锌指蛋白(zinc-finger proteins,ZFPs)是一类具有手指状结构域的转录因子[1]。锌指是一个具有特殊二级结构的小肽域,该结构由锌离子与半胱氨酸(Cys)残基和组氨酸(His)残基结合而稳定存在,是该家族的主要特征[2]。根据Cys残基和His残基的数量和位置,锌指蛋白可分为C2H2、C2HC、C2HC5、C3HC4、CCCH、C4、C4HC3、C6 和C8[3]。其 中,C2H2-ZFPs 成员在真核生物中数量最多[4-5],其锌指结构域中存在2 个Cys 和2 个His,并围绕中心锌离子组成稳定ZFP 结构域,主要参与基因的启动子调节[2]。研究发现,C2H2-ZFPs 家族参与调控拟南芥(Arabidopsis thaliana)在干旱、寒冷、盐和光胁迫响应中的耐受性[6],通过过表达OsZFP245 增强水稻(Oryza sativa)的耐旱性[7];水稻OsZFP179作为新的盐敏感基因,其过表达可以提高水稻耐盐性[8];苹果(Malus pumila Mill.)的C2H2 型 锌 指 转 录 因 子MdZAT10能显著加速叶片衰老,促进衰老相关基因的表达[9];木薯(Manihot esculenta Crantz.)C2H2 型锌指蛋白转录因子在植物生长发育和响应非生物胁迫功能方面具有重要作用[10]。
大多数植物C2H2- ZFPs 在锌指结构域(CX2-4CX3FX3QALGGHX3-5H)内含有高度保守的QALGGH 基序[6,11-13],形成了植物特有的Q 型C2H2锌指亚家族。Q 型C2H2 锌指蛋白在矮牵牛(Petunia hybrida)中首次被发现,并证明其高度保守基序QALGGH 对DNA 结合活性至关重要[14-15]。随后Q型C2H2 锌指蛋白基因在拟南芥[16]、水稻[12]、杨树(Populus trichocarpa)[17]、小 麦(Triticum aestivum)[18]、甘蓝(Brassica oleracea)、油菜(Brassica napus)、白菜(Brassica rapa)[19]和马铃薯(Solanum tuberosum)[20]中均被证明参与多种生物过程,包括植物生长和器官发育以及对逆境的反应与防御[21-24]。马铃薯Q型C2H2锌指蛋白基因参与对非生物胁迫的响应,增强其对盐和干旱胁迫的耐受性[20]。此外,在杨树和小麦中发现Q型C2H2锌指亚类响应非生物胁迫[17-18]。但在核桃中未发现对该基因家族的相关研究。
核桃(Juglans regia L.)是重要的干果经济林树种,而中国核桃主产区多分布于干旱和半干旱地区,春季及初夏降雨量少且分布不均[25],加之部分地区土壤含盐量较高,使苗木生长受到抑制[26],严重影响了核桃的产量与品质[27]。因此,鉴定核桃全基因组中Q 型C2H2 锌指蛋白基因家族,对其蛋白理化性质、染色体定位、系统进化关系和基因结构进行分析,探究其在干旱和盐胁迫下的表达情况,为研究Q型C2H2 锌指蛋白基因家族在核桃中生物学功能、利用基因工程手段提高核桃的抗逆性、扩大种植范围、提高品质与产量奠定基础。
1 材料和方法
1.1 试材处理
以甘肃农业大学园艺学院果树科学系组培室4周龄辽核4 号组培苗为材料。选取生长健壮、长势一致的核桃试管苗分别进行干旱[15%(w)聚乙二醇(polyethylene glycol,PEG)]和 盐(100 mmol · L- 1 NaCl)处理0、6、12、24 h,以正常生长的试管苗作对照,3 次生物学重复[20,28-30]。收集供试试管苗叶片用液氮冷冻,然后置于-80 ℃保存,用于后续RNA提取。
1.2 核桃Q型C2H2锌指蛋白基因家族成员的鉴定
从拟南芥数据库(https://www.arabidopsis.org/)中查找得到174 个已知C2H2 锌指蛋白基因保守蛋白序列,从核桃全基因组数据库(http://xhhuanglab.cn/data/juglans.html)[31]查询得到核桃全基因组中的保守蛋白序列。利用HMMER 3.1 软件(http://hmmer.org/download.html)将 得 到 的 拟 南 芥174 个C2H2 锌指蛋白结构域序列作为基础构建隐马尔可夫模型(hidden markov model,HMM)图谱,然后利用BLASTP 算法根据拟南芥C2H2 锌指蛋白氨基酸序列搜索核桃C2H2 锌指蛋白成员,E 植≤1×e-5。删除所有冗余序列后,将得到的候选成员提交至SMART(http://smart.embl-heidelberg.de/)和NCBI 保守域数据库(CDD),使用手动筛选的方式进一步确定Q型C2H2-ZFPs成员。对最终得到的核桃Q型C2H2-ZFPs家族成员的氨基酸数目、理论等电点、分子质量大小等理化性质数据在Expasy(https://web.expasy.org/protparam/)平台进行分析,在WoLF PSORT(https://wolfpsort.hgc.jp/)平台进行亚细胞结构定位分析。
1.3 染色体定位与基因复制
根据核桃全基因组注释信息文件利用Mapchart绘制核桃Q 型C2H2 锌指蛋白基因染色体定位图。核桃Q型C2H2锌指蛋白基因的串联重复事件是根据3 个标准:(1)2 个基因相似度大于75%;(2)比对长度大于75%(相对于较长的基因);(3)基因在同一条染色体且物理距离小于100 kb[32]。利用MCScanX分析了核桃种内Q型C2H2锌指蛋白基因片段重复事件,并用Circos软件绘图。
1.4 系统进化关系、基因结构及顺式元件调控分析
利用MEGA-X 软件分析基因家族系统进化关系,以邻接法(neighbor-joining method)构建进化树。利用MEME 5.3.2 在线网站(http://memesuite.org/)进行基因家族成员保守基序结构分析,使用TBtools 软件[33]对家族成员编码序列(coding sequence,CDS)与全基因序列进行内含子和外显子可视化,再将进化树、保守结构基序及内含子和外显子进行合并。
1.5 试材RNA提取与实时荧光定量PCR分析
通过核桃Q型C2H2锌指蛋白基因家族进化树随机挑选15个JrZFP基因,并将其核酸序列提交至上海生工生物工程股份有限公司官网进行实时荧光定量PCR(real-time quantitative PCR,qRT-PCR)引物设计(表1),使用TaKaRa公司的MiniBEST Plant RNA Extraction Kit试剂盒进行材料RNA的提取,用PrimeScript™RT reagent Kit with gDNA Eraser 试剂盒将RNA 反转录为cDNA,最后用TB Green® Premix Ex Taq™II 试剂盒进行qRT-PCR 验证各基因在不同胁迫下的表达。将所得数据利用SPSS 25 和Origin 2021进行处理并绘图。
表1 qRT-PCR 引物序列
Table 1 qRT-PCR primer sequence

