作物基因突变能够促成品种的改良,如今大多数栽培作物来源于自然发生的随机突变[1]。通常作物自然变异的概率极低,随着育种技术的发展,人工诱变可以快速得到变异植株,并且已在作物育种中广泛应用。但是,化学或物理诱变会在作物基因组中随机引入突变,导致结果无法预测,并且得到目标表型的概率很低。2013年,高效的成簇规律间隔短回文重复序列(clustered regularly interspaced short palindromic repeats, CRISPR)/CRISPR 相关蛋白9(Cas9)基因组编辑工具被开发出来,有效解决了这一难题,并迅速成为作物改良的常规手段[2]。基因组编辑技术是在基因组特定区域剪切DNA,实现DNA的特定变化并引入新的修复机制[3]。基因组编辑技术可以在作物基因组的靶点位置精确地实现删除、替换或插入特定序列的作用,最终产生新的目标性状。全基因组序列与基因组编辑工具两者的相互结合,可以改变靶基因的可控性和准确性,是研究突变植株和创造新种质的高效方法,在作物生物技术领域已发挥了变革性的效应。至今为止,CRISPR/Cas9是使用最为广泛的基因组编辑工具,在模式植物拟南芥[4]、烟草[5]、番茄[6]及水稻[7]、玉米[8]等作物中都有大量的基因突变研究报道,突出了该工具具有简单、高效、成本低和功能强大的明显优势。近年来,基因编辑技术陆续应用在柑橘[9]、葡萄[10]、苹果[11]、香蕉[12]、草莓[13]、猕猴桃[14]、可可[15]、梨[16]、荔枝[17]和蓝莓[18]等遗传转化体系稳定的果树作物中,并获得了一批目标性状显著的株系,为果树育种及研究工作提供新的工具和研究思路。
1 CRISPR/Cas9系统及作用过程
CRISPR/Cas9 系统属于CRISPR/Cas 系统中的Ⅱ型系统,为细菌和古细菌免疫系统中的一部分[19],具有避免噬菌体和质粒DNA 入侵原核生物的保护功能[20]。CRISPR/Cas系统由2部分组成:CRISPR序列和Cas 基因家族。CRISPR 序列包含了起始转录的前导序列(leader)、高度保守的重复序列(repeat)和间隔序列(spacer)[21]。Cas基因家族编码的蛋白质能够协同CRISPR序列的转录产物共同发挥作用。
CRISPR/Cas9 系统作用过程一般可分为3 个阶段:第一阶段是当外源DNA 侵染细菌时,宿主首先识别外来遗传物质并将外源DNA切割成小片段,继而将一段DNA 结合到前导区和重复区之间形成一个新的间隔序列[22]。第二阶段是CRISPR 前导序列启动转录合成CRISPR RNA 前体(pre-crRNA)和反式激活crRNA(tracrRNA),接着pre-crRNA 通过RNAase Ⅲ加工成成熟的crRNA[23]。第三阶段是成熟的crRNA激活Cas9蛋白的合成,形成一个复合物并与外源序列特异识别,接着Cas9蛋白剪切目标双链DNA,导致DNA断裂[24]。DNA被剪切后,植物体会启动自身的修复机制,主要有2种修复途径:同源重组(homologous Recombination,HR)和非同源端连接(non-homologous end joining,NHEJ)[25]。HR途径可以利用同源模板插入特定基因片段。NHEJ 途径通常有核苷酸的插入或删除,在基因的编码区内密码子阅读框发生移动,破坏基因的功能,达到基因敲除的效果[26]。CRISPR/Cas9 系统作用原理如图1所示[27]。
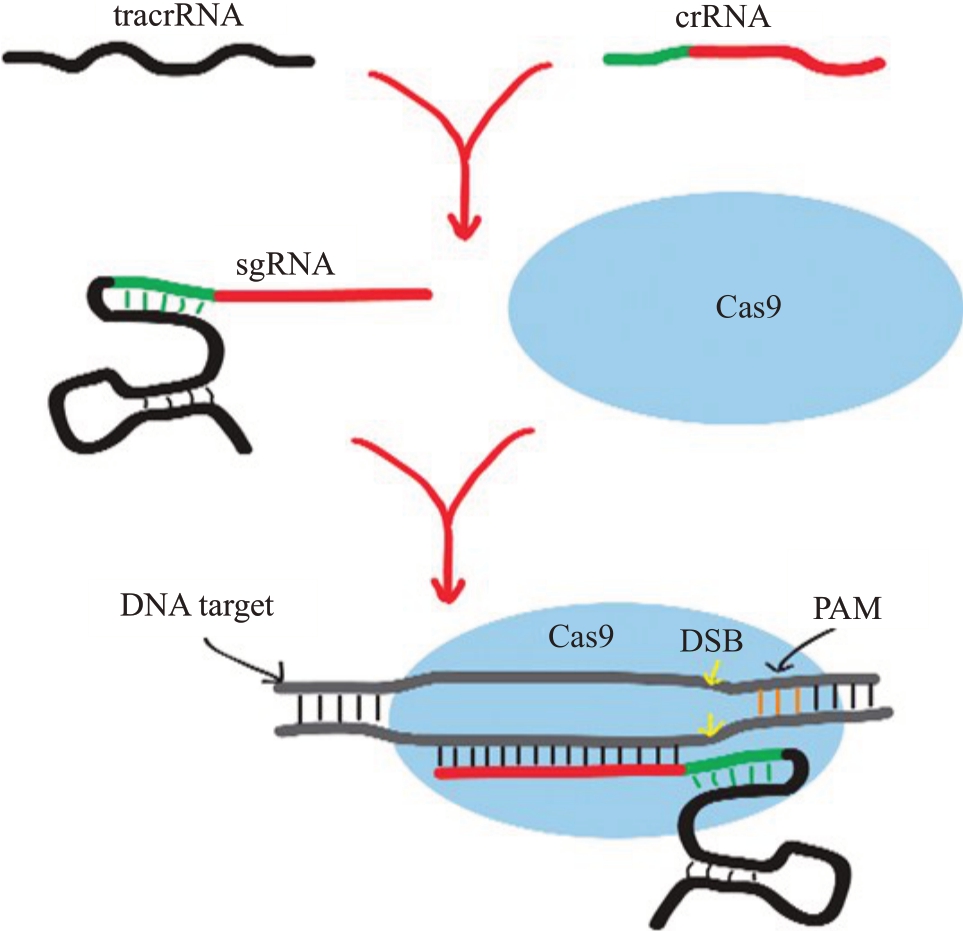
图1 CRISPR/Cas9 系统作用原理
Fig.1 Schematic diagram of the working principle of the CRISPR/Cas9 system
2 最新精确基因编辑技术
CRISPR/Cas9系统自开发以来成为了应用最为广泛的基因编辑工具。但不可否认的是,NHEJ 途径具有一定随机性,难以达到精确的单个碱基或者多个基因同时编辑的目的,并且也存在脱靶的问题;而由HR 途径完成的碱基替换或删除,效率也并不理想。于是,新的基因编辑技术被开发出来,在一定程度上达到了更加完善的效果。
2.1 单碱基编辑器
单碱基编辑器是基于CRISPR/Cas9基因编辑系统开发出的新基因编辑技术,在不产生DNA双链断裂的情况下,能够精准靶向单个碱基,产生单个碱基替换效应。该系统根据脱氨酶的不同,分为胞嘧啶碱基编辑器(CBE)和腺嘌呤碱基编辑器(ABE)。CBE 由一个失去活性的Cas9(dead Cas9,dCas9)或仅产生断裂缺口的Cas9(Cas9 nickase,nCas9)与胞嘧啶脱氨酶组成,作用过程中胞嘧啶脱氨酶将胞嘧啶(C)的氨基脱掉形成尿嘧啶(U),经过植物基因组DNA 自身的修复机制和复制将U 转化成胸腺嘧啶(T),完成C/G到A/T之间转变[28]。ABE的腺苷脱氨酶是TadA 突变体,其将腺嘌呤(A)上的氨基去除,初步转化为次黄嘌呤(I),经过基因组DNA 自身的修复和复制机制,实现A/T到C/G的转化[29]。
2.2 引导编辑
引导编辑技术(prime editing)是可以在靶标位点实现任何形式的精准基因编辑技术,例如碱基的任意替换、基因片段的删除和插入等。引导编辑器有2个部分:一是融合了nCas9(H840A)与逆转录酶(reverse transcriptase, RT)的效应蛋白;二是pegRNA(prime editing guide RNA),包含PBS(primer binding site)序列和RT 模板(RT template)序列。在pegRNA 的引导下,nCas9 剪切目标DNA 链断裂,PBS 序列与单链互补结合,并启动RT 模板的逆转录,随后经过DNA修复和复制过程,最终将RT模板上碱基序列的改变引入对应的目标靶点[30]。
2.3 CRISPR/Cpf1多基因编辑系统
CRISPR/Cpf1 系统是属于CRISPR 的2 类V-A型系统[31]。Cpf1 有双重切割酶活性的特点,能够对pre-crRNA进行加工处理,随后在crRNA引导之下,识别并剪切靶位点,使得基因双链断裂[32]。Cpf1 在同时进行多个基因的编辑方面与Cas9 相比更具有优势,与此同时脱靶效率也相对较低[33]。
3 CRISPR/Cas9基因编辑技术在果树作物中的应用
3.1 Cas9和sgRNA启动子的选择
Cas9 和sgRNA 是CRISPR/Cas9 编辑载体的核心部分。Cas9 和sgRNA 的表达量对于载体编辑效率具有决定性的意义,二者启动子的选择也就至关重要,Cas9的启动子绝大多数使用的是花椰菜花叶病毒启动子CaMV 35S 和玉米泛素启动子Pubi。sgRNA 的启动子常用的是拟南芥U3(AtU3)、U6 启动子(AtU6)和水稻U3(OsU3)、U6 启动子(OsU6)。不同的果树之间果树基因编辑的效率往往会有差异,甚至会存在差异较大的情况(表1)。目前,在苹果[34]、柑橘[53]、葡萄[58]和草莓[64]中通过使用植物内源启动子U3或U6来启动sgRNA的表达,从而有效提高编辑效率。
表1 CRISPR/Cas9 基因编辑技术在果树中的应用
