶液的吸光度值,3次重复。
1.2.2 NAC 转录因子的表达分析 采用RNAprep pure plant kit(天根,北京)提取供试材料的RNA。用1.0%凝胶电泳检测RNA 质量后,用Revert Aid First Strand cDNA Synthesis Kit 试剂盒(Thermo scientific)将RNA 反转录成cDNA,-20 ℃保存。在植物转录因子数据库PlantTFDB(http://planttfdb.gaolab.org/)下载已发表的74个NAC转录因子序列,采用NCBI 的Primer-BLAST 在线软件设计表达引物,由生工生物工程(上海)股份有限公司合成。
以红地球不同时期的cDNA为模板,葡萄Actin作为内参基因,使用ChamQ Universal SYBR qPCR Master Mix(诺唯赞,上海)进行PCR扩增。qPCR 体系为:cDNA 模板3.5 μL,上下游引物各0.4 μL,Mix10 μL,ddH2O 5.7 μL。qPCR 程序为:95 ℃预变性2 min,95 ℃变性15 s,60 ℃延伸1 min,共循环40次,反应结束后分析溶解曲线,每个样品3 次重复。基因的相对表达量按ABI7500荧光定量仪使用说明进行分析。筛选出红地球葡萄表达差异的NAC 转录因子,检测其在红巴拉多果实的表达水平。数据使用Origin 9.0软件进行处理。
1.2.3 候选NAC 基因的克隆与载体构建 利用Primer-BLAST在线设计各候选NAC基因的全长特异引物,以红地球的cDNA 为模板,采用Prime-STAR Max DNA Polymerase(TaKaRa)进行全长扩增。PCR 体系为cDNA 模板3.5 μL,PrimeSTAR Max Premix(2×)25 μL,上下游引物各0.3 μmol·L-1,无菌水加至50 μL。反应条件为:98 ℃变性10 s,55 ℃退火5 s,72 ℃延伸20 s,30 个循环,4 ℃保存。PCR产物经1%凝胶电泳后回收目的条带,并用限制性内切酶酶切PGBKT7 空质粒,利用同源重组试剂盒将目的条带胶回收产物与线性化载体进行同源重组,并转化E. coli DH5α 感受态细胞,挑取阳性克隆,送生工生物工程(上海)股份有限公司测序,测序准确后备用。
1.2.4 NAC基因的生物信息分析 利用DNAMAN软件进行NAC基因核酸序列分析;蛋白质基本性质用 Protparam(http://au.expasy.org/tools/protparam.html)预测;TMHMM 2.0(http://www.cbs.dtu.dk/services/TMHMM/)预测跨膜结构域;SignalP 4.1(http://www.cbs.dtu.dk/services/SignalP/)预测信号肽;利用Plant-mPLoc网站进行亚细胞定位预测。通过NCBI与查阅文献获得的18个NAC基因,利用MEGA 5.0软件构建系统进化树,Bootstrap 检测设置重复为1000 次。利用在线软件MEME 分析(http://memesuite.org/tools/meme)进行保守基序分析。