登录号Accession No.JreChr01G12107 JreChr02G11510 JreChr03G13269 JreChr04G11953 JreChr05G10388 JreChr05G12759 JreChr06G10479 JreChr07G11727 JreChr09G11432 JreChr10G10800 JreChr10G11227 JreChr11G12142 JreChr12G10255 JreChr13G10892 JreChr14G10249基因名称Gene name JrZFP4 JrZFP12 JrZFP21 JrZFP27 JrZFP29 JrZFP34 JrZFP35 JrZFP44 JrZFP55 JrZFP57 JrZFP59 JrZFP63 JrZFP65 JrZFP71 JrZFP77引物序列(5’-3’)Primer sequence(5’-3’)F:AGAGACAGAGCCAGGTTGAGACAG F:AGCAGTTCTTGTTGTCGTCGGATG F:GGTGGAACAGGAGAGGAGTGGAG F:TGGCAAAGCGGGCTATGAGAATG F:ACAGGGTTTGGAATGGCAAGGATG F:GAGAGGAGGCGGCTGAGGAG F:GGTATGCTGTACCTGCTGCTCAC F:CACCATCTGCCACAAGTCCTTCC F:TTCTCAAATGCTCAGGCTCTTGGG F:TCGGCATAGAATTTGGCGGTGAC F:GCGGCATAGAGCGAACGAAGG F:GAAGCTGGAAGCCGACGTGAAG F:TGGGTCGTGGTTCTGAGGCTAG F:GTGGTCACAAGGCAAGAGGGAAG F:TGAGGAAATCGGTGCTGCATCTTC R:AATGACGAGGAGGATGAGGGTGAG R:CGGAGCACTCGTGAACCTTAGAC R:AAACGAATGAGCCCTCTTGTGACC R:GGACCTCACTGATGGCACAAACC R:TGGTTCTGACGGTGGCTCTGAC R:GCGAAGCACCCTTTCACATTCTTG R:TCTGCTGGGAAGAGGGTAAAGGAG R:GTGGTGACGACGCTGCTTCC R:ATGTCCATAGGCGGCGAGTTTTC R:GTTGTGCTCCTTGGCGTCCTC R:AAGGGCGTCAAGTTCAAGTCCAAG R:CGGTGGAGCAGTTTCTAGCATAGC R:CACGGCGGTTGCTCTCTTCTTC R:GCCCGGACAAAGACATCACAGAC R:CTGCTGCTTGTCTCGGTTCAGG
2 结果与分析
2.1 核桃Q型C2H2锌指蛋白基因家族鉴定及理化性质分析
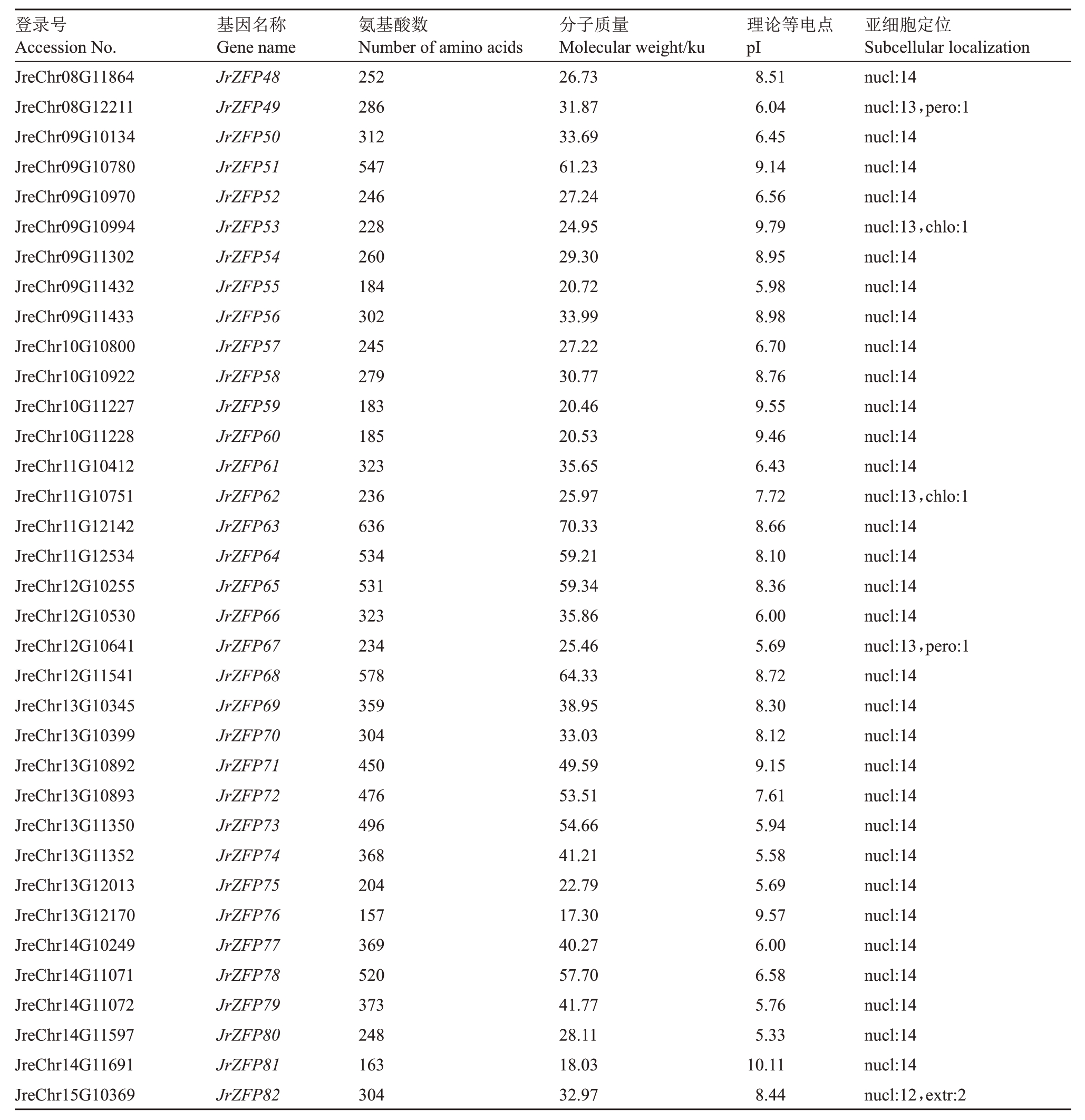
对核桃全基因组进行HMM和BLASTP 算法搜索,最后手工筛选具有核心序列CX2-4CX3FX3QALGGHX3-5H 的Q 型C2H2-ZFPs 成员,共获得82 个成员。将该基因家族成员根据其在染色体上的位置分别 编 号JrZFP1(JreChr01G10056)~JrZFP82(Jre-Chr15G10369),便于后续分析。从表2中可看出,82个Q型C2H2成员的氨基酸数目介于59(JrZFP26)~636(JrZFP63)个之间,蛋白质分子质量介于6.72(JrZFP26)~70.33(JrZFP63)ku之间,理论等电点介于5.33(JrZFP80)~10.23(JrZFP47)之间。亚细胞结构定位预测显示JrZFPs基因主要位于细胞核中,少量分布在细胞质、过氧化物酶体和叶绿体中。
表2 理化性质分析
Table 2 Physical and chemical properties analysis
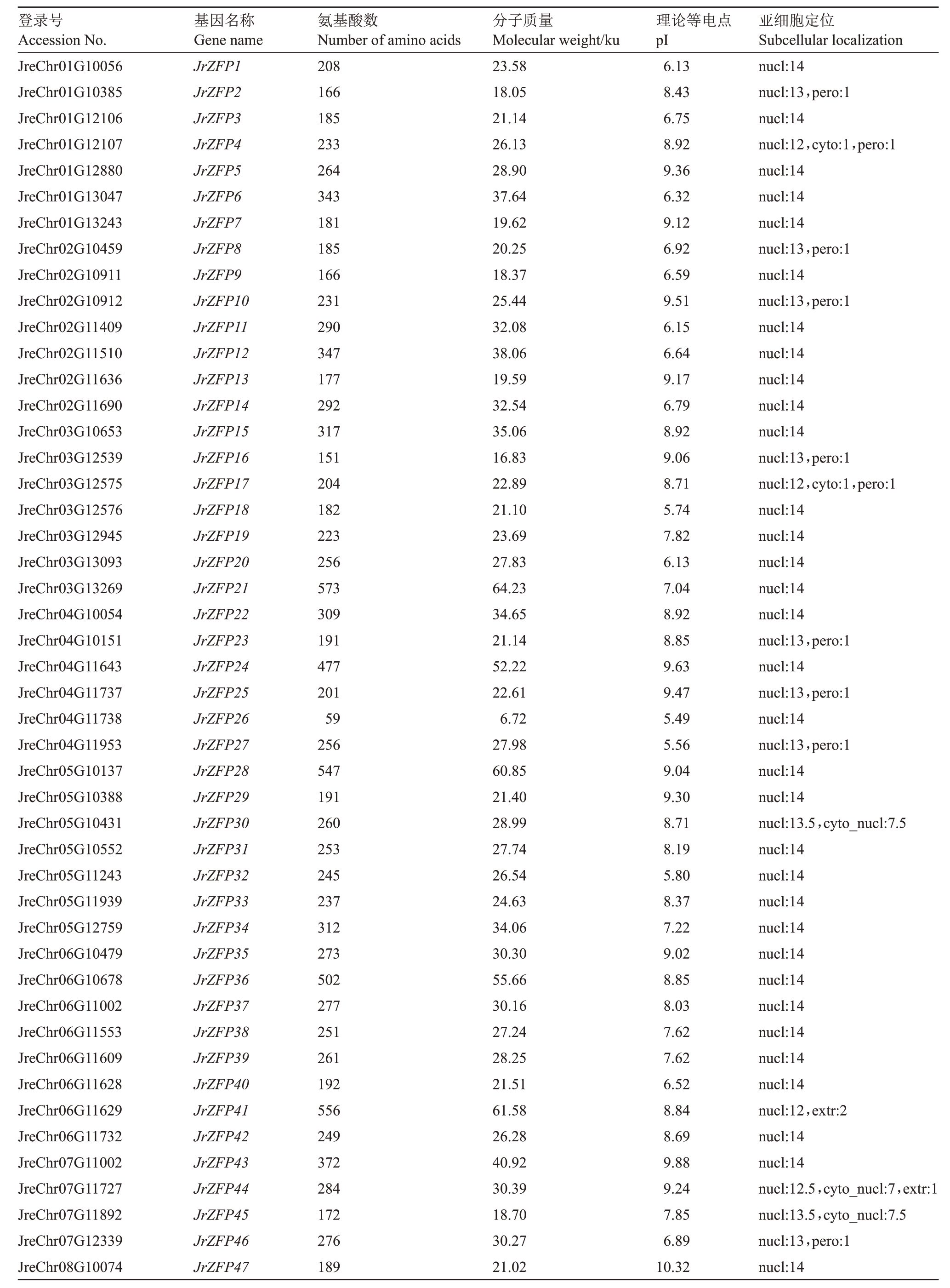
登录号Accession No.基因名称Gene name氨基酸数Number of amino acids分子质量Molecular weight/ku理论等电点pI亚细胞定位Subcellular localization JreChr01G10056 JreChr01G10385 JreChr01G12106 JreChr01G12107 JreChr01G12880 JreChr01G13047 JreChr01G13243 JreChr02G10459 JreChr02G10911 JreChr02G10912 JreChr02G11409 JreChr02G11510 JreChr02G11636 JreChr02G11690 JreChr03G10653 JreChr03G12539 JreChr03G12575 JreChr03G12576 JreChr03G12945 JreChr03G13093 JreChr03G13269 JreChr04G10054 JreChr04G10151 JreChr04G11643 JreChr04G11737 JreChr04G11738 JreChr04G11953 JreChr05G10137 JreChr05G10388 JreChr05G10431 JreChr05G10552 JreChr05G11243 JreChr05G11939 JreChr05G12759 JreChr06G10479 JreChr06G10678 JreChr06G11002 JreChr06G11553 JreChr06G11609 JreChr06G11628 JreChr06G11629 JreChr06G11732 JreChr07G11002 JreChr07G11727 JreChr07G11892 JreChr07G12339 JreChr08G10074 JrZFP1 JrZFP2 JrZFP3 JrZFP4 JrZFP5 JrZFP6 JrZFP7 JrZFP8 JrZFP9 JrZFP10 JrZFP11 JrZFP12 JrZFP13 JrZFP14 JrZFP15 JrZFP16 JrZFP17 JrZFP18 JrZFP19 JrZFP20 JrZFP21 JrZFP22 JrZFP23 JrZFP24 JrZFP25 JrZFP26 JrZFP27 JrZFP28 JrZFP29 JrZFP30 JrZFP31 JrZFP32 JrZFP33 JrZFP34 JrZFP35 JrZFP36 JrZFP37 JrZFP38 JrZFP39 JrZFP40 JrZFP41 JrZFP42 JrZFP43 JrZFP44 JrZFP45 JrZFP46 JrZFP47 208 166 185 233 264 343 181 185 166 231 290 347 177 292 317 151 204 182 223 256 573 309 191 477 201 59 256 547 191 260 253 245 237 312 273 502 277 251 261 192 556 249 372 284 172 276 189 23.58 18.05 21.14 26.13 28.90 37.64 19.62 20.25 18.37 25.44 32.08 38.06 19.59 32.54 35.06 16.83 22.89 21.10 23.69 27.83 64.23 34.65 21.14 52.22 22.61 6.72 27.98 60.85 21.40 28.99 27.74 26.54 24.63 34.06 30.30 55.66 30.16 27.24 28.25 21.51 61.58 26.28 40.92 30.39 18.70 30.27 