Table 1 Application of CRISPR/Cas9 gene editing technology in fruit trees
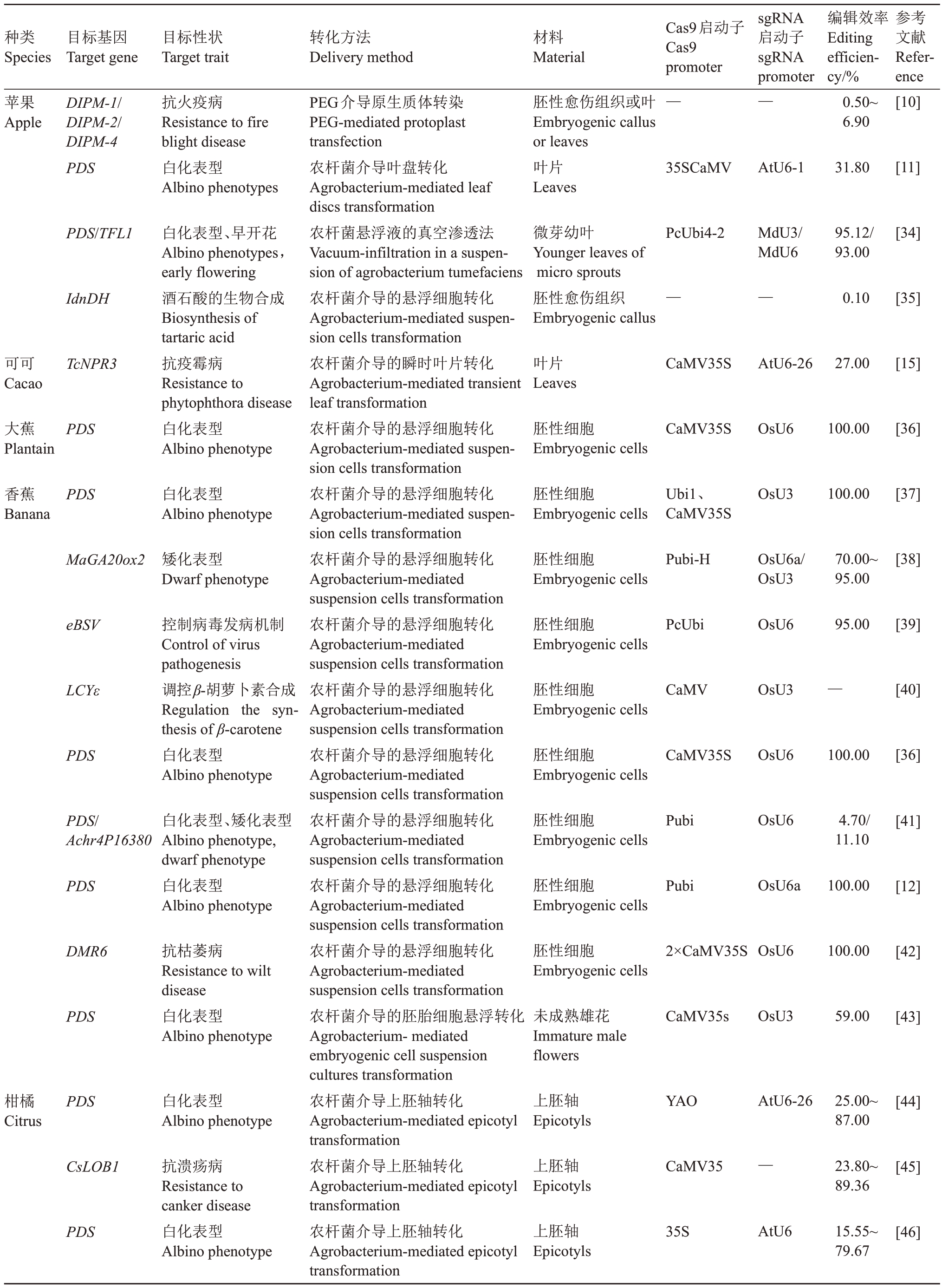
种类Species目标基因Target gene目标性状Target trait转化方法Delivery method材料Material Cas9启动子Cas9 promoter sgRNA启动子sgRNA promoter编辑效率Editing efficiency/%参考文献Reference苹果Apple DIPM-1/DIPM-2/DIPM-4 PDS抗火疫病Resistance to fire blight disease白化表型Albino phenotypes胚性愈伤组织或叶Embryogenic callus or leaves叶片Leaves——PEG介导原生质体转染PEG-mediated protoplast transfection农杆菌介导叶盘转化Agrobacterium-mediated leaf discs transformation农杆菌悬浮液的真空渗透法Vacuum-infiltration in a suspension of agrobacterium tumefaciens农杆菌介导的悬浮细胞转化Agrobacterium-mediated suspension cells transformation农杆菌介导的瞬时叶片转化Agrobacterium-mediated transient leaf transformation农杆菌介导的悬浮细胞转化Agrobacterium-mediated suspension cells transformation农杆菌介导的悬浮细胞转化Agrobacterium-mediated suspension cells transformation农杆菌介导的悬浮细胞转化Agrobacterium-mediated suspension cells transformation农杆菌介导的悬浮细胞转化Agrobacterium-mediated suspension cells transformation农杆菌介导的悬浮细胞转化Agrobacterium-mediated suspension cells transformation农杆菌介导的悬浮细胞转化Agrobacterium-mediated suspension cells transformation农杆菌介导的悬浮细胞转化Agrobacterium-mediated suspension cells transformation农杆菌介导的悬浮细胞转化Agrobacterium-mediated suspension cells transformation农杆菌介导的悬浮细胞转化Agrobacterium-mediated suspension cells transformation农杆菌介导的胚胎细胞悬浮转化Agrobacterium-mediated embryogenic cell suspension cultures transformation农杆菌介导上胚轴转化Agrobacterium-mediated epicotyl transformation农杆菌介导上胚轴转化Agrobacterium-mediated epicotyl transformation农杆菌介导上胚轴转化Agrobacterium-mediated epicotyl transformation 0.50~6.90[10]35SCaMV AtU6-1 31.80[11]PDS/TFL1 PcUbi4-2 MdU3/MdU6 95.12/93.00[34]IdnDH微芽幼叶Younger leaves of micro sprouts胚性愈伤组织Embryogenic callus——0.10[35]可可Cacao TcNPR3叶片Leaves CaMV35S AtU6-26 27.00[15]大蕉Plantain PDS白化表型、早开花Albino phenotypes,early flowering酒石酸的生物合成Biosynthesis of tartaric acid抗疫霉病Resistance to phytophthora disease白化表型Albino phenotype胚性细胞Embryogenic cells CaMV35S OsU6 100.00[36]香蕉Banana PDS 白化表型Albino phenotype胚性细胞Embryogenic cells Ubi1、CaMV35S OsU3 100.00[37]MaGA20ox2矮化表型Dwarf phenotype胚性细胞Embryogenic cells Pubi-H OsU6a/OsU3 70.00~95.00[38]eBSV 胚性细胞Embryogenic cells PcUbi OsU6 95.00[39]LCYε胚性细胞Embryogenic cells CaMV OsU3—[40]PDS控制病毒发病机制Control of virus pathogenesis调控β-胡萝卜素合成Regulation the synthesis of β-carotene白化表型Albino phenotype胚性细胞Embryogenic cells CaMV35S OsU6 100.00[36]PDS/Achr4P16380胚性细胞Embryogenic cells Pubi OsU6 4.70/11.10[41]PDS白化表型、矮化表型Albino phenotype,dwarf phenotype白化表型Albino phenotype胚性细胞Embryogenic cells Pubi OsU6a 100.00[12]DMR6胚性细胞Embryogenic cells 2×CaMV35SOsU6 100.00[42]PDS抗枯萎病Resistance to wilt disease白化表型Albino phenotype未成熟雄花Immature male flowers CaMV35s OsU3 59.00[43]柑橘Citrus PDS 白化表型Albino phenotype上胚轴Epicotyls YAO AtU6-26 25.00~87.00[44]CsLOB1上胚轴Epicotyls CaMV35—23.80~89.36[45]PDS抗溃疡病Resistance to canker disease白化表型Albino phenotype上胚轴Epicotyls 35S AtU6 15.55~79.67[46]
续表Continued Table
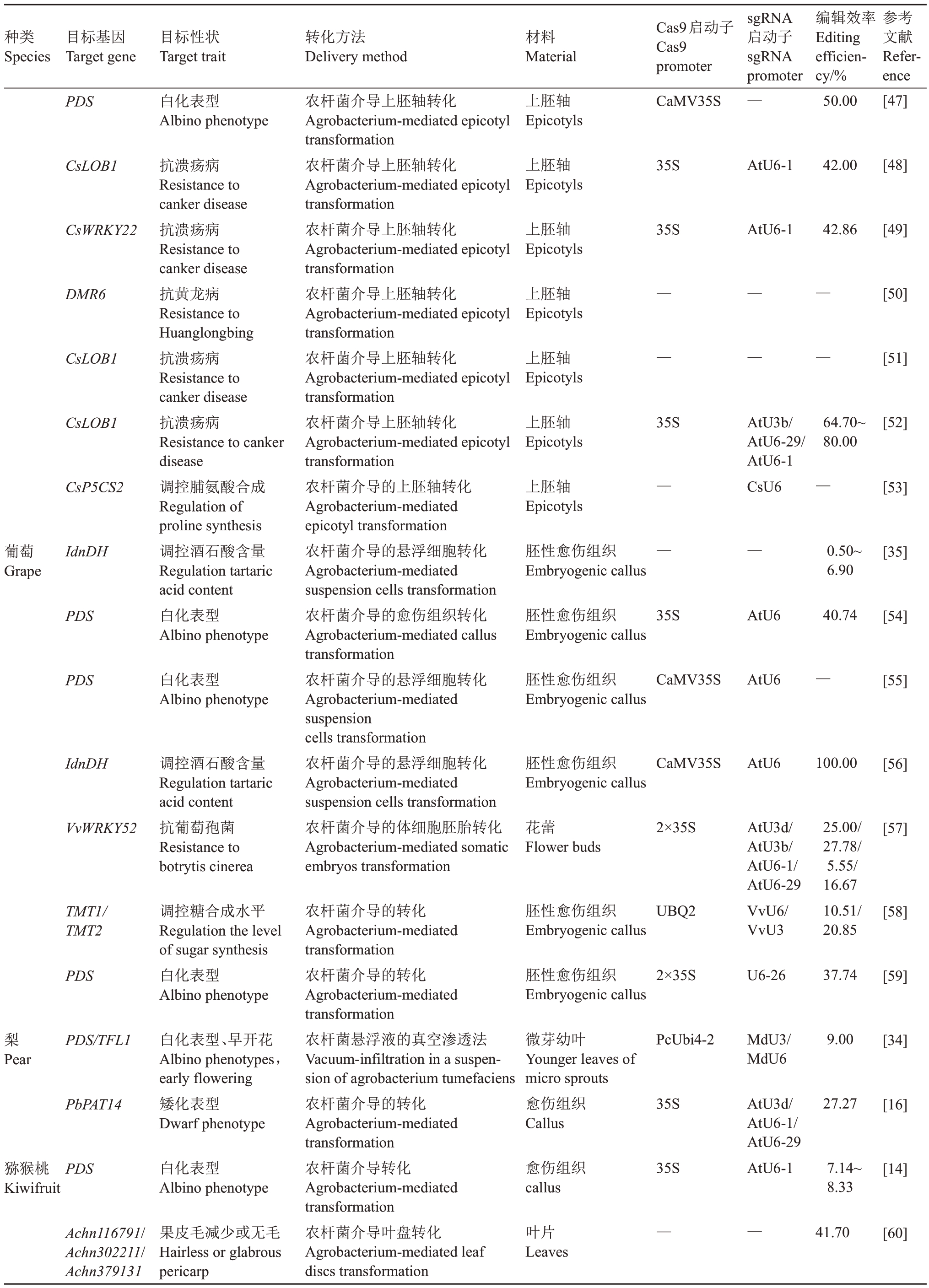