1.2.5 候选NAC 转录因子在葡萄不同组织中的表达 采集红地球的根部、茎部、成熟叶片、花序、果实、卷须,提取各部位的RNA,反转录成cDNA,-20 ℃保存。使用ChamQ Universal SYBR qPCR Master Mix(诺唯赞,上海)进行PCR 扩增,PCR 体系如上。数据使用Origin 9.0软件进行处理。
1.2.6 候选NAC 基因转录自激活分析 将构建好并测序验证的pGBKT7- VvNAC5、pGBKT7-VvNAC11、pGBKT7-VvNAC13、pGBKT7-VvNAC18与pGBKT7空质粒转化至酵母AH109菌株中,然后涂布在SD-Trp、SD-Trp-His-Ade、SD-Trp-His-Ade(X-α-gal)缺陷培养基上,再后恒温培养箱避光培养,29 ℃培养2~3 d,观察酵母菌株生长情况。
2 结果与分析
2.1 果实的理化性质分析
如图1 所示,在红地球与红巴拉多果实从坐果到成熟过程中,单果质量呈逐渐增加的趋势,直至成熟。葡萄果实中检测到的可溶性糖分物质主要为葡萄糖、果糖,含量随着果实发育逐渐提高。红地球中果糖略高于葡萄糖,而红巴拉多果实中葡萄糖含量略高。随着成熟果实逐渐着色,果皮花色苷含量随之升高,直至成熟。
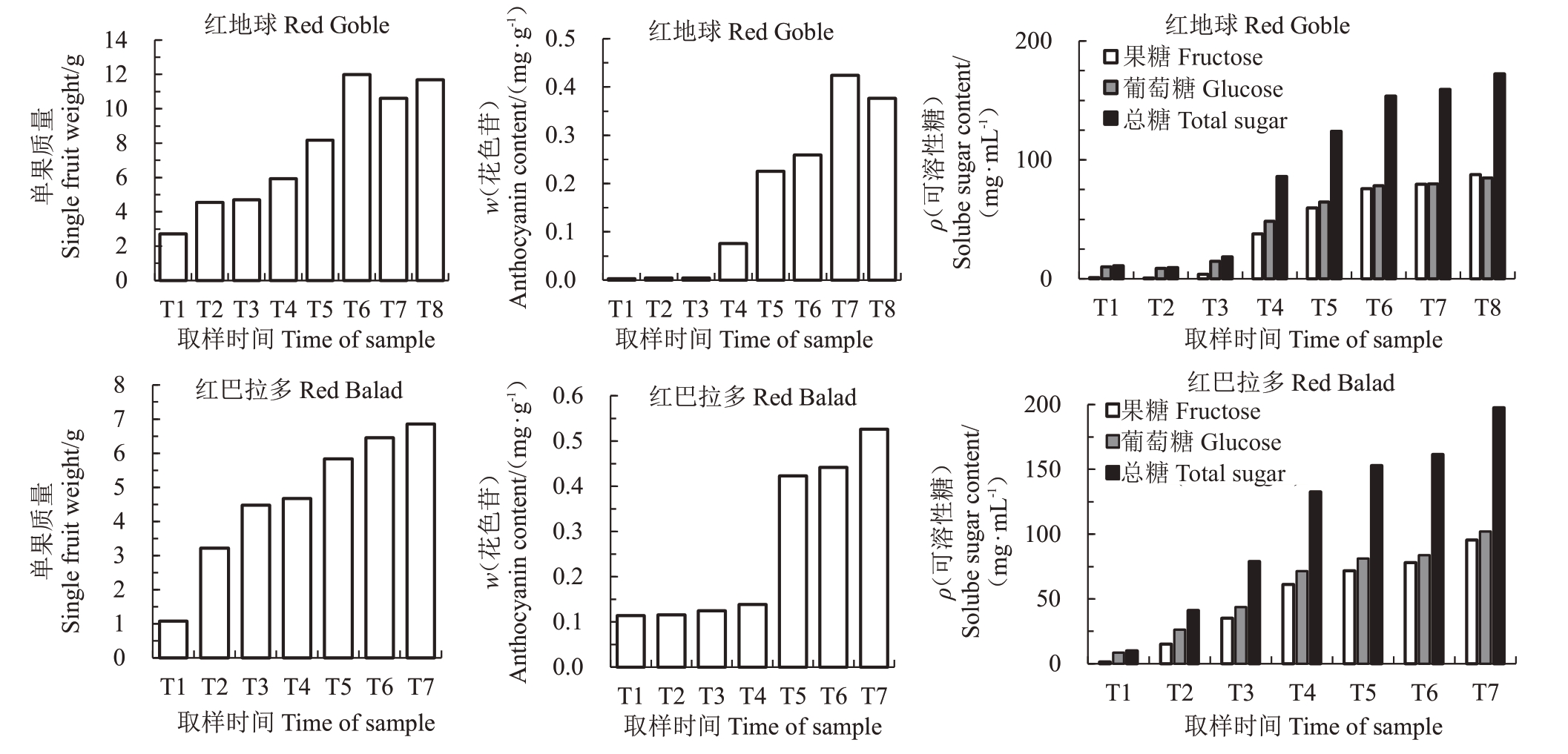
图1 红地球与红巴拉多葡萄果实发育过程的品质分析
Fig.1 Analysis of quality of Red Globe and Red Balad during fruit development
2.2 NAC转录因子的表达分析
为明确NAC转录因子与果实发育成熟的关系,利用实时荧光定量PCR技术检测NAC家族的74个转录因子的表达情况。结果表明(图2)红地球发育成熟过程中,11 个转录因子出现表达差异。其中,NAC74 表达量变化最大,其次为VvNAC13、VvNAC11、VvNAC47 等转录因子。11 个转录因子主要是2 种不同的表达模式。部分NAC 转录因子(VvNAC17、 VvNAC26、 VvNAC33、 VvNAC47、
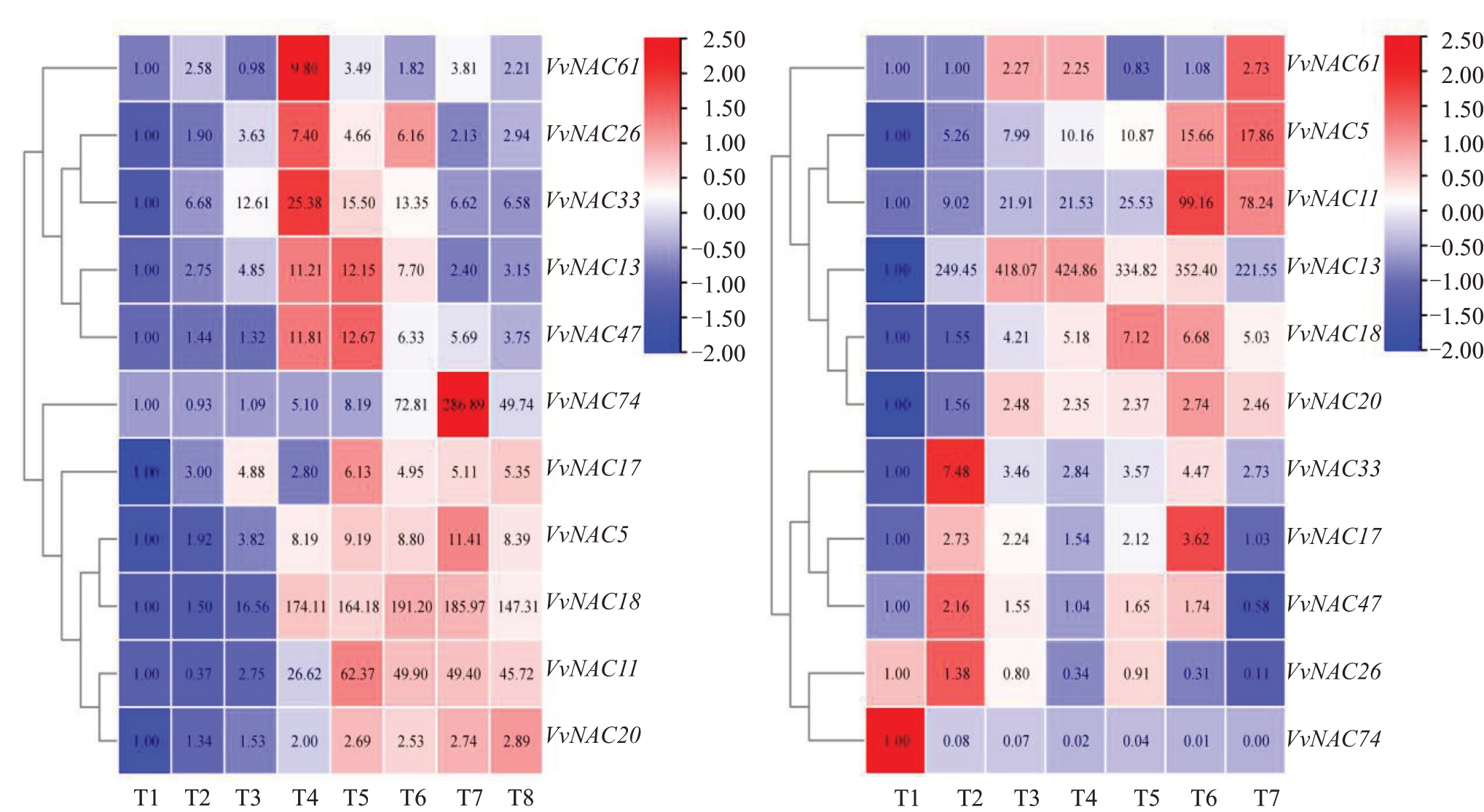
图2 红地球(左)和红巴拉多(右)葡萄不同发育时期的NAC 基因表达
Fig.2 NAC gene expression in different periods of Red Globe(left)and Red Balad(right)