21.02 6.13 8.43 6.75 8.92 9.36 6.32 9.12 6.92 6.59 9.51 6.15 6.64 9.17 6.79 8.92 9.06 8.71 5.74 7.82 6.13 7.04 8.92 8.85 9.63 9.47 5.49 5.56 9.04 9.30 8.71 8.19 5.80 8.37 7.22 9.02 8.85 8.03 7.62 7.62 6.52 8.84 8.69 9.88 9.24 7.85 6.89 10.32 nucl:14 nucl:13,pero:1 nucl:14 nucl:12,cyto:1,pero:1 nucl:14 nucl:14 nucl:14 nucl:13,pero:1 nucl:14 nucl:13,pero:1 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:13,pero:1 nucl:12,cyto:1,pero:1 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:13,pero:1 nucl:14 nucl:13,pero:1 nucl:14 nucl:13,pero:1 nucl:14 nucl:14 nucl:13.5,cyto_nucl:7.5 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:12,extr:2 nucl:14 nucl:14 nucl:12.5,cyto_nucl:7,extr:1 nucl:13.5,cyto_nucl:7.5 nucl:13,pero:1 nucl:14
续表Continued Table
注:nucl. 细胞核;cyto. 细胞质;extr. 细胞外;pero. 过氧化物酶体;cysk. 细胞骨架;chlo. 叶绿体;cyto_nucl. 细胞质_细胞核。
Note:nucl.Nucleus;cyto.Cytoplasm;extr.Extracellular;pero.Peroxisome;cysk.Cytoskeleton;chlo.Chloroplast;cyto_nucl.Cytoplasm_nucleus.
登录号Accession No.基因名称Gene name氨基酸数Number of amino acids分子质量Molecular weight/ku理论等电点pI亚细胞定位Subcellular localization JreChr08G11864 JreChr08G12211 JreChr09G10134 JreChr09G10780 JreChr09G10970 JreChr09G10994 JreChr09G11302 JreChr09G11432 JreChr09G11433 JreChr10G10800 JreChr10G10922 JreChr10G11227 JreChr10G11228 JreChr11G10412 JreChr11G10751 JreChr11G12142 JreChr11G12534 JreChr12G10255 JreChr12G10530 JreChr12G10641 JreChr12G11541 JreChr13G10345 JreChr13G10399 JreChr13G10892 JreChr13G10893 JreChr13G11350 JreChr13G11352 JreChr13G12013 JreChr13G12170 JreChr14G10249 JreChr14G11071 JreChr14G11072 JreChr14G11597 JreChr14G11691 JreChr15G10369 JrZFP48 JrZFP49 JrZFP50 JrZFP51 JrZFP52 JrZFP53 JrZFP54 JrZFP55 JrZFP56 JrZFP57 JrZFP58 JrZFP59 JrZFP60 JrZFP61 JrZFP62 JrZFP63 JrZFP64 JrZFP65 JrZFP66 JrZFP67 JrZFP68 JrZFP69 JrZFP70 JrZFP71 JrZFP72 JrZFP73 JrZFP74 JrZFP75 JrZFP76 JrZFP77 JrZFP78 JrZFP79 JrZFP80 JrZFP81 JrZFP82 252 286 312 547 246 228 260 184 302 245 279 183 185 323 236 636 534 531 323 234 578 359 304 450 476 496 368 204 157 369 520 373 248 163 304 26.73 31.87 33.69 61.23 27.24 24.95 29.30 20.72 33.99 27.22 30.77 20.46 20.53 35.65 25.97 70.33 59.21 59.34 35.86 25.46 64.33 38.95 33.03 49.59 53.51 54.66 41.21 22.79 17.30 40.27 57.70 41.77 28.11 18.03 32.97 8.51 6.04 6.45 9.14 6.56 9.79 8.95 5.98 8.98 6.70 8.76 9.55 9.46 6.43 7.72 8.66 8.10 8.36 6.00 5.69 8.72 8.30 8.12 9.15 7.61 5.94 5.58 5.69 9.57 6.00 6.58 5.76 5.33 10.11 8.44 nucl:14 nucl:13,pero:1 nucl:14 nucl:14 nucl:14 nucl:13,chlo:1 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:13,chlo:1 nucl:14 nucl:14 nucl:14 nucl:14 nucl:13,pero:1 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:14 nucl:12,extr:2
2.2 核桃Q型C2H2锌指蛋白基因家族染色体定位与基因复制
核桃Q型C2H2锌指蛋白基因家族成员不均匀分布在15条染色体上,其中15号染色体上只有1个基因JrZFP82。研究JrZFPs基因家族的串联重复和片段重复事件,以阐述核桃Q 型C2H2 锌指蛋白基因家族基因重复事件。共鉴定出11 对(22/82,26.83%)串联重复基因(图1),其中1、2、3、4、6、10和14 号染色体各有1 对串联重复基因,9 和13 号染色体中各有2 对串联重复基因。除串联重复基因外,利用MCScanX鉴定出核桃基因组内关于JrZFPs基因家族发生节段重复事件基因共有63 个(63/82,76.83%)(图2)。
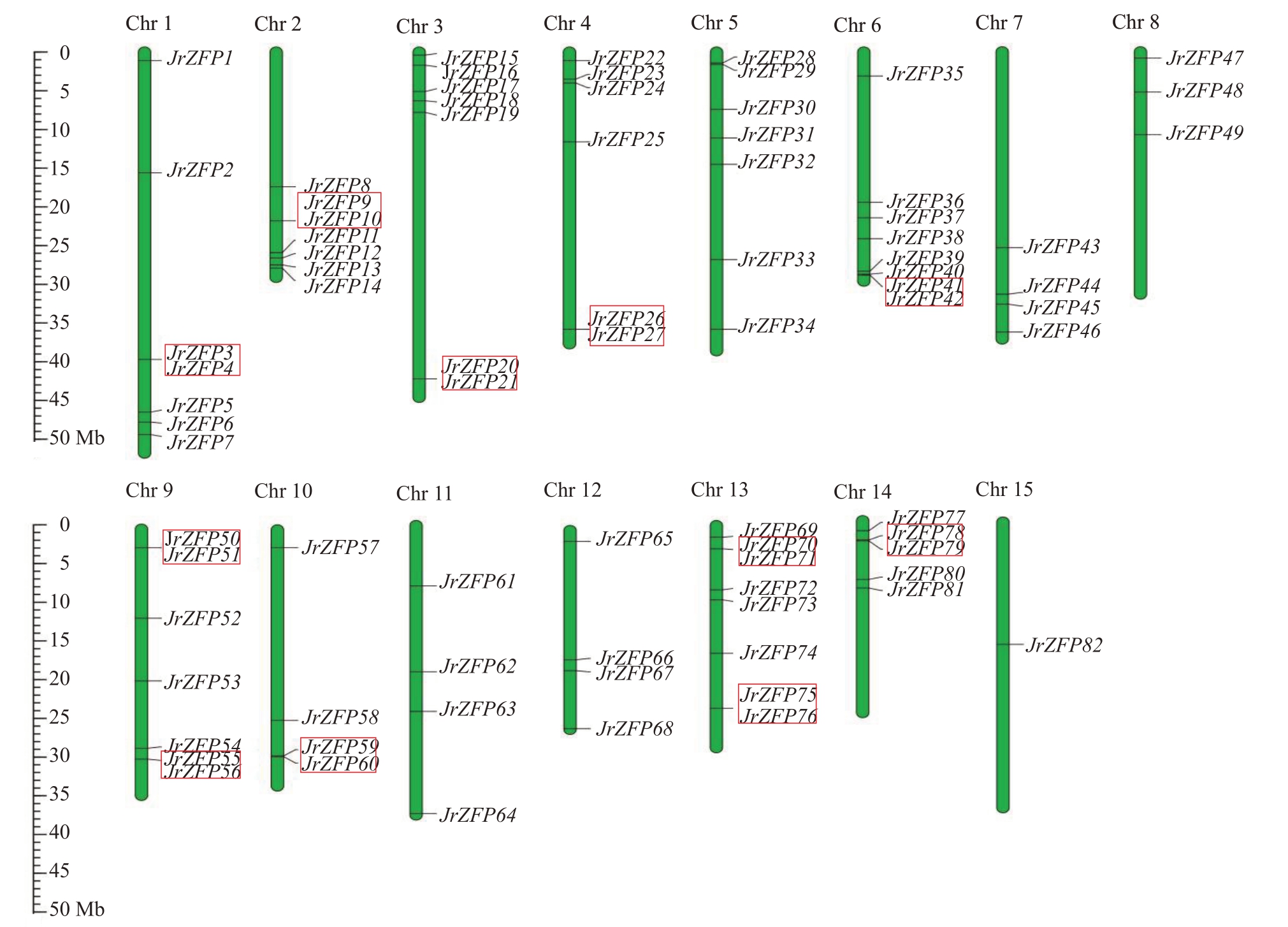
图1 核桃Q 型C2H2 锌指蛋白基因家族成员染色体定位
Fig.1 Chromosomal location of members of walnut Q-type C2H2 gene family
红色框中基因为串联重复基因。
The genes in the red box are tandem repeat genes.