种类Species目标基因Target gene目标性状Target trait转化方法Delivery method材料Material Cas9启动子Cas9 promoter sgRNA启动子sgRNA promoter编辑效率Editing efficiency/%参考文献Reference PDS 白化表型Albino phenotype上胚轴Epicotyls CaMV35S —50.00[47]CsLOB1上胚轴Epicotyls 35S AtU6-1 42.00[48]CsWRKY22上胚轴Epicotyls 35S AtU6-1 42.86[49]DMR6上胚轴Epicotyls[50]CsLOB1上胚轴Epicotyls——————[51]CsLOB1上胚轴Epicotyls 35S 64.70~80.00[52]CsP5CS2上胚轴Epicotyls AtU3b/AtU6-29/AtU6-1 CsU6—[53]葡萄Grape IdnDH 胚性愈伤组织Embryogenic callus———0.50~6.90[35]PDS抗溃疡病Resistance to canker disease抗溃疡病Resistance to canker disease抗黄龙病Resistance to Huanglongbing抗溃疡病Resistance to canker disease抗溃疡病Resistance to canker disease调控脯氨酸合成Regulation of proline synthesis调控酒石酸含量Regulation tartaric acid content白化表型Albino phenotype胚性愈伤组织Embryogenic callus 35S AtU6 40.74[54]PDS 白化表型Albino phenotype胚性愈伤组织Embryogenic callus CaMV35S AtU6—[55]IdnDH 胚性愈伤组织Embryogenic callus CaMV35S AtU6 100.00[56]VvWRKY52调控酒石酸含量Regulation tartaric acid content抗葡萄孢菌Resistance to botrytis cinerea农杆菌介导上胚轴转化Agrobacterium-mediated epicotyl transformation农杆菌介导上胚轴转化Agrobacterium-mediated epicotyl transformation农杆菌介导上胚轴转化Agrobacterium-mediated epicotyl transformation农杆菌介导上胚轴转化Agrobacterium-mediated epicotyl transformation农杆菌介导上胚轴转化Agrobacterium-mediated epicotyl transformation农杆菌介导上胚轴转化Agrobacterium-mediated epicotyl transformation农杆菌介导的上胚轴转化Agrobacterium-mediated epicotyl transformation农杆菌介导的悬浮细胞转化Agrobacterium-mediated suspension cells transformation农杆菌介导的愈伤组织转化Agrobacterium-mediated callus transformation农杆菌介导的悬浮细胞转化Agrobacterium-mediated suspension cells transformation农杆菌介导的悬浮细胞转化Agrobacterium-mediated suspension cells transformation农杆菌介导的体细胞胚胎转化Agrobacterium-mediated somatic embryos transformation花蕾Flower buds 2×35S [57]TMT1/TMT2胚性愈伤组织Embryogenic callus UBQ2 AtU3d/AtU3b/AtU6-1/AtU6-29 VvU6/VvU3 25.00/27.78/5.55/16.67 10.51/20.85[58]PDS调控糖合成水平Regulation the level of sugar synthesis白化表型Albino phenotype胚性愈伤组织Embryogenic callus 2×35S U6-26 37.74[59]梨PDS/TFL1 PcUbi4-2 9.00[34]Pear MdU3/MdU6 PbPAT14白化表型、早开花Albino phenotypes,early flowering矮化表型Dwarf phenotype微芽幼叶Younger leaves of micro sprouts愈伤组织Callus 35S 27.27[16]猕猴桃Kiwifruit PDS 白化表型Albino phenotype愈伤组织callus 35S AtU3d/AtU6-1/AtU6-29 AtU6-1 7.14~8.33[14]Achn116791/Achn302211/Achn379131果皮毛减少或无毛Hairless or glabrous pericarp农杆菌介导的转化Agrobacterium-mediated transformation农杆菌介导的转化Agrobacterium-mediated transformation农杆菌悬浮液的真空渗透法Vacuum-infiltration in a suspension of agrobacterium tumefaciens农杆菌介导的转化Agrobacterium-mediated transformation农杆菌介导转化Agrobacterium-mediated transformation农杆菌介导叶盘转化Agrobacterium-mediated leaf discs transformation叶片Leaves——41.70[60]
续表Continued Table
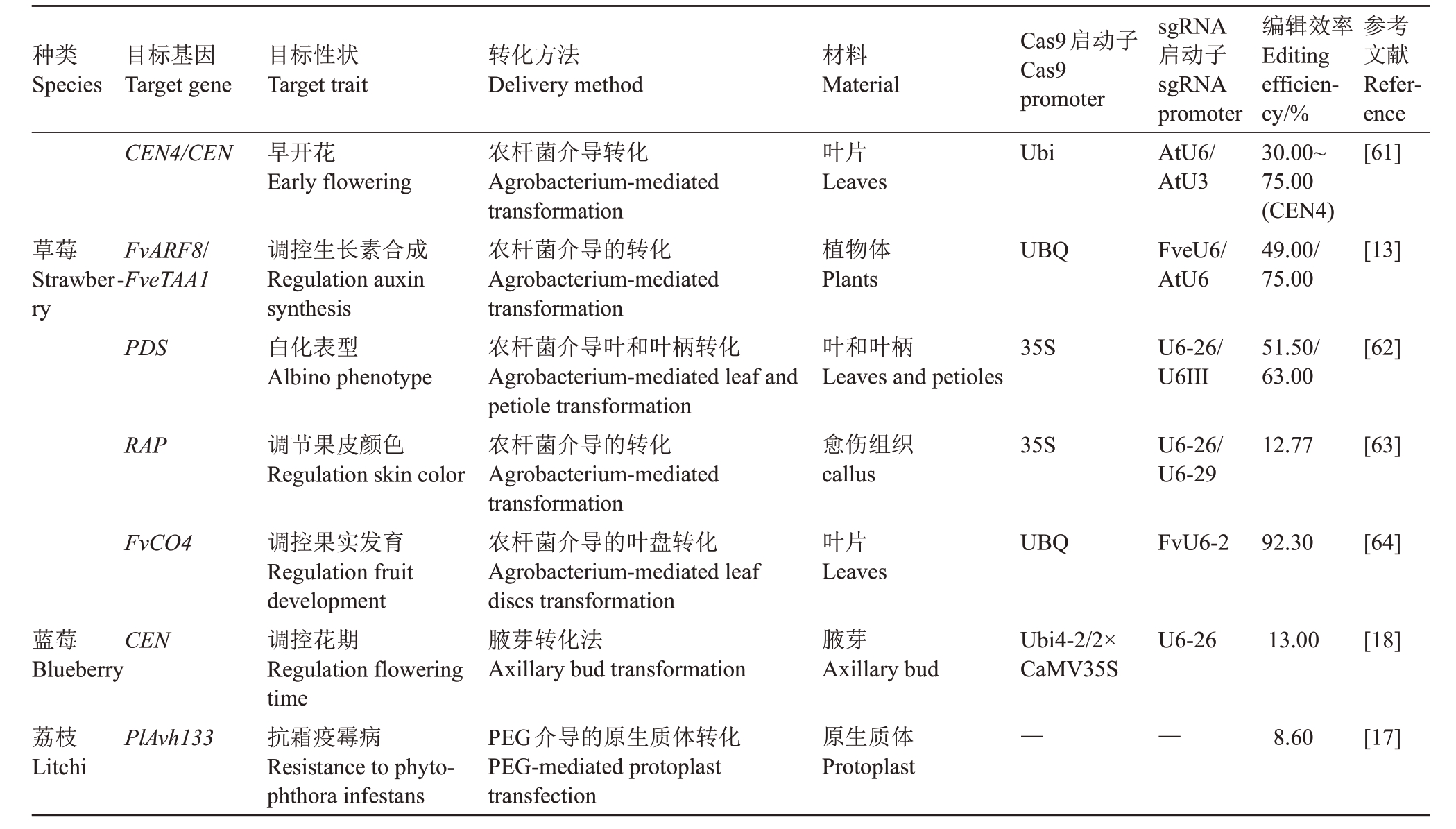
注:“—”表示参考文献中无相关信息。
Note:“—”means no relevant information in the references.