VvNAC61)于发育中期出现表达高峰,但随着成熟逐渐降低;另一种(VvNAC5、VvNAC13、VvNAC20、VvNAC74)随着果实发育成熟逐渐升高或者持续高表达。检测了11 个表达差异基因在红巴拉多果实发育成熟过程的基因表达情况。结果表明9个NAC基因出现表达水平差异,尤其是VvNAC5、VvNAC11、VvNAC13、VvNAC18、VvNAC33 表达水平相对较高。VvNAC74、VvNAC26在红地球果实中表达量较高,但在红巴拉多中表达却极低。
由表1可知,葡萄果实品质指标与NAC基因的表达水平存在不同的相关性。其中,红地球中,单果质量、果糖、葡萄糖、花色苷均与VvNAC5、VvNAC11、VvNAC13、VvNAC20 基因表达显著相关。另外,单果质量与VvNAC18 基因显著相关,花色苷与VvNAC74基因显著相关;红巴拉多中单果质量、果糖、葡萄糖、花色苷均与VvNAC5、VvNAC11、VvNAC18基因显著相关,单果质量、果糖、葡萄糖与VvNAC20 基因显著相关,单果质量与VvNAC13 基因显著相关。
表1 果实品质与NAC 基因表达的相关性
Table 1 The correlation analysis between fruit quality and expression of NAC genes
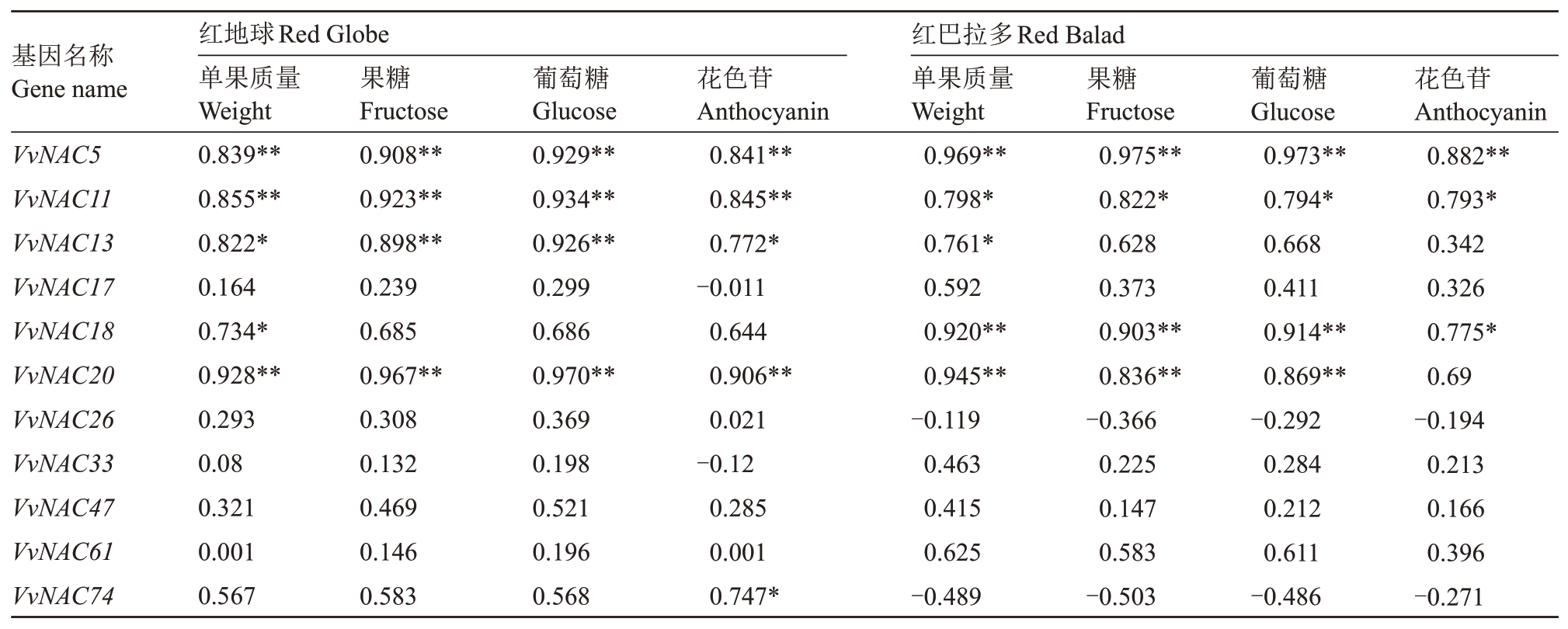
注:**与*代表在0.01 与0.05 水平显著相关。
Note:**and*indicate significant correlation at p <0.01 and p <0.05.
?
2.3 候选NAC基因的克隆及生物信息分析
综合NAC基因在红地球、红巴拉多中的表达分析,确定VvNAC5、VvNAC11、VvNAC13、VvNAC18 作为后续研究的NAC 转录因子。以红地球的cDNA为模板,克隆得到4 个NAC 基因核苷酸序列。DNAMAN 软件分析核苷酸序列显示VvNAC5、VvNAC11、VvNAC13、VvNAC18 基因的ORF 长度分别为1083、1092、1098、1062 bp,分别编码360、363、365、353 个氨基酸。经NCBI-CDD(Conserved Domain Database)比对氨基酸序列,结果显示4个NAC蛋白在氨基酸N 端均有1 个高度保守的NAM 结构域,均属于NAC家族转录因子。4个NAC蛋白分子质量、等电点等如表2 所示。4 个NAC 蛋白总平均亲水性(GRAVY)均为负值,说明蛋白均具有亲水性,为亲水性蛋白。从蛋白质稳定性上看,4个NAC蛋白均为不稳定蛋白质。另外,4 个NAC 蛋白均不存在跨膜区域,不含信号肽。Plant-mPLoc进行亚细胞定位预测显示最有可能定位于细胞核中。
表2 葡萄4 个NAC 转录因子的基本信息
Table 2 Basic information of 4 NAC transcription factors

?