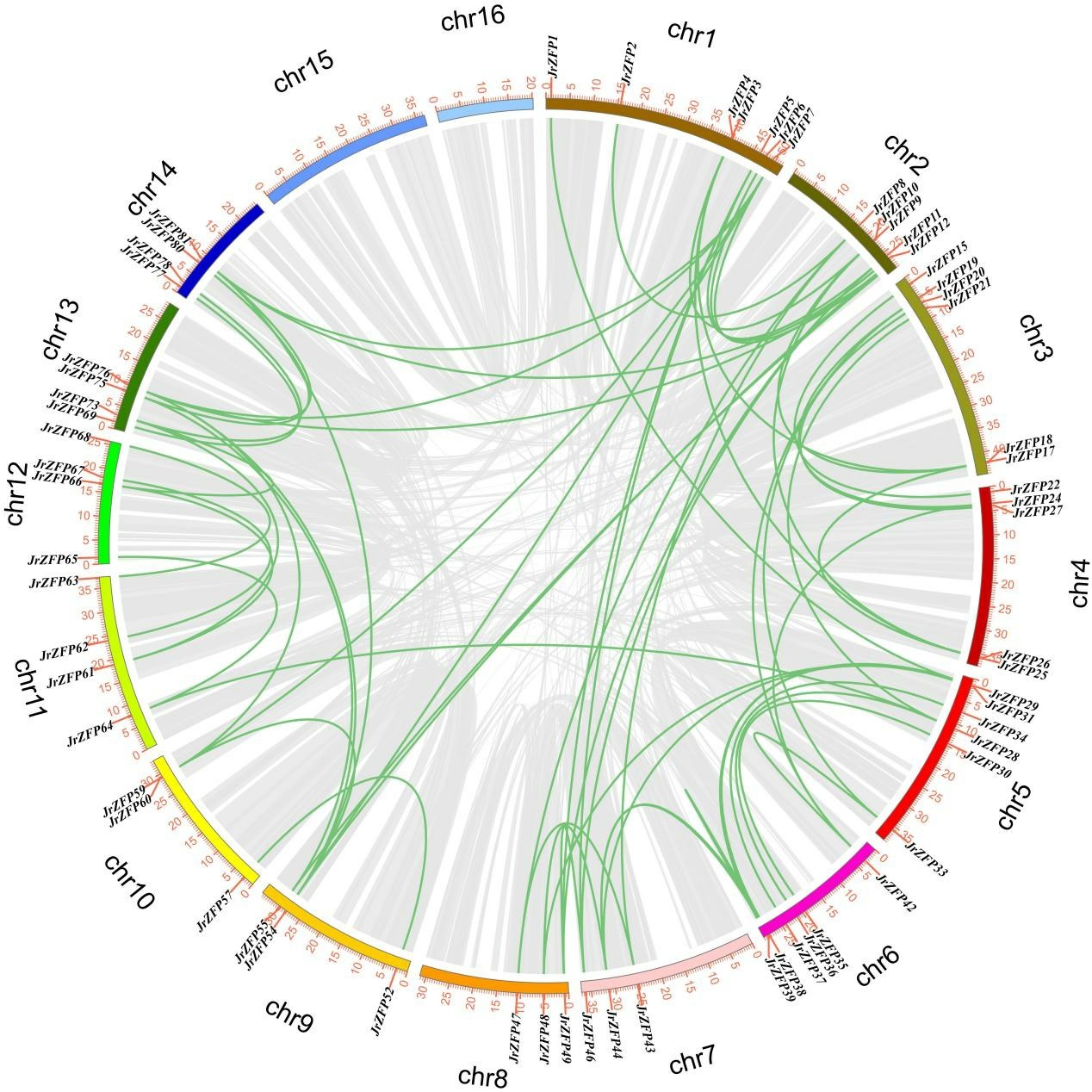
图2 核桃Q 型C2H2 锌指蛋白基因片段复制事件
Fig.2 The segmental replication events of Q-type C2H2 genes in walnut
2.3 Q型C2H2锌指蛋白基因家族进化关系分析
为研究核桃和拟南芥Q型C2H2锌指蛋白基因之间的进化关系,对140 个Q 型C2H2 锌指蛋白(包括58个拟南芥和82个核桃)氨基酸序列进行系统发育树分析(图3)。根据序列相似性和拓扑结构,将进化树分为7 个ZFP 亚家族,分别为C1、C2-Ⅰ、C2-Ⅱ、C2-Ⅲ、C2-Ⅳ、C3-Ⅰ和C3-Ⅱ。C1 子类有3 个JrZFP 氨基酸序列和2 个ATZFP 氨基酸序列,C2 子类有34 个JrZFP 氨基酸序列和27 个ATZFP 氨基酸序 列,C3 子 类 有44 个JrZFP 氨 基 酸 序 列 和30 个ATZFP氨基酸序列。通过图3中核桃和拟南芥的系统发育分析,82 个JrZFP 氨基酸序列被划分为7 个亚家族。构建的JrZFPs 基因系统发育树如图4-A所示。
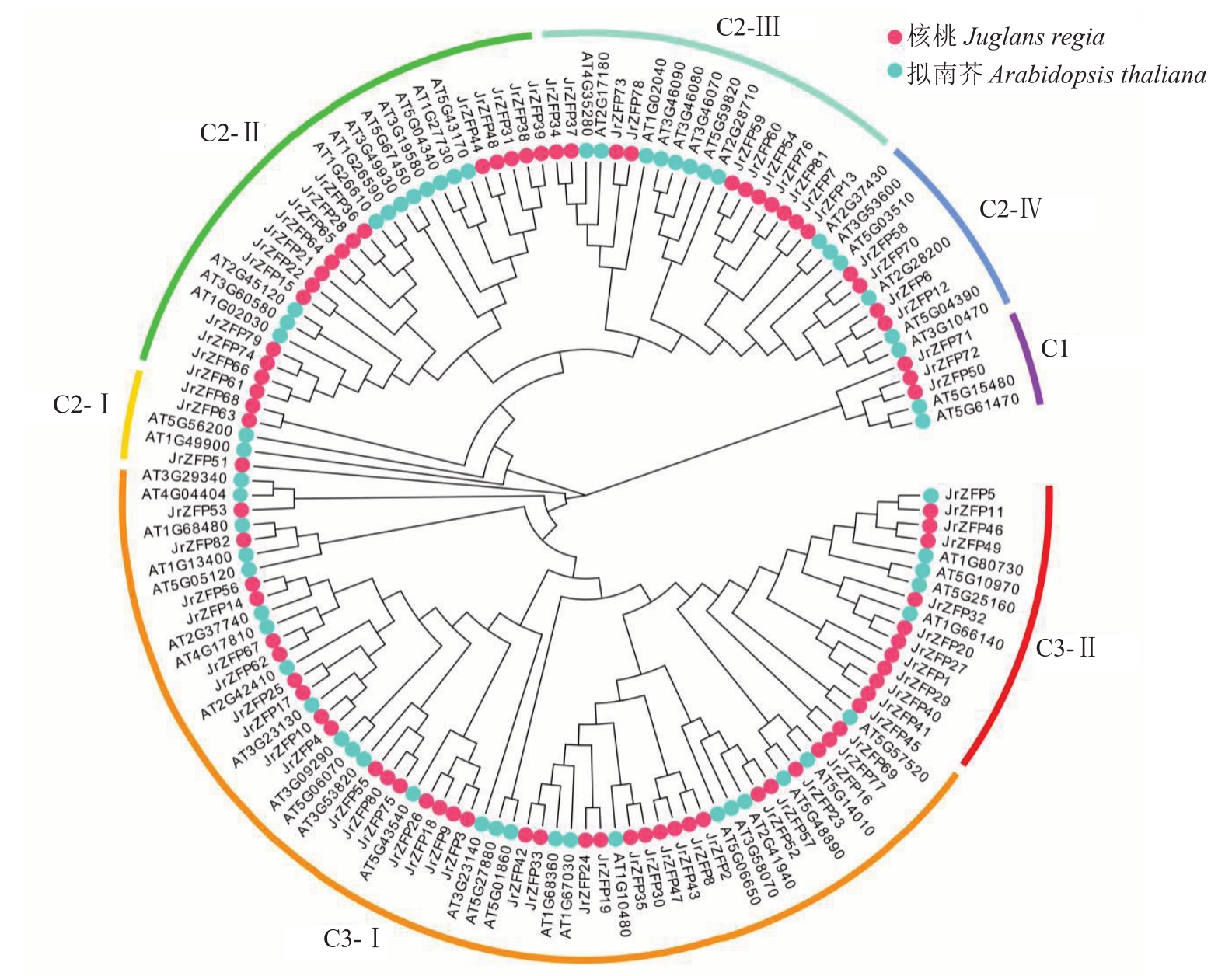
图3 拟南芥(AT)和核桃(Jr)Q 型C2H2 锌指蛋白进化树
Fig.3 Phylogenetic tree of Q-type C2H2 in Arabidopsis thaliana(AT)and Juglans regia(Jr)
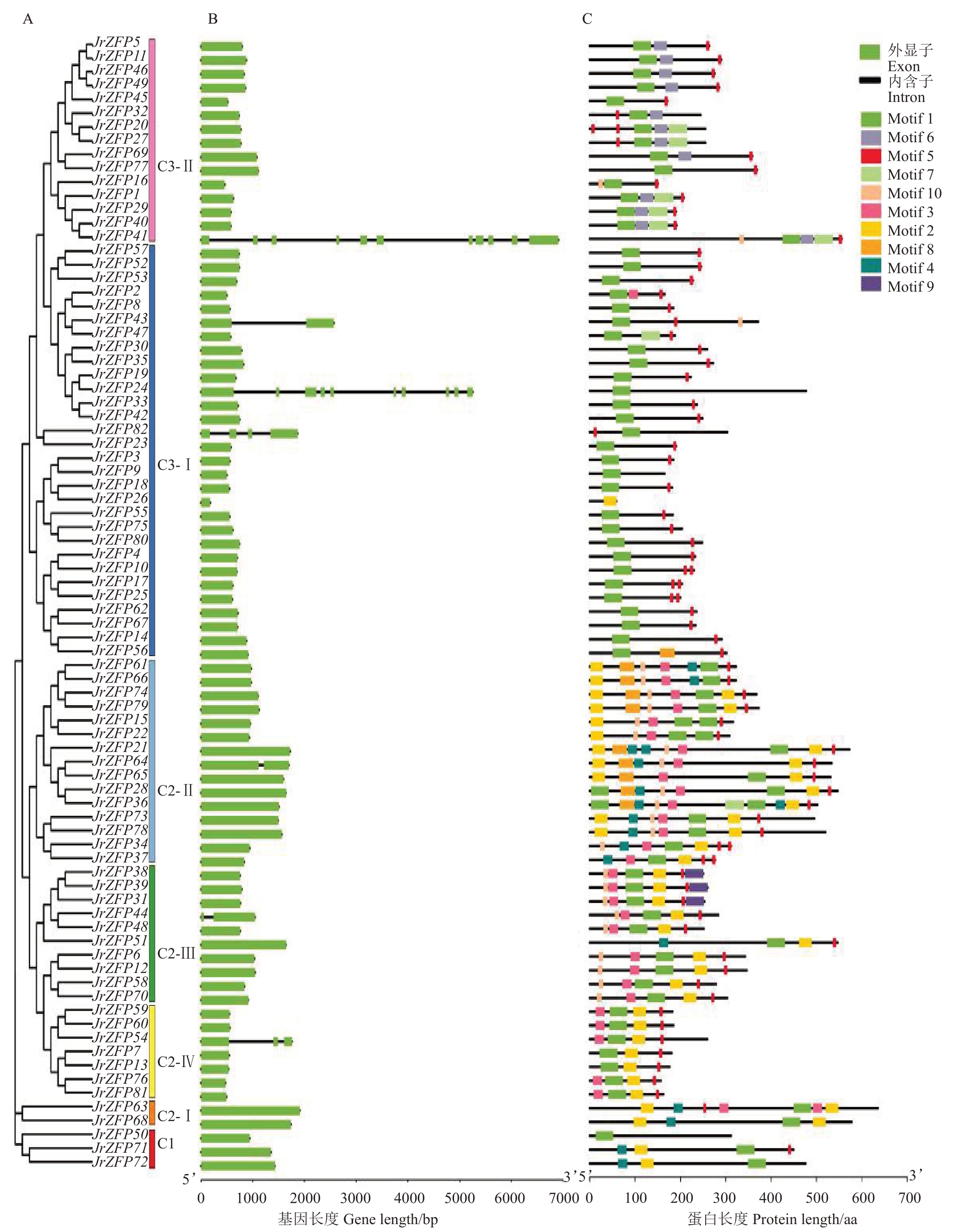
图4 核桃Q 型C2H2 锌指蛋白基因家族的系统发育树、基因结构和保守基序
Fig.4 Phylogenetic tree,gene structure and conserved motif in Q-type C2H2 gene family
A. 基于邻接法构建的系统进化树;B.JrZFP 的基因结构;C.JrZFP 的保守基序。
A. Phylogenetic tree was constructed using the Neighbor-joining method; B. The composition and position of Exon, introns and Intron in JrZFP genes;C.Conserved motifs in JrZFP proteins.