草莓Strawberry蓝莓Blueberry荔枝Litchi CEN4/CEN FvARF8/FveTAA1 PDS RAP FvCO4 CEN PlAvh133早开花Early flowering调控生长素合成Regulation auxin synthesis白化表型Albino phenotype调节果皮颜色Regulation skin color调控果实发育Regulation fruit development调控花期Regulation flowering time抗霜疫霉病Resistance to phytophthora infestans农杆菌介导转化Agrobacterium-mediated transformation农杆菌介导的转化Agrobacterium-mediated transformation农杆菌介导叶和叶柄转化Agrobacterium-mediated leaf and petiole transformation农杆菌介导的转化Agrobacterium-mediated transformation农杆菌介导的叶盘转化Agrobacterium-mediated leaf discs transformation腋芽转化法Axillary bud transformation PEG介导的原生质体转化PEG-mediated protoplast transfection叶片Leaves植物体Plants叶和叶柄Leaves and petioles愈伤组织callus叶片Leaves腋芽Axillary bud原生质体Protoplast Ubi UBQ 35S 35S UBQ Ubi4-2/2×CaMV35S—AtU6/AtU3 FveU6/AtU6 U6-26/U6III U6-26/U6-29 FvU6-2 U6-26—30.00~75.00(CEN4)49.00/75.00 51.50/63.00 12.77 92.30 13.00 8.60[61][13][62][63][64][18][17]种类Species目标基因Target gene目标性状Target trait转化方法Delivery method材料Material Cas9启动子Cas9 promoter sgRNA启动子sgRNA promoter编辑效率Editing efficiency/%参考文献Reference
3.2 不同果树所适用的转化方法和材料存在一定差异
在果树遗传转化的过程中,CRISPR/Cas9 载体几乎都是通过农杆菌介导的方法侵染植物的组织或器官,除了农杆菌侵染外,聚乙二醇(polyethylene glycol ,PEG)介导的原生质体转化法在苹果[10]和荔枝[16]中也有应用。
不同的物种所适用的转化材料存在一定差异(表1),如香蕉[37]和葡萄[35]通常使用悬浮胚性细胞,也有使用花器官的报道[43,57],柑橘[44]适用上胚轴,草莓[62]和猕猴桃[61]普遍利用叶片进行叶盘的转化等。
3.3 CRISPR/Cas9编辑技术在果树作物中的应用
目前,CRISPR/Cas9 编辑技术主要应用在柑橘[9]、葡萄[10]、苹果[11]、香蕉[12]和草莓[13]等果树作物上,猕猴桃[14]、荔枝[17]和蓝莓[18]等果树也相继被报道(表1)。八氢番茄红素脱氢酶(phytoene desaturase,PDS)作为指示基因,容易获得白化目标性状,在绝大多数果树中已被敲除。在果树花期调控的研究中,敲除苹果[34]和猕猴桃[61]FTL 和CEN 基因可以获得提早开花的突变植株。通过基因编辑培育抗病虫害的品种在果树中应用最为广泛,特别是敲除柑橘CsLOB1 基因进行抗柑橘溃疡病的研究[45];另外,香蕉抗枯萎病[42]、葡萄抗灰霉病[57]、可可抗疫霉病[15]和荔枝抗霜疫霉病[17]的植株培育效果显著。株型改良方面,在香蕉[38]和梨[16]中,CRISPR/Cas9 编辑技术也被用于创制矮化表型新品种植株。果实品质方面,利用基因编辑技术在香蕉[40]、苹果[35]、葡萄[35,56,58]中实现了果实营养物质合成的调控,也对果实外观品质进行改善和优化,如产生白果皮草莓[63]、表皮毛减少或无毛猕猴桃[60]。
3.3.1 产生白化表型 植株白化表型是检验和判断CRISPR/Cas9体系是否有效的经典和指示表型。自2013 年CRISPR/Cas9 系统开始应用于植物基因组编辑至今,已成功实现了苹果、大蕉、可可、香蕉、柑橘、葡萄、猕猴桃、草莓、蓝莓、梨和荔枝多种果树的基因组编辑[65]。敲除PDS 基因,能够阻止叶绿素和类胡萝卜素的产生,从而导致植株形成白化表型[66]。该目标性状相对容易获得,因此往往用来判断基因组编辑系统在果树中的有效性。设计针对香蕉中2个PDS基因的sgRNA引导Cas9剪切靶基因,突变效率达到59%,获得了完全白化和杂色表型的植株[43]。利用经过改良后的CRISPR/Cas9多靶点载体系统编辑香蕉MaPDS基因,结果发现,61.2%的植株表现出白化突变表型[12]。此外,利用CRISPR/Cas9 基因编辑系统在香蕉中递送多顺反子gRNAs靶向PDS 基因外显子的编辑效率高达100%[37]。同时,在草莓中实现了高效的PDS 基因突变结果,获得明显白化表型的目标植株[62]。另外,在柑橘中用AtYAO 基因启动子代替常用的花椰菜花叶病毒35S启动子驱动Cas9 基因时,突变效率明显提高,甚至达到100%[44]。使用SaCas9/sgRNA 系统替代Sp-Cas9/sgRNA 系统敲除PDS 基因也能成功进行柑橘基因组编辑[46]。CRISPR/Cas9 编辑技术也在苹果[11,34]、葡萄[54-55,59]、猕猴桃[14]、梨[34]、金橘[47]和大蕉[36]中成功应用,实现PDS基因的敲除。
3.3.2 调控童期和花期 一般多年生果树童期长,限制了品种通过传统育种方法进行优化[67]。童期的长短与终端开花(terminal flower,TFL)蛋白相关。TFL 通过抑制多种开花蛋白的表达,发挥着开花的负调控作用[68]。利用针对TFL1 基因的不同sgRNAs,通过CRISPR/Cas9 系统编辑苹果和梨,苹果中有93%早花表型的突变株系,而梨中只有9%具有早花性状,突变率相对苹果显得极低[34]。培育早花早熟品种,可以满足市场需求,增加生产者收益。CEN 和与其密切相关的基因TFL1 是拟南芥的开花抑制因子,CEN 的敲除可能导致早熟开花。通过CRISPR/Cas9系统介导的蓝莓基因组编辑最终获得含有CEN 区域突变的6 个株系,但是这些株系不仅没有得到早开花的目标性状,反而植株生长明显受到抑制,可能是开花调节物质不仅影响生殖发育,还影响营养生长[18]。同时,CEN 同源基因是潜在的开花抑制因子。以猕猴桃CEN的同源基因AcCEN4和AcCEN 为靶点,利用CRISPR/Cas9 系统同时进行敲除,发现突变植株由原来的多年生木本攀缘植物转变为株型紧凑的植株,并且产生顶花和果实迅速发育的现象[61]。
3.3.3 改善果实品质 果实品质由外观品质和内在品质组成,是衡量果品商品价值的重要指标。在香蕉中,以LCYε基因的第5外显子为靶点,利用CRISPR/Cas9技术提高香蕉中β胡萝卜素的含量,创制了富含β 胡萝卜素的香蕉品种;编辑株系的果肉与未编辑株系相比,果肉中β 胡萝卜素积累量提高了6倍[40]。在葡萄中,分别用VvU6和UBQ2启动子替代AtU6 和35S 启动子对CRISPR/Cas9 系统进行了优化,开发了一个包含传统多个sgRNA表达盒或多顺反子tRNA-sgRNA 盒的多重基因组编辑系统;以渗透性单糖转运体TMT 基因家族成员TMT1 和TMT2基因为靶点,进行了葡萄多基因组编辑,整体编辑效率提高了10%,同时糖合成水平下降,表明这2个基因在葡萄糖积累中起重要作用[58]。以葡萄中调控酒石酸(tartaric acid,TA)生物合成的IdnDH 基因为靶基因进行敲除沉默,TA在葡萄悬浮细胞及其再生植株中的生物合成或积累失败[35,56]。通过CRISPR/Cas9 系统定点突变山金柑中脯氨酸合成限速酶编码基因CsP5CS2,得到了突变的阳性植株[53]。