对4个NAC蛋白序列进行相似性对比,结果显示(图3)4个NAC蛋白虽然在氨基酸种类与长度上不一致,但在N端都包含高度保守的NAM结构域,C端序列高度变异预示其蛋白功能存在一定差异。
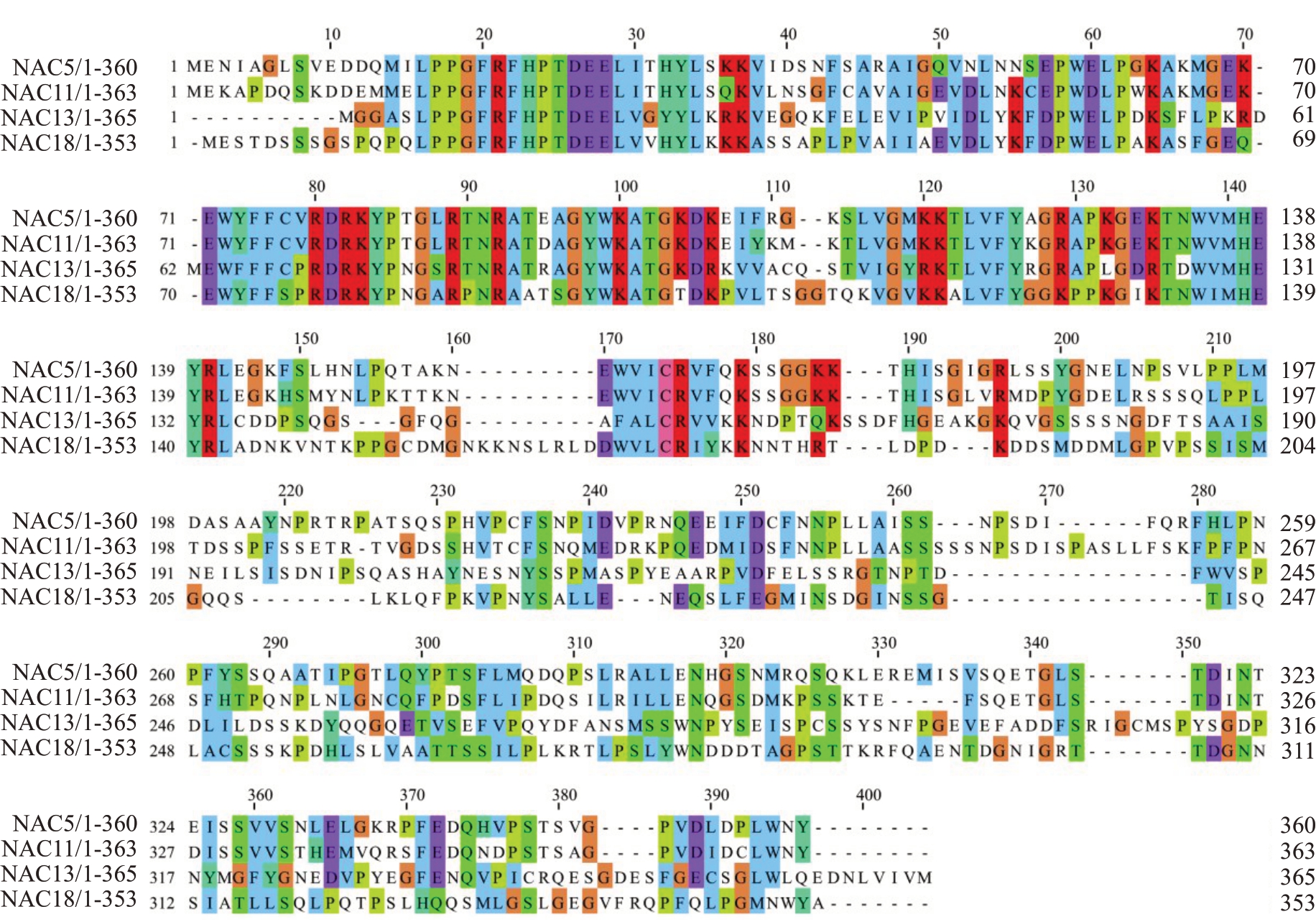
图3 葡萄NAC 氨基酸序列相似性比对
Fig.3 Homology alignment of the deduced amino acid sequences of grape NACs
为研究葡萄NAC转录因子的生物学功能,本研究通过NCBI与查阅文献获得18个NAC基因,利用MEGA X构建了包括拟南芥、水稻、番茄、矮牵牛、马铃薯、橙、苹果、葡萄等多种植物的系统进化树。结果如图4所示,大体分为3个分支。
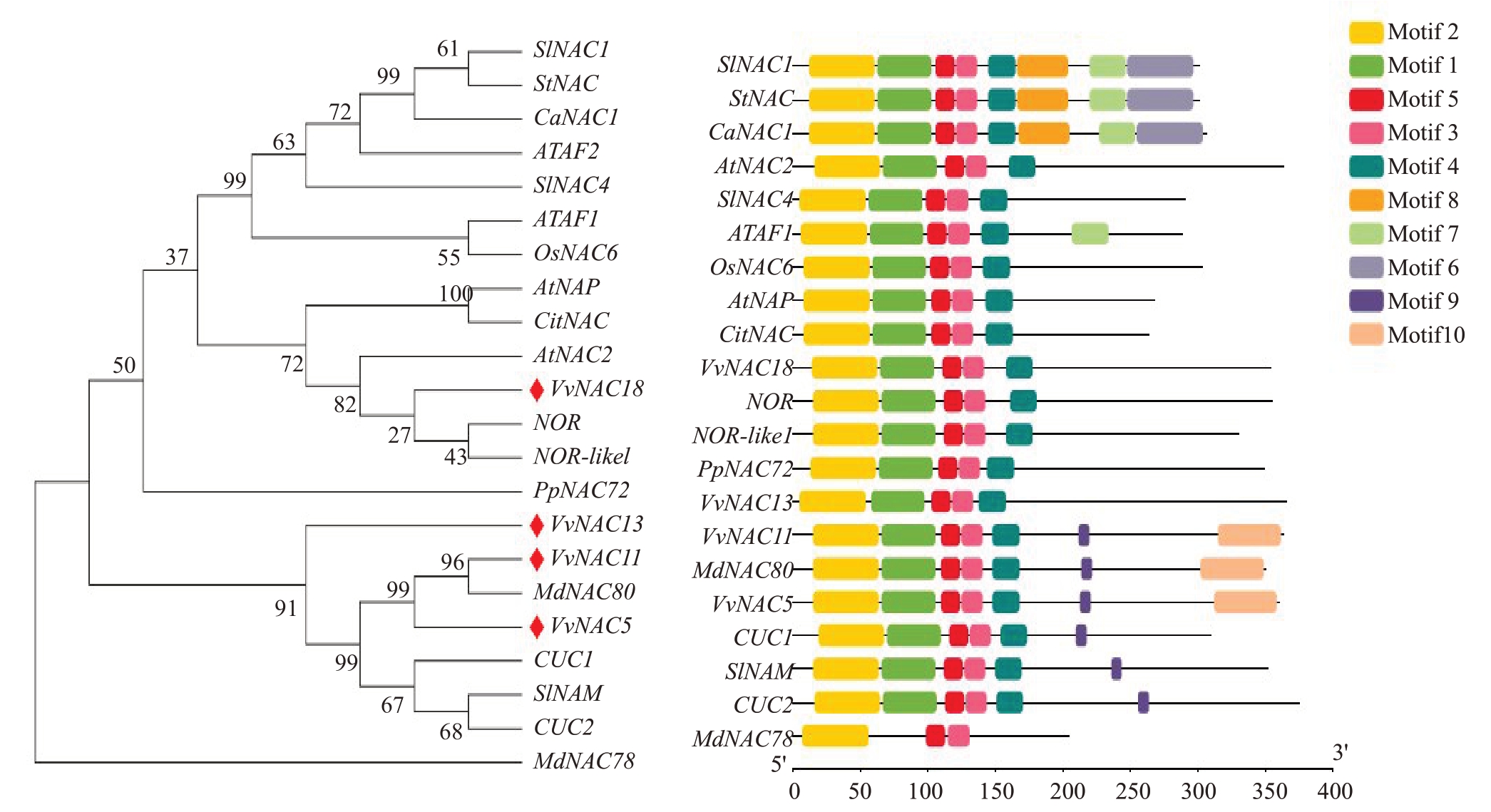
图4 葡萄NAC 转录因子与其他物种NAC 转录因子系统进化分析(左)与保守基序分析(右)
Fig.4 Phylogenetic and conserved motif analyses of the amino acid sequences of 4 NAC transcription factors in grape and in other species
分支一为众多参与胁迫响应的NAC转录因子,例如ATAF1 能够增强对白粉病的抗性,却不同程度地减弱了对细菌性叶斑病灰霉病以及黑斑病的抗性[11];过量表达OsNAC6 可明显增强水稻对稻瘟病的抗性[23];SlNAC1受盐胁迫诱导且在番茄中过量表达而提高耐寒性[24]。
分支二为VvNAC18 与众多调控果实成熟衰老及叶片脱落相关基因NOR[25]、NOR-like1[13]、Cit-NAC[17]、AtNAP[5]等蛋白,由此推测VvNAC18 参与葡萄果实的成熟调控过程。
分支三主要是分生组织形成与器官发育相关的NAC蛋白,其中,CUC1和CUC2在子叶边缘和茎顶端分生组织的形态建成中发挥重要作用[26]。结果显示,VvNAC5、VvNAC11、VvNAC13 均处于这个分支,推测可能同时在组织形成发育过程中发挥作用。另外,VvNAC5、VvNAC11 与苹果果实乙烯生成的正调控因子MdNAC80聚到一起[15],推测在果实成熟过程中有重要作用。
为了进一步研究NAC蛋白质的结构多样性,利用MEME在线平台对上述22个NAC蛋白进行了保守基序分析。结果(图4)显示22 个NAC 蛋白都具有NAC类转录因子保守的亚结构域,但也具有独特的基序。聚类相近的NAC蛋白具有共同的基序,比如SlNAC1、StNAC、CaNAC1 具有一致的基序组成,且单独具有motif9 基序。另外,VvNAC5、VvNAC11与MdNAC80 也具有一致的基序组成,且单独具有motif10基序。
2.4 相关NAC转录因子的组织表达分析
选取红地球的根部、茎部、花序、成熟叶片、果实、卷须为试材,利用荧光定量PCR 技术检测了4个NAC 基因在不同组织的表达情况,结果表明(图5),4个NAC基因表达存在组织特异性。VvNAC5、VvNAC18 主要在果实中表达,且显著高于其他部位,推测主要参与果实的发育成熟。另外,VvNAC11、VvNAC13分别在根、叶片表达量最高,但在果实中也都有较高的表达量。