2.4 核桃Q型C2H2锌指蛋白基因家族基因结构
为深入了解JrZFPs基因结构,对其进行内含子和外显子及保守基序分析。结果显示,75 个JrZFP基因(91.46%)无内含子,其余成员中有3 个含有1个内含子,4 个含有2 个及2 个以上的内含子(图4-B)。亚家族中C1 和C2-Ⅰ所有成员均无内含子;C2-Ⅱ、C2-Ⅲ、C2-Ⅳ和C3-Ⅱ中均只有1个成员有内含子,其余成员均无内含子;C3-Ⅰ中3 个成员内含子。使用MEME 分析核桃Q型C2H2锌指蛋白基因家族成员保守基序分布情况(图4-C),共鉴定出10个Motif,主要的3个Motif如图5所示。其中,Motif 1 和Motif 2 均含有Q 型C2H2 锌指蛋白核心序列CX2-4CX3FX3QALGGHX3-5H,Motif 5为EAR-motif。每一亚族中大多数JrZFPs 基因通常具有相似的母序构成。由图4-C 可知,C1 和C2 亚族中的JrZFPs基因包含2个锌指结构域,而C3亚族中的成员只包含1 个锌指结构域。Motif 1 和Motif 2 为Q 型锌指结构域,广泛分布于82 个JrZFP 基因中。76 个JrZFP基因都含有EAR-motif,其余成员无EAR-motif。
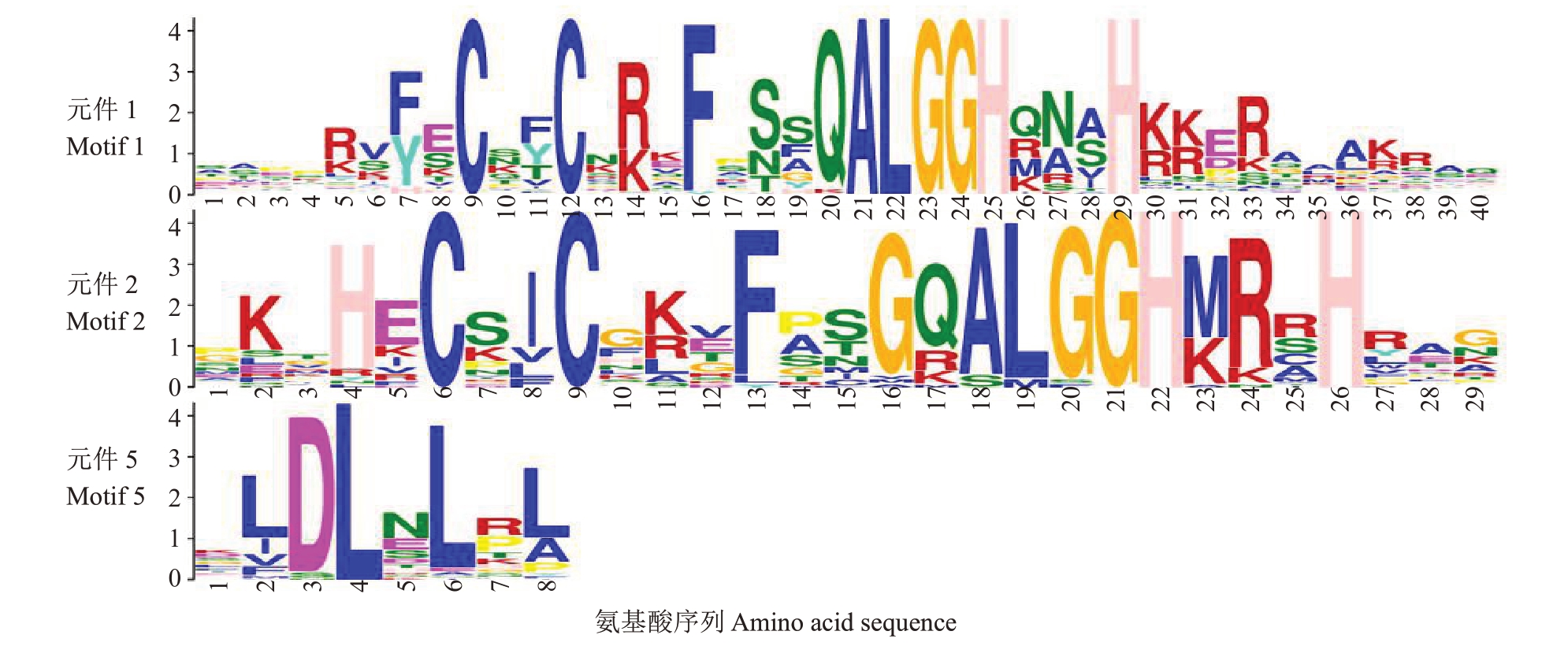
图5 核桃Q 型C2H2 锌指蛋白基因家族主要保守基序
Fig.5 The conserved motifs of walnut Q-type C2H2 gene family
2.5 核桃Q型C2H2锌指蛋白基因家族顺式调控元件分析
根据82 个JrZFP 基因家族成员上游2000 bp 碱基序列预测其在启动子区顺式作用元件的种类和数量,研究该基因家族转录调控因子(图6)。核桃Q型C2H2锌指蛋白基因家族中存在与非生物胁迫响应相关的顺式作用元件,如82个JrZFP基因成员的启动子区共包含415个ACE(光)元件、261个ABRE(脱落酸)元件、87 个CGTCA-motif(MeJA)元件、81个GARE-motif(赤霉素)元件、65 个LTR(低温)元件、56个MBS(干旱)元件、46个TCA-element(水杨酸)元件、46 个TGA-element(生长素)元件和30 个TC-rich repeats(防御和压力)元件。该基因家族中98%的成员具有光响应元件,93%的成员具有脱落酸响应元件,50%的成员具有低温响应元件,44%的成员具有干旱响应元件。因此,推测JrZFPs基因家族在核桃树体响应干旱环境中起重要作用。