草莓果实的颜色是草莓育种的重要性状。叶柄花青素还原基因(reduced anthocyanins in petioles,RAP)编码1 种谷胱甘肽S 转移酶(glutathione Stransferase,GST),该酶结合花青素,促进其从细胞质运输到液泡。CRISPR/Cas9系统在T0代同时敲除6个RAP基因拷贝,得到了绿茎白果表型。经检测,果实的总花青素含量较野生型果实显著降低[63]。草莓的FvCO4 基因在果实的髓部和皮层都有特异性表达,CRISPR/Cas9 基因编辑草莓FvCO4 基因,获得12株基因编辑株系,突变株系的果实发育受到显著影响,证明该基因与草莓果实发育相关[64]。以拟南芥表皮毛起始和发育基因的候选基因Achn116791、Achn302211 和Achn379131 为靶基因,在猕猴挑中进行CRISPR/Cas9 基因编辑,分别获得9、1和0个株系的基因编辑植株,且突变植株出现叶片表皮毛密度降低、长度变短的表型[60]。
3.3.4 产生矮化表型 CRISPR/Cas9系统诱导调控香蕉矮小性状的MaGA20ox2 基因突变,共得到7 个半矮化突变系,检测显示赤霉素含量与突变表型变化一致[38]。另外,CRISPR/Cas9 系统敲除香蕉候选矮化基因Achn379131,获得了87 个抗性转基因株系[41]。培育具有矮化、紧凑特性的植株可显著提高梨果实产量。设计3 种不同的sgRNAs,通过CRISPR/Cas9 系统敲除梨中与植物发育相关的S 酰基转移酶基因PbPAT14,得到6个矮黄、叶片数量减少表型的株系[16]。
3.3.5 培育抗病虫害植株 果树生长发育过程中受到的虫害和病害,会直接影响到果品的产量和品质。病虫害的发生通常会增加生产成本,也增大造成重大损失的风险。解决这些问题最有效和经济的办法之一是培育抗病虫害品种。柑橘溃疡病由细菌病原体柑橘黄单胞菌(Xanthomonas citri subsp.citri,Xcc)引起,Citrus sinensis LATERAL ORGAN BOUNDARIES(CsLOB1)是该病的易感基因[69]。研究显示,通过使用CRISPR/Cas9 系统修饰CsLOB1的2 个等位基因启动子中PthA4 效应子顺式元件,可以明显提高突变体植株对Xcc 的抗性[9]。通过构建5 个CRISPR/Cas9 编辑载体修饰柑橘CsLOB1 启动子中的PthA4 结合元件,有4 个品系表现出对柑橘溃疡病具有较强的抗性[48]。由CRISPR/Cas9系统衍生的双组分系统Cas9/sgRNA,只需要对sgRNA进行微小的改变就可以特异性靶向目的基因,实现在柑橘中突变CsLOB1 基因[51]。利用CRISPR/Cas9系统靶向晚锦橙基因组的CsLOB1 启动子,在突变体植株中也实现了较大DNA 片段的删除[52]。在柑橘中分别敲除CsWRKY22[49]和DMR6[50]基因也得到了抗溃疡病和抗黄龙病的突变植株。
以苹果DIPM-1、DIPM-2 和DIPM-4 基因为靶点,将CRISPR/Cas9系统直接递送到原生质体系统,从而实现定向基因编辑,提高植株对火疫病的抗性[10]。针对葡萄的WRKY52 基因(编码与生物胁迫响应相关的转录因子)的第一个外显子设计4 个gRNAs,CRISPR/Cas9系统介导敲除WRKY52基因,增强了对灰霉病菌的抗性[57]。CRISPR/Cas9系统编辑可可的NON-EXPRESSOR OF PATHOGENESISRELATED GENES3(NPR3)基因,该基因编码防御反应抑制因子。先在叶片中进行瞬时表达测试,随后生成CRISPR/Cas9系统突变的体细胞胚胎[15]。
香蕉条纹病毒病(banana streak virus,BSV)对香蕉有很大的危害,利用CRISPR/Cas9 系统对整合到香蕉基因组的BSV基因序列进行突变。水分胁迫下,75%的基因组编辑再生植株没有出现病害症状[39]。香蕉黄单胞菌病(banana xanthomonas wilt,BXW)是由香蕉黄单胞菌引起的病害,只有野生种具有抗BXW 的能力。Downy mildew resistance 6(DMR6)是编码2-氧戊二酸铁(II)依赖性加氧酶(2-oxoglutarate oxygenase,2OGO)的易感基因,该基因在病原体感染期间上调,抑制植物的抗性免疫功能。CRISPR/Cas9 系统敲除香蕉MusaDMR6 基因,大多数突变体对病害BXW 的抗性增强,植株延迟感病时间甚至未有感病症状,并且能够正常生长,没有形态上的变异[42]。荔枝霜疫病严重危害荔枝产业的发展,该病的RXLR (Arg-X- Leu-Arg,X 代表任意氨基酸)效应蛋白编码PlAvh133基因,通过借鉴大豆的CRISPR/Cas9 编辑体系,获得了5 个突变植株,进一步验证显示该基因在突变体中未表达[17]。
4 展 望
CRISPR/Cas9 系统是一项革命性的技术,可以通过引导和控制基因组的变化用于作物育种和生物研究。果树的世代周期长、杂合度高,采用传统育种手段培育新品种难度高且耗时,CRISPR/Cas9 基因编辑技术的开发能够在很大程度上解决这一问题。但是,CRISPR/Cas9 系统在应用的过程中仍然有许多障碍需要克服。首先,高效的组织培养再生系统和遗传转化技术是开展基因编辑技术工作的基础。虽然许多果树都有自身的遗传转化和再生流程体系,但是遗传转化方法通常具有基因型特异性,在一个物种中建立高效的遗传转化体系需要对多个因素进行探索优化,研究者关注度较高的柑橘、葡萄、苹果和梨等大宗果树遗传转化体系趋于成熟,香蕉、荔枝等热带果树遗传转化体系也逐步建立起来,但研究不够深入的油梨、番石榴、莲雾和杨桃等优稀果树则稍显滞后。其次,脱靶效应是CRISPR/Cas9 基因编辑存在的普遍问题。再者,需要选择更加合适的Cas9和sgRNA启动子以提高编辑效率。
高效的编辑效率是应用CRISPR/Cas9系统过程中追求的重要目标,因此,持续不断地优化遗传转化体系和再生系统,对于将CRISPR/Cas9 系统应用于果树中意义重大。在降低脱靶效率方面,基因靶位点的选择和sgRNA的设计都显得格外重要,通常编辑载体设计多个sgRNAs同时靶向一个目标基因来提高编辑靶基因的准确性[70]。近年来,利用拟南芥YAO启动子和植物泛素UBQ启动子驱动Cas9的表达,克隆本物种内源U3和U6启动子驱动sgRNA的表达,编辑的效率明显提高。由此可见,选择合适的启动子同时表达Cas9 和sgRNA 是果树高频率突变的重要因素。新出现的基因编辑技术也能够弥补CRISPR/Cas9系统的不足。如单碱基编辑器能够精准替换单个碱基,实现C/G 到A/T 和C/G 到A/T 的转变,该技术已在水稻和玉米应用,获得了性状改良的植株[71]。引导编辑系统可以实现所有类型的碱基编辑,目前在水稻和小麦上已建立起适用于植物的引导编辑系统[72]。近年来发现的新基因编辑系统CRISPR/Cpf1,在多重基因编辑和染色体片段删除方面更有优势。利用该技术在水稻中实现了多重基因编辑[73],在大豆中也实现8 个靶基因的编辑并且实现染色体大、中、小片段的删除[74]。从成功应用新编辑系统的作物中可以推测,单碱基编辑、引导编辑以及CRISPR/Cpf1 系统都有应用于果树的可能性,但是不同的编辑系统所产生的目的效应不同,选择的编辑系统应根据具体研究情况而定。