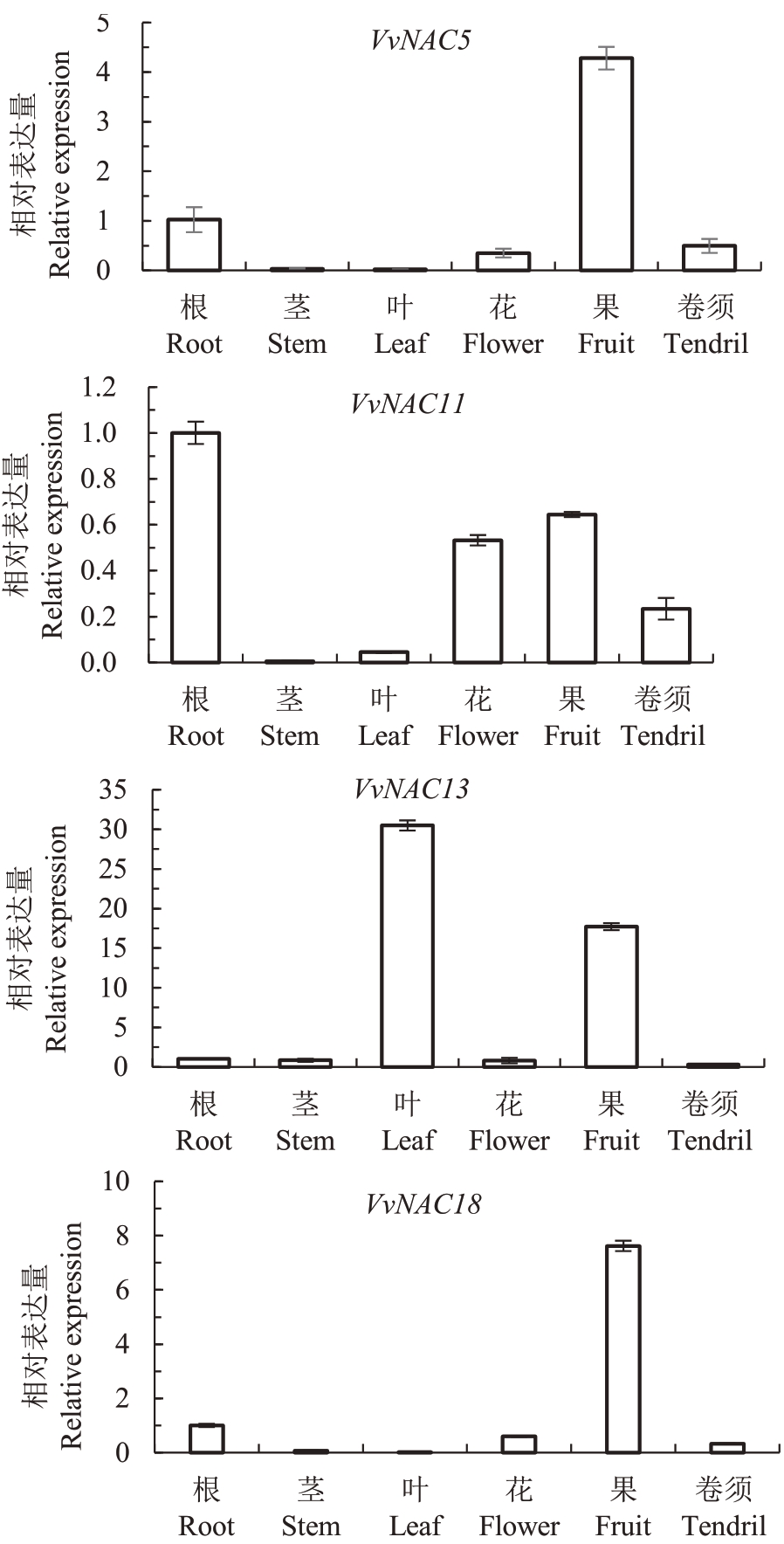
图5 NAC 基因在葡萄多个组织中的表达
Fig.5 Expression of NAC genes in different tissues of grape
2.5 相关NAC转录因子转录活性分析
将重组质粒与pGBKT7 空质粒转化进入酵母菌AH109 中,结果如图6 所示,发现转化重组质粒与pGBKT7 空质粒的酵母菌在SD-Trp 单缺陷培养基上均能正常生长;转化pGBKT7空质粒酵母菌不能在SD-Trp-His-Ade 缺陷培养基平板生长,转化pGBKT7-NACs 重组质粒的酵母菌可以在SD-Trp-His-Ade 缺陷培养基平板生长,且能够分解X-α-gal产生蓝色底物,说明4个NAC转录因子均具有转录自激活能力。
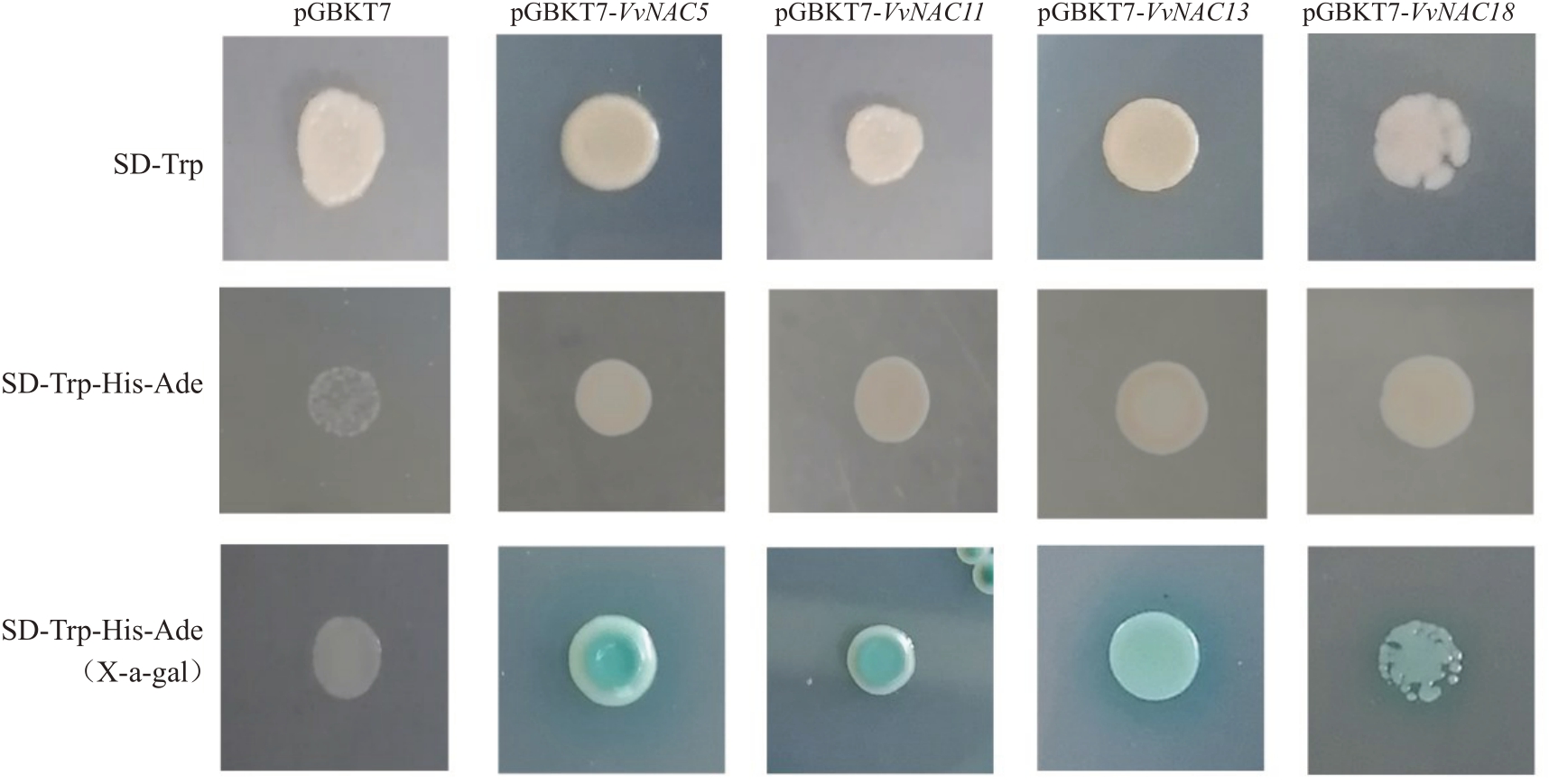
图6 NAC 基因的转录激活活性检测
Fig.6 Transcriptional activation assay of NAC genes
3 讨 论
NAC 转录因子是特异性存在于植物中的一类转录因子,家族成员多,为最大转录因子家族之一。NAC 转录因子在不同发育时期和多种环境因素诱导下,激活特定目的蛋白发挥着各种重要生物功能,主要涉及植物体生长发育的调控和环境胁迫的响应。该家族已成为当前植物基因功能及表达网络调控研究的热点之一。
研究表明,葡萄NAC转录因子在响应外界环境胁迫、生长发育等方面有重要的调控作用。VvNAC1在拟南芥中的异源表达修饰了防御基因的转录,增强了其对非生物和生物胁迫的耐受性[27]。VvDRL1不仅调控植株形态发育,还能响应干旱胁迫,过量表达的烟草植株发育迟缓、形态矮小,且对干旱的抗性降低[28-29]。山葡萄VaNAC26 能响应干旱和茉莉酸,在拟南芥中过量表达增强了对干旱的耐受性[30]。然而,NAC转录因子在葡萄果实成熟中的功能尚不明确。本研究通过分析NAC基因在红地球、红巴拉多不同发育时期的表达水平,筛选出4个NAC基因在果实发育后期表达上升明显,且与果实品质指标显著相关。组织特异性分析表明,4 个NAC 基因在果实中均有较高的表达,暗示可能参与葡萄果实的成熟过程。拟南芥中,AtNAP 转录因子在调控植物生长发育、叶片衰老等方面发挥重要作用,AtNAP转录因子不仅与叶片衰老有关,也与果实衰老紧密相关。番茄NOR-like1 能直接结合乙烯生物合成(SlACS2、SlACS4)、颜色形成(SlGgpps2、SlSGR1)和细胞壁代谢(SlPG2a、SlPL、SlCEL2 和SlEXP1)的基因,促进果实转色成熟。苹果MdNAC80能够结合乙烯合成途径中MdACS1、MdACS3a基因启动子,正向调控乙烯的合成,促进果实成熟。本研究中系统进化树分析比表明,VvNAC18与众多调控果实成熟衰老及叶片脱落相关基因NOR、NOR-like1、AtNAP 聚为一类,推测VvNAC18直接参与葡萄果实的成熟调控过程。