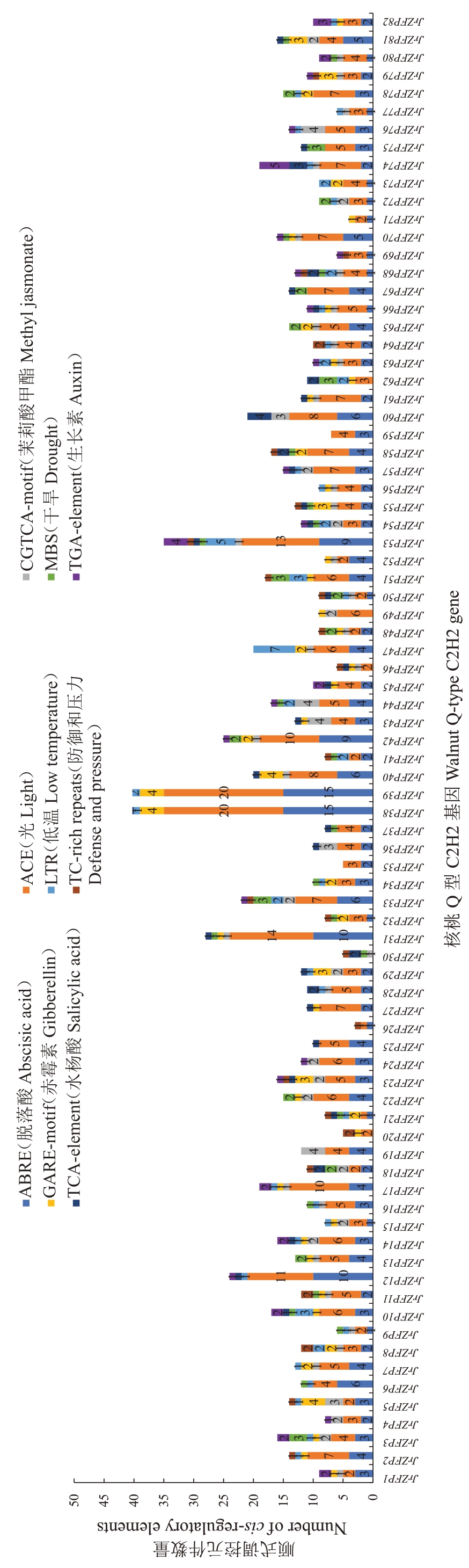
图6 核桃Q型C2H2 锌指蛋白基因家族顺式调控元件
Fig. 6 Cis-regulatory elements of walnut Q-type ZFP gene family
2.6 核桃Q型C2H2锌指蛋白基因家族成员的表达分析
2.6.1 核桃Q型C2H2锌指蛋白基因家族在NaCl胁迫下的表达分析 对核桃组培苗进行NaCl胁迫(图7),15 个JrZFP 基因在不同时期表现出不同应答模式,均为显著上调。与S0 相比,JrZFP21、JrZFP44、JrZFP55 和JrZFP65 能迅速响应NaCl 胁迫,其相对表达量显著上调,在S1 期就已达到峰植,其中JrZFP55 上 调 最 高,是S0 的14.85 倍;JrZFP12、JrZFP27、JrZFP34、JrZFP35、JrZFP57和JrZFP71应答较为缓慢,其相对表达量在S2 达到峰植,其中JrZFP71 上调最高,显著高于S0,是S0 的13.97 倍;JrZFP4、JrZFP29、JrZFP59、JrZFP63 和JrZFP77 应答最为缓慢,其相对表达量在S3 达到峰植,其中JrZFP59 上调最高,显著高于S0,是S0 的13.89 倍。所有时期中,JrZFP55 相对表达量和S0 相比最高,推测JrZFP55在核桃NaCl胁迫中起正向调节作用。
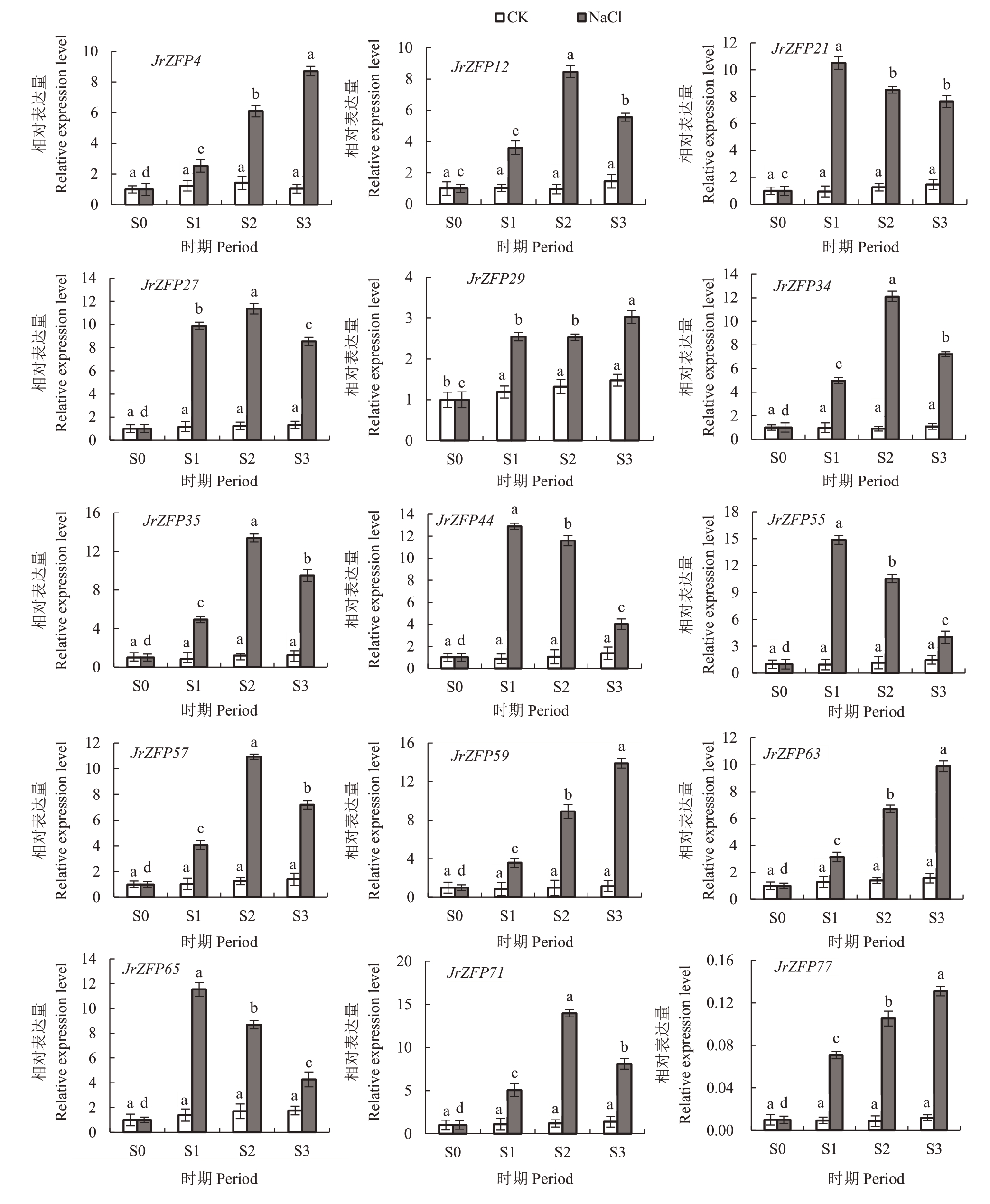
图7 在NaCl 处理下核桃Q 型C2H2 锌指蛋白基因家族表达分析
Fig.7 Expression analysis of Q-type C2H2 gene family under NaCl treatments in walnut
15 个JrZFPs 基因在0(S0)、6(S1)、12(S2)和24 h(S3)NaCl 胁迫下的表达分析。不同小写字母表示不同时期差异显著(p <0.05)。下同。
The expression analyses of fifteen JrZFPs under NaCl stress 0(S0),6(S1),12(S2)and 24 h(S3).Different small letters represent significant difference among different periods(p <0.05).The same below.