[1] SIKORA P,CHAWADE A,LARSSON M,OLSSON J,OLSSON O,TALON M. Mutagenesis as a tool in plant genetics,functional genomics,and breeding[J]. International Journal of Plant Genomics,2011,2011:314829.
[2] PETOLINO J F,SRIVASTAVA V,DANIELL H. Editing Plant Genomes:A new era of crop improvement[J].Plant Biotechnology Journal,2016,14(2):435-436.
[3] VOYTAS D F. Plant genome engineering with sequence-specific nucleases[J].Annual Review of Plant Biology,2013,64:327-350.
[4] LI J F,NORVILLE J E,AACH J,MCCORMACK M,ZHANG D,BUSH J,CHURCH G M,SHEEN J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9[J]. Nature Biotechnology,2013,31(8):688-691.
[5] NEKRASOV V,STASKAWICZ B,WEIGEL D,JONES J D G,KAMOUN S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease[J]. Nature Biotechnology,2013,31(8):691-693.
[6] MARTINELLI F,GRILLONE G,SGROI F. Proposal of a genome editing system for genetic resistance to tomato spotted wilt virus[J]. American Journal of Applied Sciences,2014,11(11):1904-1913.
[7] SHAN Q W,WANG Y P,LI J,ZHANG Y,CHEN K L,LIANG Z,ZHANG K,LIU J X,XI J Z J,QIU J L,GAO C X.Targeted genome modification of crop plants using a CRISPR-Cas system[J].Nature Biotechnology,2013,31(8):686-688.
[8] SCHUSTER M,SCHWEIZER G,REISSMANN S,REGINE K.Genome editing in Ustilago maydis using the CRISPR-Cas system[J].Fungal Genetics and Biology,2016,89:3-9.
[9] JIA H G,ORBOVIC V,JONES J B,WANG N. Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:DCsLOB1.3 infection[J]. Plant Biotechnology Journal,2016,14(5):1291-1301.
[10] MALNOY M,VIOLA R,JUNG M H,KOO O J,KIM S,KIM J S,VELASCO R,KANCHISWAMY C N. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins[J].Frontiers in Plant Science,2016,7:1904.
[11] NISHITANI C,HIRAI N,KOMORI S,WADA M,OKADA K,ASAKABE K,YAMAMOTO T,OSAKABE Y. Efficient genome editing in apple using a CRISPR/Cas9 system[J].Scientific Reports,2016,6:31481.
[12] 胡春华,邓贵明,孙晓玄,左存武,李春雨,邝瑞彬,杨乔松,易干军.香蕉CRISPR/Cas9 基因编辑技术体系的建立[J].中国农业科学,2017,50(7):1294-1301.HU Chunhua,DENG Guiming,SUN Xiaoxuan,ZUO Cunwu,LI Chunyu,KUANG Ruibin,YANG qiaosong,YI Ganjun. Establishment of an efficient CRISPR/Cas9-mediated gene editing system in banana[J]. Scientia Agricultura Sinica,2017,50(7):1294-1301.
[13] ZHOU J H,WANG G M,LIU Z C. Efficient genome editing of wild strawberry genes,vector development and validation[J].Plant Biotechnology Journal,2018,16(11):1868-1877.
[14] WANG Z P,WANG S B,LI D W,ZHANG Q,LI L,ZHONG C H,LIU Y F HUANG H W.Optimized paired-sgRNA/Cas9 cloning and expression cassette triggers high-efficiency multiplex genome editing in kiwifruit[J].Plant Biotechnology Journal,2018,16(8):1424-1433.
[15] FISTER A S,LANDHERR L,MAXIMOVA S N,GUILTINAN M J. Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao[J].Frontiers in Plant Science,2018,9:268.
[16] PANG H G,YAN Q,ZHAO S L,HE F,XU J F,QI B X,ZHANG Y X. Knockout of the S-acyltransferase gene,Pb-PAT14,confers the dwarf yellowing phenotype in first generation pear by ABA accumulation[J]. International Journal of Molecular Sciences,2019,20(24):6347.