VvNAC5、VvNAC11、VvNAC13 与众多参与组织形成与器官发育相关的NAC蛋白聚为一类,同时与MdNAC80 聚类更近,推测3 个NAC 基因同时在调控组织形态发育、果实成熟进程。此外,葡萄VvNAC17基因能上调ABA和应激相关基因的表达,响应干旱胁迫[31]。本研究中VvNAC17 基因在葡萄果实发育成熟过程中也呈差异表达,猜测VvNAC17基因也参与调控果实的成熟,是否通过调控脱落酸途径来实现,还有待进一步研究。
4 结 论
研究首次系统性地检测NAC 家族在果实发育成熟过程的表达模式,以红地球和红巴拉多葡萄的果实为试材,筛选出了4 个NAC 转录因子(VvVvNAC5、VvNAC11、VvNAC13、VvNAC18),克隆并分析蛋白质的理化性质与组织表达模式,推测4个NAC 转录因子在果实成熟过程中起重要调控作用,这将为后续研究NAC 基因生物学功能奠定基础。
[1] 杨菲颖,申艳红,耿姣姣,吴用,李科,陈晓静.番木瓜NAC 转录因子的克隆与表达分析[J]. 热带作物学报,2016,37(5):895-900.YANG Feiying,SHEN Yanhong,GENG Jiaojiao,WU Yong,LI Ke,CHEN Xiaojing. Cloning and expression analysis of NAC transcription factor from Carica papaya[J]. Chinese Journal of Tropical Crops,2016,37(5):895-900.
[2] SABLOWSKI R W M,MEYEROWITZ E M.A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA[J].Cell,1998,92(1):93-103.
[3] HE X J,MU R L,CAO W H,ZHANG Z G,ZHANG J S,CHEN S Y.AtNAC2,a transcription factor downstream of ethylene and auxin signaling pathways,is involved in salt stress response and lateral root development[J]. Plant Journal,2005,44(6):903-916.
[4] KIM H J,NAM H G,LIM P O. Regulatory network of NAC transcription factors in leaf senescence[J]. Current Opinion in Plant Biology,2016,33:48-56.
[5] GUO Y F,GAN S S.AtNAP,a NAC family transcription factor,has an important role in leaf senescence[J].Plant Journal,2006,46(4):601-612.