2.6.2 核桃Q型C2H2锌指蛋白基因家族在干旱胁迫下的表达分析 对核桃组培苗进行PEG处理(图8),15 个JrZFP 基因在不同时期表现出不同的应答模式,既有显著上调又有显著下调。JrZFP12、JrZFP27、JrZFP29、JrZFP35、JrZFP59和JrZFP63均在S3 中相对表达量最高,其中JrZFP12 上调最高,是S0 的13.97 倍;与S0 相 比,JrZFP4、JrZFP21、JrZFP34、JrZFP44、JrZFP55、JrZFP57、JrZFP65、JrZFP71 和JrZFP77 均 显 著 下 调,其 中JrZFP4、JrZFP21、JrZFP44和JrZFP55的相对表达量在S1最低;S2 相对表达量最低的是JrZFP77;JrZFP34、JrZFP57、JrZFP65和JrZFP77的相对表达量在S3达到最低植。JrZFP4、JrZFP55 和JrZFP57 在盐胁迫中下调幅度最大,推测这3 个基因在核桃干旱胁迫中起负调控作用。

图8 在PEG 处理下核桃Q 型C2H2 锌指蛋白基因家族表达分析
Fig.8 Expression analysis of Q-type C2H2 gene family under PEG treatments in walnut
15 个JrZFPs 在0(S0)、6(S1)、12(S2)和24 h(S3)PEG 胁迫下表达分析。
The expression analyses of fifteen JrZFPs under PEG stress 0(S0),6(S1),12(S2)and 24 h(S3).
3 讨 论
Q 型C2H2-ZFPs 是植物特有的C2H2 锌指蛋白亚家族[16]。已有研究表明,Q 型C2H2-ZFPs 在拟南芥、水稻、小麦、马铃薯和杨树等植物中具有特异抗氧化活性[20]。笔者在本研究中利用生物信息学方法在核桃全基因组范围内共鉴定出82个Q型C2H2锌指蛋白基因,该家族成员在核桃中的理论等电点介于5.33~10.23 之间,其等电点范围与马铃薯[20]和扫帚黍(Dichanthelium oligosanthes)[34]相似;核桃Q 型C2H2锌指蛋白基因家族75个(91.46%)JrZFP基因无内含子,这一数量与水稻[12]、杨树[35]和马铃薯[20]相比明显增加。顺式调控元件分析结果表明,核桃Q型C2H2锌指蛋白基因家族启动子包含多个对植物激素和应激信号响应的顺式调控元件,包括ABRE(脱落酸)元件和CGTCA-motif(MeJA)元件等。拟南芥[16]中C2H2-ZFPs 启动子中也发现类似的顺式元件,需要对这些调控区域进行进一步的分析,以验证其在逆境胁迫中的作用。在植物基因组进化[36]中,基因重复事件有助于新基因家族成员的扩增。在本研究中共鉴定出11 对(26.83%)串联重复JrZFPs 基因,63 个(76.83%)JrZFPs 基因发生节段重复事件。重复事件的发生导致一些进化中的新成员可能会失去原有的功能,或获得新的功能以增强植物的适应性,或成为假基因[37]。
据前人报道,在植物中很多ZFPs 特征都是转录抑制因子[38]。例如,LATE(LATE FLOWERING)作为抑制开花的C2H2 锌指蛋白,在所有组织中的异位表达均会导致植物出现晚开花、花器官特性改变和不育花的现象[39];KNU(KNUCKLES)作为细胞增殖的锌指蛋白转录因子,沿着拟南芥雌核发育的近远轴调控花的确定性和基本模式元件的相对大小[40];SUPERMAN 是活跃的抑制因子,其对拟南芥正常发育及开花至关重要[41]。以上蛋白均含有乙烯响应元件结合因子相关的两亲抑制基序(EAR-motif),是植物中发现的最主要转录抑制基序,LxLxL 和DLNxxP 是最常见的类型[42]。蛋白基序是高度保守的氨基酸残基,被认为可能在活性蛋白中具有功能或结构作用[43]。本研究中,Motif 5包含LxLxL和DLNxxP。82个JrZFP中有76个(93%)含有EAR-motif,其中有69个含EAR-motif LxLxL,有7 个含EAR-motif DLNxxP。这表明核桃Q 型C2H2锌指蛋白基因富集了潜在的转录抑制因子。
植物Q型C2H2锌指蛋白转录因子在植物对干旱、寒冷、渗透胁迫、伤害和机械负荷等各种环境胁迫的耐受性中发挥重要作用[17]。本研究中,NaCl处理下JrZFPs基因均为上调基因,其中JrZFP55相对表达量显著上调,推测JrZFP55参与植物NaCl胁迫并起正向调节作用。TaZFP33 在水稻中同源率最高的蛋白是ZOS8-13,主要在种子中表达,盐胁迫下在叶片中表达上调[21],这与JrZFP34 表达结果一致。已有研究报道,过表达ZAT18提高了拟南芥的耐旱性,说明ZAT18 在拟南芥的耐旱性中发挥了积极作用[44]。在本研究中,JrZFP12 相对表达量显著上调,而且随着干旱胁迫时间的延长不断增加,说明其可能与干旱响应相关;JrZFP4、JrZFP55 和JrZFP57 显著下调,推测其在干旱胁迫中起负调控作用。
4 结 论
笔者在本研究中采用生物信息学、实时荧光定量等方法在核桃中鉴定了82个JrZFP基因,并对其进行了理化性质、染色体定位、进化关系、基因结构分析以及干旱胁迫和盐胁迫处理的qRT-PCR 表达。研究表明,JrZFP55在核桃NaCl胁迫中起正向调节作用,JrZFP4、JrZFP55和JrZFP57这3个基因在核桃干旱胁迫中起负调控作用,JrZFP12 在核桃干旱胁迫中起正调控作用。初步获得这些参与盐胁迫和干旱胁迫响应的成员,为下一步解析这些响应基因的功能奠定了基础。
[1] CORBI N,LIBRI V,PASSANANTI C. Artificial zinc finger peptides:A promising tool in biotechnology and medicine[J].Transcription Factors,2004,166:491-507.
[2] TAKATSUJI H. Zinc-finger proteins:The classical zinc finger emerges in contemporary plant science[J].Plant Molecular Biology,1999,39(6):1073-1078.
[3] KIEŁBOWICZ- MATUK A. Involvement of plant C2H2- type zinc finger transcription factors in stress responses[J]. Plant Science,2012,185/186:78-85.
[4] CIFTCI-YILMAZ S,MITTLER R. The zinc finger network of plants[J]. Cellular and Molecular Life Sciences,2008,65(7/8):1150-1160.
[5] MUTHAMILARASAN M,BONTHALA V S,MISHRA A K,KHANDELWAL R,KHAN Y,ROY R,PRASAD M. C2H2 type of zinc finger transcription factors in foxtail millet define response to abiotic stresses[J]. Functional & Integrative Genomics,2014,14(3):531-543.
[6] SAKAMOTO H,MARUYAMA K,SAKUMA Y,MESHI T,IWABUCHI M,SHINOZAKI K,YAMAGUCHI S K.Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought,cold,and high-salinity stress conditions[J].Plant Physiology,2004,136(1):2734-2746.
[7] HUANG J,SUN S J,XU D Q,YANG X,BAO Y M,WANG Z F,TANG H J,ZHANG H S.Increased tolerance of rice to cold,drought and oxidative stresses mediated by the overexpression of a gene that encodes the zinc finger protein ZFP245[J]. Biochemical and Biophysical Research Communications,2009,389(3):556-561.
[8] SUN S J,GUO S Q,YANG X,BAO Y M,TANG H J,SUN H,HUANG J,ZHANG H S. Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice[J].Journal of Experimental Botany,2010,61(10):2807-2818.
[9] YANG K,AN J P,LI C Y,SHEN X N,LIU Y J,WANG D R,JI X L,HAO Y J,YOU C X. The apple C2H2-type zinc finger transcription factor MdZAT10 positively regulates JA-induced leaf senescence by interacting with MdBT2[J]. Horticulture Research,2021,8:159.
[10] 井建玲,张鹏,王振宇,马秋香.木薯C2H2 型锌指蛋白转录因子家族全基因组鉴定及表达分析[J].植物生理学报,2020,56(12):2664-2676.JING Jianling,ZHANG Peng,WANG Zhenyu,MA Qiuxiang.Genome-wide identification and expression analysis of C2H2-type zinc finger protein transcription factor family in Cassava[J].Plant Physiology Journal,2020,56(12):2664-2676.
[11] PABO C O,PEISACH E,GRANT R A. Design and selection of novel Cys2His2 zinc finger proteins[J]. Annual Review of Biochemistry,2001,70:313-340.
[12] AGARWAL P,ARORA R,RAY S,SINGH A K,SINGH V P,TAKATSUJI H,KAPOOR S,TYAGI A K.Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis[J]. Plant Molecular Biology,2007,65(4):467-485.