[17] 司徒俊键,江立群,邵毅,孔广辉,习平根,姜子德. CRISPR/Cas9 介导的荔枝霜疫霉基因组编辑系统的建立[J]. 菌物研究,2020,18(3):181-188.SITU Junjian,JIANG Liqun,SHAO Yi,KONG Guanghui,XI Pinggen,JIANG Zide. Establishment of CRISPR/Cas9 genome editing system in Peronophythora litchii[J]. Journal of Fungal Research,2020,18(3):181-188.
[18] OMORI M,YAMANE H,OSAKABE K,OSAKABE Y,TAO R. Targeted mutagenesis of CENTRORADIALIS using CRISPR/Cas9 system through the improvement of genetic transformation efficiency of tetraploid highbush blueberry[J]. The Journal of Horticultural Science and Biotechnology,2021,96(2):153-161.
[19] CONG L,RAN F A,COX D,LIN S,BARRETTO R,HABIB N,HSU P D,WU X,JIANG W,MARRAFFINI L A,ZHANG F. Multiplex genome engineering using CRISPR/cas systems[J].Science,2013,339(6121):819-823.
[20] BARRANGOU R,FREMAUX C,DEVEAU H,RICHARDS M,BOYAVAL P,MOINEAU S,ROMERO D A,HORVATH P.CRISPR provides acquired resistance against viruses in prokaryotes[J].Science,2007,315(5819):1709-1712.
[21] JINEK M,CHYLINSKI K,FONFARA I,HAUER M,DOUDNA J A.A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity[J]. Science,2012,337(6096):816-821.
[22] WIEDENHEFT B,STERNBERG S H,DOUDNA J A. RNAguided genetic silencing systems in bacteria and Archaea[J].Nature,2012,482(7385):331-338.
[23] KARGINOV F V,HANNON G J. The CRISPR system:Small RNA- guided defense in bacteria and Archaea[J]. Molecular Cell,2010,37(1):7-19.
[24] GARNEAU J E,DUPUIS M È,VILLION M,ROMERO D A,BARRANGOU R,BOYAVAL P,FREMAUX C,HORVATH P,MAGADÁN A H,MOINEAU S.The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA[J]. Nature,2010,468(7320):67-71.
[25] KISHI-KABOSHI M,AIDA R,SASAKI K. Genome engineering in ornamental plants:Current status and future prospects[J].Plant Physiology and Biochemistry,2018,131:47-52.
[26] KARKUTE S G,SINGH A K,GUPTA O P,SINGH P M,SINGH B. CRISPR/Cas9 mediated genome engineering for improvement of horticultural crops[J]. Frontiers in Plant Science,2017,8:1635.
[27] ZHOU J H,LI D D,WANG G M,WANG F X,KUNJAL M,JOLDERSMA D,LIU Z C. Application and future perspective of CRISPR/Cas9 genome editing in fruit crops[J]. Journal of Integrative Plant Biology,2020,62(3):269-286.
[28] KOMOR A C,KIM Y B,PACKER M S,ZURIS J A,LIU D R.Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage[J]. Nature,2016,533(7603):420-424.
[29] GAUDELLI N M,KOMOR A C,REES H A,APACKER M S,BADRAN A H,BRYSON D I,LIU D R.Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage[J].Nature,2017,551(7681):464-471.
[30] LIN Q P,JIN S,ZONG Y,YU H,ZHU Z X,LIU G W,KOU L Q,WANG Y P,QIU J H,LI J Y,GAO C X. High-efficiency prime editing with optimized,paired pegRNAs in plants[J]. Nature Biotechnology,2021,39(8):923-927.
[31] MAKAROVA K S,WOLF Y I,ALKHNBASHI O S,COSTA F,SHAH S A,SAUNDERS S J,BARRANGOU R,BROUNS S J J,CHARPENTIER E,HAFT D H,HORVATH P,MOINEAU S,MOJICA F J M,TERNS B M,TERNS M P,WHITE M F,YAKUNIN A F,GARRETT G A,OOST J V D,BACKOFEN R B,KOONIN E V. An updated evolutionary classification of CRISPR-Cas systems[J]. Nature Reviews Microbiology,2015,13(11):722-736.
[32] FONFARA I,RICHTER H,BRATOVIČ M,RHUN A L,CHARPENTIER E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA[J]. Nature,2016,532(7600):517-521.
[33] 郭婷,安新民. 多重基因组编辑中CRISPR-Cas9 系统和CRISPR-Cpf1 系统的应用和比较[J]. 中国细胞生物学学报,2019,41(11):2234-2244.GOU Ting,AN Xinmin. Application and comparison of CRISPR-Cas9 system and CRISPR-Cpf1 system in multigenome editing[J]. Chinese Journal of Cell Biology,2019,41(11):2234-2244.
[34] CHARRIER A,VERGNE E,DOUSSET N J,RICHER A,PETITEAU A,CHEVREAU E. Efficient targeted mutagenesis in apple and first time edition of pear using the CRISPR-Cas9 system[J].Frontiers in Plant Science,2019,10:40.
[35] OSAKABE Y,LIANG Z C,REN C,NISHITANI C,OSAKABE K,WADA W,KOMORI S,MALNOY M,VELASCO R,POLI M,JUNG M H,KOO O J,VIOLA R,KANCHISWAMY C N. CRISPR-Cas9-mediated genome editing in apple and grapevine[J].Nature Protocols,2018,13(12):2844-2863.
[36] NTUI V O,TRIPATHI J N,TRIPATHI L. Robust CRISPR/Cas9 mediated genome editing tool for banana and plantain (Musa spp.)[J].Current Plant Biology,2020,21:100128.
[37] NAIM F,DUGDALE B,KLEIDON J,BRININ A,SHAND K,WATERHOUSE P,DALE J. Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9[J]. Transgenic Research,2018,27(5):451-460.
[38] SHAO X H,WU S P,DOU T X,ZHU H C,HU C H,HUO H Q,HE W D,DENG G M,SHENG O,BI F C,GAO H J,DONG T,LI C Y,YANG Q S,YI G.Using CRISPR/Cas9 genome editing system to create MaGA20ox2 gene-modified semi-dwarf banana[J].Plant Biotechnology Journal,2020,18(1):17-19.
[39] TRIPATHI J N,NTUI V O,RON M,MUIRURI S K,BRITT A,TRIPATHI L. CRISPR/Cas9 editing of endogenous Banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding[J]. Communications Biology,2019,2:46.
[40] KAUR N,ALOK A,SHIVANI,KUMAR P,KAUR N,AWASTHI P,CHATURVEDI S,PANDEY P,PANDEY A,PANDEY A K,TIWARI S. CRISPR/Cas9 directed editing of lycopene Epsilon-cyclase modulates metabolic flux for β-carotene biosynthesis in banana fruit[J]. Metabolic Engineering,2020,59:76-86.
[41] 孙晓玄.香蕉CRISPR/Cas9 基因编辑技术体系的建立[D].广州:华南农业大学,2017.SUN Xiaoxuan.Establishment of an efficient CRISPR/Cas9-mediated gene editing system in banana[D].Guangzhou:South China Agricultural University,2017.
[42] TRIPATHI J N,NTUI V O,SHAH T,TRIPATHI L. CRISPR/Cas9-mediated editing of DMR6 orthologue in banana (Musa spp.) confers enhanced resistance to bacterial disease[J]. Plant Biotechnology Journal,2021,19(7):1291-1293.
[43] KAUR N,ALOK A,SHIVANI,KAUR N,PANDEY P,AWASTHI P,TIWARI S. CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome[J]. Functional & Integrative Genomics,2018,18(1):89-99.
[44] ZHANG F,LEBLANC C,IRISH V F,JACOB Y.Rapid and efficient CRISPR/Cas9 gene editing in Citrus using the YAO promoter[J].Plant Cell Reports,2017,36(12):1883-1887.
[45] JIA H G,ZHANG Y Z,ORBOVIĆ V,XU J,WHITE F F,JONES J B,WANG N. Genome editing of the disease susceptibility gene CsLOB1 in Citrus confers resistance to Citrus canker[J].Plant Biotechnology Journal,2017,15(7):817-823.
[46] JIA H G,XU J,ORBOVIĆ V,ZHANG Y Z,WANG N. Editing Citrus genome via SaCas9/sgRNA system[J]. Frontiers in Plant Science,2017,8:2135.