[6] MICHAEL W C,COLETTE M,DAGMARA P S,CHARLOTTE O S,SOREN L,NIELS E M,INGER B H,KIM H,KAREN S,PER L G. Barley plants over-expressing the NAC transcription factor gene HvNAC005 show stunting and delay in development combined with early senescence[J]. Journal of Experimental Botany,2016,67(17):5259-5273.
[7] YANG S D,SEO P J,YOON H K,PARK C M.The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes[J]. Plant Signaling&Behavior,2011,23(6):2155-2168.
[8] XIE Q,FRUGIS G,COLGAN D,CHUA N H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development[J]. Genes & Development,2000,14(23):3024-3036.
[9] SHAO H B,WANG H Y,TANG X L.NAC transcription factors in plant multiple abiotic stress responses:progress and prospects[J].Frontiers in Plant Science,2015,6(902):1-8.
[10] ASLAM M,GROVER A,SINHA V B,FAKHER B,PANDE V,YADAV P V,GUPTA S M,ANANDHAN S,AHMED Z. Isolation and characterization of cold responsive NAC gene from Lepidium latifolium[J]. Molecular Biology Reports,2012,39(10):9629-9638.
[11] WANG X E,BASNAYAKE B M,ZHANG H J,LI G J,LI W,VIRK N,MENGISTE T,SONG F M.The Arabidopsis ATAF1,a NAC transcription factor,is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens[J].Molecular Plant-Microbe Interactions,2009,22(10):1227-1238.
[12] NURUZZAMAN M,SHARONI A M,KIKUCHI S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants[J]. Frontiers in Microbiology,2013,4(248):1-16.