[13] FARAJI S,RASOULI S H,KAZEMITABAR S K. Genomewide exploration of C2H2 zinc finger family in durum wheat(Triticum turgidum ssp.durum):Insights into the roles in biological processes especially stress response[J]. Biometal,2018,31(6):1019-1042.
[14] TAKATSUJI H,MORI M,BENFEY P N,REN L,CHUA N H.Characterization of a zinc finger DNA- binding protein expressed specifically in Petunia petals and seedlings[J].The EMBO Journal,1992,11(1):241-249.
[15] KUBO K,SAKAMOTO A,KOBAYASHI A,RYBKA Z,KANNO Y,NAKAGAWA H,NISHINO T,TAKATSUJI H. Cys2/His2 zinc-finger protein family of Petunia:Evolution and general mechanism of target-sequence recognition[J]. Nucleic Acids Research,1998,26(2):608-615.
[16] ENGLBRECHT C C,SCHOOF H,BÖHM S. Conservation,diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome[J]. BMC Genomics,2004,5(1):39.
[17] GOURCILLEAU D,LENNE C,ARMENISE C,MOULIA B,JULIEN J L,BRONNER G,FOURNIER N L. Phylogenetic study of plant Q-type C2H2 zinc finger proteins and expression analysis of poplar genes in response to osmotic,cold and mechanical stresses[J].DNA Research,2011,18(2):77-92.
[18] CHEUK A,HOUDE M. Genome wide identification of C1-2i zinc finger proteins and their response to abiotic stress in hexaploid wheat[J]. Molecular Genetics and Genomics,2016,291(2):873-890.
[19] LAWRENCE S D,NOVAK N G. Comparative analysis of the genetic variability within the Q-type C2H2 zinc-finger transcription factors in the economically important cabbage,canola and Chinese cabbage genomes[J].Hereditas,2018,155:29.
[20] LIU Z,JEFFREY A C,LI Y M,ZHANG X J,MENG J G,ZHANG J L,LIU Y H. Genome-wide identification and analysis of the Q-type C2H2 gene family in potato (Solanum tuberosum L.)[J]. International Journal of Biological Macromolecules,2020,153:327-340.
[21] KAM J,GRESSHOFF P M,SHORTER R,XUE G P. The Qtype C2H2 zinc finger subfamily of transcription factors in Triticum aestivum is predominantly expressed in roots and enriched with members containing an EAR repressor motif and responsive to drought stress[J]. Plant Molecular Biology,2008,67(3):305-322.
[22] KIM S H,AHN Y O,AHN M J,JEONG J C,LEE H L,KWAK S S.Cloning and characterization of an orange gene that increases carotenoid accumulation and salt stress tolerance in transgenic sweetpotato cultures[J]. Plant Physiology and Biochemistry,2013,70:445-454.
[23] WANG L J,HE S Z,ZHAI H,LIU D G,WANG Y N,LIU Q C.Molecular cloning and functional characterization of a salt tolerance-associated gene IbNFU1 from sweetpotato[J].Journal of Integrative Agriculture,2013,12(1):27-35.
[24] WANG F B,TONG W J,ZHU H,KONG W L,PENG R H,LIU Q C,YAO Q H. A novel Cys2/His2 zinc finger protein gene from sweetpotato,IbZFP1,is involved in salt and drought tolerance in transgenic Arabidopsis[J].Planta,2016,243(3):783-797.
[25] 刘杜玲,彭少兵,孙红梅,张博勇,朱海兰.早实核桃不同品种抗旱性综合评价[J].园艺学报,2014,41(5):967-974.LIU Duling,PENG Shaobing,SUN Hongmei,ZHANG Boyong,ZHU Hailan. Comprehensive evaluation on drought resistance of early fruiting walnut cultivars[J]. Acta Horticulturae Sinica,2014,41(5):967-974.
[26] 高娅,梁玉,董智,李红丽,刘炳花,范小莉,周晓莹.盐胁迫下印度梨形孢对核桃幼苗生长的影响[J].干旱区资源与环境,2019,33(8):194-198.GAO Ya,LIANG Yu,DONG Zhi,LI Hongli,LIU Binghua,FAN Xiaoli,ZHOU Xiaoying. Effects of Piriformospora indica on the growth of walnut seedlings under salt stress[J].Journal of Arid Land Resources and Environment,2019,33(8):194-198.
[27] ZHOU L,QUAN S W,MA L,XU H,YANG J P,NIU J X.Molecular characterization of SBP-box gene family during floral induction in walnut (Juglans regia L.)[J]. Tree Genetics & Genomes,2019,16(1):1-16.
[28] 李晓斐,张舒婷,陈晓慧,申序,蒋梦琦,刘蒲东,陈裕坤,林玉玲,赖钟雄.龙眼HDAC 家族成员的全基因组鉴定及表达分析[J].果树学报,2020,37(6):793-807.LI Xiaofei,ZHANG Shuting,CHEN Xiaohui,SHEN Xu,JIANG Mengqi,LIU Pudong,CHEN Yukun,LIN Yuling,LAI Zhongxiong. Genome-wide identification and expression analysis of HDAC gene family in Dimocarpus longan Lour. [J]. Journal of Fruit Science,2020,37(6):793-807.
[29] 杨亚明,丁毓端,陈丽娟,田雪婷,殷伟杰,彭红慧,杜薇,梁丽平,任小林.苹果液泡铁离子转运蛋白基因家族的鉴定与表达分析[J].园艺学报,2021,48(2):205-218.YANG Yaming,DING Yuduan,CHEN Lijuan,TIAN Xueting,YIN Weijie,PENG Honghui,DU Wei,LIANG Liping,REN Xiaolin. Identification and expression analysis of the vacuolar iron transporter gene family in apple[J].Acta Horticulturae Sinica,2021,48(2):205-218.
[30] 王洁,吴晓宇,杨柳,段巧红,黄家保.大白菜ACA 基因家族的全基因组鉴定与表达分析[J]. 中国农业科学,2021,54(22):4851-4868.WANG Jie,WU Xiaoyu,YANG Liu,DUAN Qiaohong,HUANG Jiabao. Genome-wide identification and expression analysis of ACA gene family in Brassica rapa[J]. Scientia Agricultura Sinica,2021,54(22):4851-4868.
[31] ZHANG J P,ZHANG W T,JI F Y,QIU J,SONG X B,BU D C,PAN G,MA Q G,CHEN J X,HUANG R M,CHANG Y Y,PEI D. A high-quality walnut genome assembly reveals extensive gene expression divergences after whole-genome duplication[J].Plant Biotechnology Journal,2020,18(9):1848-1850.
[32] VATANSEVER R,KOC I,OZYIGIT I I,SEN U,URAS M E,ANJUM N,PEREIRA E,FILIZ E. Genome-wide identification and expression analysis of sulfate transporter (SULTR) genes in potato (Solanum tuberosum L.)[J]. Planta,2016,244(6):1167-1183.
[33] CHEN C J,CHEN H,ZHANG Y,HANNAH R T,MARGARET H F,HE Y H,XIA R.TBtools:an integrative toolkit developed for interactive analyses of big biological data[J].Molecular Plant,2020,13(8):1194-1202.
[34] MAHAPATRA M,MAHANTY B,JOSHI R K. Genome wide identification and functional assignments of C2H2 Zinc-finger family transcription factors in Dichanthelium oligosanthes[J].Bioinformation,2019,15(9):689-696.
[35] LIU Q G,WANG Z C,XU X M,ZHANG H Z,LI C H. Genome-wide analysis of C2H2 zinc-finger family transcription factors and their responses to abiotic stresses in poplar (Populus trichocarpa)[J].PLoS One,2015,10(8):134753.
[36] CANNON S B,MITRA A,BAUMGARTEN A,YOUNG N D,MAY G.The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana[J].BMC Plant Biology,2004,4:10.
[37] DIAS A P,BRAUN E L,MCMULLEN M D,GROTEWOLD E. Recently duplicated maize R2R3 Myb genes provide evidence for distinct mechanisms of evolutionary divergence after duplication[J].Plant Physiology,2003,131(2):610-620.
[38] UEHARA Y,TAKAHASHI Y,BERBERICH T,MIYAZAKI A,TAKAHASHI H,MATSUI K,OHME T M,SAITOH H,TERAUCHI R,KUSANO T.Tobacco ZFT1,a transcriptional repressor with a Cys2/His2 type zinc finger motif that functions in spermine-signaling pathway[J]. Plant Molecular Biology,2005,59(3):435-448.
[39] WEINGARTNER M,SUBERT C,SAUER N. LATE,a C2H2 zinc-finger protein that acts as floral repressor[J].The Plant Journal,2011,68(4):681-692.
[40] PAYNE T,JOHNSON S D,KOLTUNOW A M. KNUCKLES(KNU)encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium[J].Development,2004,131(15):3737-3749.
[41] HIRATSU K,OHTA M,MATSUI K,OHME T M.The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers[J].FEBS Letters,2002,514(2/3):351-354.
[42] KAGALE S,ROZWADOWSKI K. EAR motif-mediated transcriptional repression in plants[J]. Epigenetics,2011,6(2):141-146.
[43] VATANSEVER R,FILIZ E,EROGLU S.Genome-wide exploration of metal tolerance protein (MTP) genes in common wheat(Triticum aestivum):Insights into metal homeostasis and biofortification[J].Biometals,2017,30(2):217-235.
[44] YIN M Z,WANG Y P,ZHANG L H,LI J Z,QUAN W L,LI Y,WANG Q F,CHAN Z L. The Arabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress[J]. Journal of Experimental Botany,2017,68(11):2991-3005.