[47] ZHU C Q,ZHENG X J,HUANG Y,YE J L,CHEN P,ZHANG C L,ZHAO F,XIE Z Z,ZHANG S Q,WANG N,LI H,WANG L,TANG X M,CHAI L J,XU Q,DENG X X.Genome sequencing and CRISPR/Cas9 gene editing of an early flowering Mini-Citrus (Fortunella hindsii) [J]. Plant Biotechnology Journal,2019,17(11):2199-2210.
[48] PENG A H,CHEN S C,LEI T G,XU L Z,HE Y G,WU L,YAO L X,ZOU X P. Engineering canker- resistant plants through CRISPR/Cas9- targeted editing of the susceptibility gene CsLOB1 promoter in Citrus[J]. Plant Biotechnology Journal,2017,15(12):1509-1519.
[49] WANG L J,CHEN S C,PENG A H,XIE Z,HE Y R,ZHOU X P. CRISPR/Cas9-mediated editing of CsWRKY22 reduces susceptibility to Xanthomonas citri subsp. citri in Wanjincheng orange [Citrus sinensis (L.) Osbeck][J]. Plant Biotechnology Reports,2019,13(5):501-510.
[50] ZHANG S J,SHI Q C,DUAN Y P,HALL D,GUPTA G,STOVER E. Regulation of citrus DMR6 via RNA interference and CRISPR/Cas9-mediated gene editing to improve Huanglongbing tolerance[C]// International Congress of Plant Pathology,Plant Health in A Global Economy.APSNET,2018:13.
[51] JIA H G,ZOU X P,ORBOVIC V,WANG N. Genome editing in Citrus tree with CRISPR/Cas9[J].Methods in Molecular Biology,2019,1917:235-241.
[52] 邹修平,范迪,彭爱红,何永睿,许兰珍,雷天刚,姚利晓,李强,罗克明,陈善春.CRISPR/Cas9 介导柑橘CsLOB1 基因启动子的多位点编辑[J].园艺学报,2019,46(2):337-344.ZOU Xiuping,FAN Di,PENG Aihong,HE Yongrui,XU Lanzhen,LEI Tiangang,YAO Lixiao,LI Qiang,LUO Keming,CHEN Shanchun. CRISPR/Cas9-mediated editing of multiple sites in the Citrus CsLOB1 promoter[J].Acta Horticulturae Sinica,2019,46(2):337-344..
[53] 徐旋.柑橘原生质体瞬时转化体系优化及利用CRISPR/Cas9技术定点突变山金柑基因[D].武汉:华中农业大学,2018.XU Xuan. Optimization of Citrus protoplast transient transformation system and using CRISPR/cas9 genome editing system to modify gene in fortuella hindsii[D]. Wuhan:Huazhong Agricultural University,2018.
[54] NAKAJIMA I,BAN Y,AZUMA A,ONOUE N,MORIGUCHI T,YAMAMOTO T,TOKI S,ENDO M. CRISPR/Cas9-mediated targeted mutagenesis in grape[J]. PLoS One,2017,12(5):e0177966.
[55] REN F R,REN C,ZHANG Z,DUAN W,LECOURIEUX D,LI S H,LIANG Z C.Efficiency optimization of CRISPR/Cas9-mediated targeted mutagenesis in grape[J]. Frontiers in Plant Science,2019,10:612.
[56] REN C,LIU X J,ZHANG Z,WANG Y,DUAN W,LI S H,LIANG Z C. CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay(Vitis vinifera L.)[J].Scientific Reports,2016,6:32289.
[57] WANG X H,TU M X,WANG D J,LIU J W,LI Y J,LI Z,WANG Y J,WANG X P. CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation[J]. Plant Biotechnology Journal,2018,16(4):844-855.
[58] REN C,LIU Y F,GUO Y C,DUAN W,FAN P G,LI S H,LIANG Z C.Optimizing the CRISPR/Cas9 system for genome editing in grape by using grape promoters[J]. Horticulture Research,2021,8(1):52.
[59] 郭晔,万东艳,柴壮壮,王跃进,文颖强.利用CRISPR/Cas9 敲除葡萄VviPDS1 基因的研究[J].园艺学报,2019,46(4):623-634.GUO Ye,WAN Dongyan,CHAI Zhuangzhuang,WANG Yuejin,WEN Yingqiang. Knock-out analysis of VviPDS1 gene using CRISPR/Cas9 in grapevine[J].Acta Horticulturae Sinica,2019,46(4):623-634.
[60] 李创.猕猴桃表皮毛发育基因的筛选及其在海沃德猕猴桃中的基因编辑研究[D].杨凌:西北农林科技大学,2019.LI Chuang. Screening the genes regulating trichome development in Actinidia and their genome editing in‘Hayward’Kiwifruit[D].Yangling:Northwest A&F University,2019.
[61] VARKONYI-GASIC E,WANG T C,VOOGD C,JEON S,DRUMMOND R S,GLEAVE A P,ALLAN A C. Mutagenesis of kiwifruit CENTRORADIALIS-like genes transforms a climbing woody perennial with long juvenility and axillary flowering into a compact plant with rapid terminal flowering[J].Plant Biotechnology Journal,2019,17(5):869-880.
[62] WILSON F M,HARRISON K,ARMITAGE A D,SIMKKIN A J,HARRISON R. CRISPR/Cas9-mediated mutagenesis of phytoene desaturase in diploid and octoploid strawberry[J]. Plant Methods,2019,15(1):1-13.
[63] GAO Q,LUO H F,LI Y P,LIU Z C,KANG C Y.Genetic modulation of RAP alters fruit coloration in both wild and cultivated strawberry[J]. Plant Biotechnology Journal,2020,18(7):1550-1561.
[64] 陈文君.FvCO4在草莓果实早期发育中的功能鉴定[D].沈阳:沈阳农业大学,2019.CHE Wenjun. Function analysis of FvCO4 in the early development of strawberry fruit[D]. Shenyang:Shenyang Agricultural University,2019.
[65] LI J F,NORVILLE J E,AACH J,MCCORMACK M,ZHANG D D,BUSH J,CHURCH G M,SHEEN J.Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9[J]. Nature Biotechnology,2013,31(8):688-691.
[66] GAO J P,WANG G H,MA S Y,XIE X D,WU X W,ZHAGN X T,WU Y Q,ZHAO P,XIA Q Y. CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum[J]. Plant Molecular Biology,2015,87(1/2):99-110.
[67] FURR J R,COOPER W C,REECE P C. An investigation of flower formation in adult and juvenile Citrus trees[J].American Journal of Botany,1947,34(1):1-8.
[68] PILLITTERI L J,LOVATT C J,WALLING L L. Isolation and characterization of a TERMINAL FLOWER homolog and its correlation with juvenility in Citrus[J]. Plant Physiology,2004,135(3):1540-1551.
[69] HU Y,ZHANG J L,JIA H G,SOSSO D,LI T,FROMMER W B,YANG B,WHITE F F,WANG N,JONES J B. Lateral organ boundaries 1 is a disease susceptibility gene for Citrus bacterial canker disease[J]. Proceedings of the National Academy of Sciences,2014,111(4):E521-E529.
[70] HASHIMOTO R,UETA R,ABE C,OSAKABE Y,OSAKABE K. Efficient multiplex genome editing induces precise,and selfligated type mutations in tomato plants[J].Frontiers in Plant Science,2018,9:916.
[71] ZONG Y,WANG Y P,LI C,ZHANG R,CHEN K L,RAN Y D,QIU J L,WANG D W,GAO C X. Precise base editing in rice,wheat and maize with a Cas9-cytidine deaminase fusion[J]. Nature Biotechnology,2017,35(5):438-440.
[72] TANG X,SRETENOVIC S,REN Q R,JIA X Y,LI M K,FAN T T,YIN D S,XIANG S Y,GUO Y C,LIU L,ZHENG X L,QI Y P,ZHANG Y. Plant prime editors enable precise gene editing in rice cells[J].Molecular Plant,2020,13(5):667-670.
[73] WANG M G,MAO Y F,LU Y M,TAO X P,ZHU J K. Multiplex gene editing in rice using the CRISPR-Cpf1 system[J].Molecular Plant,2017,10(7):1011-1013.
[74] DUAN K X,CHENG Y Y,JI J,WANG C C,WEI Y S,WANG Y C. Large chromosomal segment deletions by CRISPR/Lb-Cpf1-mediated multiplex gene editing in soybean[J]. Journal of Integrative Plant Biology,2021,63(9):1620-1631.