[13] GAO Y,WEI W,ZHAO X D,TAN X L,FAN Z Q,ZHANG Y P,JING Y,MENG L H,ZHU B Z,ZHU H L,CHEN J Y,JIANG C Z,GRIERSON D,LUO Y B,FU D Q.A NAC transcription factor,NOR-like1,is a new positive regulator of tomato fruit ripening[J].Horticulture Research,2018,5(1):1-18.
[14] ZHU M K,CHEN G P,ZHOU S,TU Y,WANG Y,DONG T T,HU Z L.A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor,SlNAC4,functions as a positive regulator of fruit ripening and carotenoid accumulation[J]. Plant Cell Physiology,2014,55(1):119-135.
[15] 张琪静.苹果果实成熟相关NAC 转录因子的筛选与功能鉴定[D].沈阳:沈阳农业大学,2018.ZHANG Qijing. Screening and functional identification of NAC transcription factors related with apple fruit ripening[D].Shenyang:Shenyang Agricultural University,2018.
[16] 安建平,宋来庆,赵玲玲,由春香,王小非,郝玉金.苹果愈伤组织超表达MdNAC029 促进花青苷积累[J].园艺学报,2018,45(5):845-854.AN Jianping,SONG Laiqing,ZHAO Lingling,YOU Chunxiang,WANG Xiaofei,HAO Yujin. Overexpression of Md-NAC029 promotes anthocyanin accumulation in Apple Calli[J].Acta Horticulturae Sinica,2018,45(5):845-854.
[17] LIU Y Z,Baig M N R,FAN R,YE J L,CAO Y C,DENG X X.Identification and expression pattern of a novel NAM,ATAF,and CUC-like gene from Citrus sinensis Osbeck[J].Plant Molecular Biology Reporter,2009,27(3):292-297.
[18] 武肖琦,冯涛,刘艳军,焦思鹏,孙攀,杨静慧.桃PpNAC72 的克隆及表达分析[J].生物技术通报,2019,35(6):16-23.WU Xiaoqi,FENG Tao,LIU Yanjun,JIAO Sipeng,SUN Pan,YANG Jinghui. Cloning and expression analysis of the Pp-NAC72 in pach[J].Biotechnology Bulletin,2019,35(6):16-23.
[19] ZHOU H,WANG K L,WANG H L,GU C,DARE A P,ESPLEY R V,HE H P,ALLAN A C,HAN Y P. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors[J]. Plant Journal,2015,82(1):105-121.
[20] WANG N,ZHENG Y,XIN H P,FANG L C,LI S H. Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera[J].Plant Cell Reports,2013,32(1):61-75.
[21] 钟海霞,潘明启,张付春,张雯,谢辉,韩守安,艾尔买克·才卡斯木,伍新宇.不同砧木对克瑞森葡萄果实可溶性糖含量的影响[J].新疆农业科学,2018,55(9):1633-1638.ZHONG Haixia,PAN Mingqi,ZHANG Fuchun,ZHANG Wen,XIE Hui,HAN Shou’an,Ermek Chaikasimu,WU Xinyu. Effects of different rootstocks on soluble sugar content of Crimson grape fruit[J].Xinjiang Agricultural Sciences,2018,55(9):1633-1638
[22] 杨夫臣,吴江,程建徽,徐凯,陈俊伟.葡萄果皮花色素的提取及其理化性质[J].果树学报,2007,24(3):287-292.YANG Fuchen,WU Jiang,CHENG Jianhui,XU Kai,CHEN Junwei. Studies on extraction and physical-chemical properties of anthocyanin from Red Globe grape peel[J]. Journal of Fruit Science,2007,24(3):287-292.
[23] NAKASHIMA K,TRAN L S,VAN N D,FUJITA M,MARUYAMA K,TODAKA D,ITO Y,HAYASHI N,SHINOZAKI K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice[J].Plant Journal,2007,51(4):617-630.
[24] MA N N,ZUO Y Q,LIANG X Q,YIN B,WANG G D,MENG Q W. The multiple stress-responsive transcription factor Sl-NAC1 improves the chilling tolerance of tomato[J]. Physiologia Plantarum,2013,149(4):474-486.
[25] MARTEL C,VREBALOV J,TAFELMEYER P,GIOVANNONI J J. The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner[J].Plant Physiology,2011,157(3):1568-1579.
[26] TAKEDA S,HANANO K,KARIYA A,SHIMIZU S,LI Z,MATSUI M,TASAKA M,AIDA M. CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3,two members of the ALOG gene family,in shoot organ boundary cells[J].Plant Journal,2011,66(6):1066-1077.
[27] GAELLE L H,CAMILLE P,BARBARA C,FANJA R,CLEMENTINE G,CHRISTOPHE C,FABIENNE B,SYLVAIN C,SANDRINE D C.Grapevine NAC1 transcription factor as a convergent node in developmental processes,abiotic stresses,and necrotrophic/biotrophic pathogen tolerance[J].Journal of Experimental Botany,2013,64(16):4877-4893.
[28] 闫朝辉,李桂荣,穆金燕,娄航通,朱自果.‘粉红亚都蜜’葡萄NAC 转录因子基因VvDRL1 的功能初步分析[J].园艺学报,2016,43(4):643-652.YAN Chaohui,LI Guirong,MU Jinyan,LOU Hangtong,ZHU Ziguo. Subcellular localization and functional analysis of a NAC gene VvDRL1 from Vitis vinifera‘Yatomi Rosa’[J].Acta Horticulturae Sinica,2016,43(4):643-652.
[29] 朱自果,阴启忠,张庆田,韩真,张倩,李勃.欧洲葡萄‘粉红亚都蜜’NAC 基因DRL1 负向调节植物抗旱性[J]. 园艺学报,2020,47(12):2290-2300.ZHU Ziguo,YIN Qizhong,ZHANG Qingtian,HAN Zhen,ZHANG Qian,LI Bo. DRL1,a NAC gene from Vitis vinifera‘Yatomo Rose’,negatively regulates the drought tolerance[J].Acta Horticulturae Sinica,2020,47(12):2290-2300.
[30] FANG L C,SU L Y,SUN X M,LI X B,SUN M X,FANG S,CHU J F,LI S H,XIN H P. Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis[J].Journal of Experimental Botany,2016,67(9):2829-2845
[31] JU Y L,YUE X F,MIN Z,WANG X H,FANG Y L,ZHANG J X. VvNAC17,a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor,increases sensitivity to abscisic acid and enhances salinity,freezing,and drought tolerance in transgenic Arabidopsis[J]. Plant Physiology and Biochemistry,2020,146:98-111.