葡萄炭疽病又名晚腐病,是一种主要由胶孢炭疽菌(C.gloeosporioides)引起的真菌性病害[1],其在世界范围内广泛分布。主要危害成熟期的果实,染病的果实和叶片上出现黑褐色的炭疽病斑点,果实表面溃烂、汁液外渗、诱发霉菌污染,严重影响葡萄的产量和品质。据统计,葡萄属中对炭疽病的抗性在种间和种内表现出较大差异,东亚种群对葡萄炭疽病抗性普遍较强[2],欧亚种葡萄的整体抗病能力较弱[3]。
人们曾认为胶孢炭疽菌是引起炭疽病的唯一病原菌,但随后在澳大利亚、日本、韩国、美国等多个地区又陆续发现因尖孢炭疽菌(C.acutatum)感染而显现葡萄炭疽病病症的果实[4-6]。此后又在部分地区发病葡萄果实上分离出少量其他种类炭疽病菌。截至目前,炭疽菌属已经超过100 多个种[7],已证实能在葡萄上引起病症的病原菌包括C. gloeosporioides、C. acutatum、C. fructicola、C. viniferum、C. crassipes(Speg.)Arx、C. vitis 和C. hebeiense 等[8-14],其中报道最多、最具有代表性的是胶孢炭疽菌。此外,由炭疽病病菌引起的植物炭疽病还是一种广谱性的病害,其可以在多种果树或作物上引起炭疽病症状[8]。
目前,已在多种植物上利用RNA-seq测序技术,对炭疽病病原菌侵染引起的炭疽病响应机制和差异表达基因进行了研究报道。陈哲等[15]利用转录组测序分析,发现炭疽病病原菌侵染影响了类黄酮生物合成和植物激素信号传导等通路中关键基因的表达,并选出了19个与草莓应答炭疽病相关的差异表达基因。Sudheeran等[16]对C.gloeosporioides侵染杧果的红色面和青色面进行转录组研究,认为杧果对炭疽病抗性的产生同类黄酮化合物合成和光照的诱导相关,并受到了22个转录因子和33个信号相关转录产物的影响。Hong等[17]利用转录组测序分析,从杧果中筛选到35个候选基因,可能参与C.gloeosporioides侵染引起的防御响应,包括17个乙烯响应因子(ERF)、6个核苷酸结合位点富含亮氨酸重复序列(NBS LRR)、6个致病相关基因非表达子(NPR)和6个致病相关蛋白(PRs)。茶树受炭疽病病原菌诱导的转录组分析发现,差异表达基因主要富集在植物激素传导和植病原体互作通路,而PR1 蛋白和内源水杨酸在炭疽病感染过程中对免疫激活起关键作用[18]。李静[19]检测了草莓28 个NBS-LRR 基因在接种炭疽病后的表达量,发现15个基因表达量发生了变化,其中有2 个基因表达量上调。在被侵染的梨叶片不同代谢通路分析中,各种免疫模式和类黄酮、苯丙烷类代谢等均参与了梨叶片对炭疽病病原菌的抵抗过程,SA 信号通路在抗病响应中发挥重要作用,而JA、ET信号通路等则受到了抑制作用[20]。
笔者利用感病欧洲葡萄里扎马特和抗病中国野生刺葡萄黑珍珠杂交后代中极感病株系和极抗病株系进行RNA-seq 测序分析,比对其转录组水平的差异,筛选葡萄抗炭疽病相关候选基因,为葡萄炭疽病抗性分子机制研究提供理论基础。
1 材料和方法
1.1 材料
以感炭疽病欧亚种里扎马特和抗炭疽病刺葡萄黑珍珠的杂交后代感炭疽病7-2-6 株系和抗炭疽病7-1-8株系为材料;以上材料均定植于中国农业科学院郑州果树研究所的国家果树种质郑州葡萄圃。
用菌饼接种法在叶片上接种胶孢炭疽菌,置于28 ℃恒温箱培养。同时以刺伤后清水处理的叶片做对照,分别于接种后0,24,48,72 h收集叶片,设分组:R0、R1、R2、R3分别为抗病株系接种胶孢炭疽菌0、24、48、72 h 的样本,R0_M0、R1_M1、R2_M2、R3_M3为对应时期清水对照;S代表感病株系,命名方式同上,所有时期样本均设3 个生物学重复。将样本液氮速冻,放置在-80 ℃超低温冰箱保存备用。
1.2 RNA-seq 文库制备与测序及差异表达基因的筛选
转录组测序委托深圳华大基因科技服务有限公司进行。对获得的Raw reads进行过滤,获得高质量的Clean reads,用于后续的信息分析。以葡萄基因组12×PN40024 为参考基因进行比对。使用Bowtie2 将clean reads 比对到参考序列以统计基因比对率,RSEM计算基因和转录本的表达水平。DEGseq算法进行差异表达基因检测,将差异倍数为2 倍以上且Q-value ≤0.001 的基因,作为显著差异表达基因。
1.3 差异表达基因的富集、转录因子家族表达和WGCNA分析
根据KEGG 注释结果以及官方分类,将差异表达基因进行生物通路分类,使用R 软件中的Phyper函数进行富集分析。计算P 值,并进行FDR 校正,将FDR ≤0.01 的通路视为显著富集;用Getorf 检测差异表达基因的ORF,用Hmmsearch将ORF比对到转录因子蛋白结构域,根据转录因子家族特征对差异表达基因进行TF编码能力鉴定;使用百迈客数据分析平台进行Weighted gene co-expression network analysis(WGCNA);使用TBtools-HeatMap做热图表达模式分析。
1.4 实时荧光定量PCR分析
基于分析挑取9个差异表达基因进行qRT-PCR定量。每个样品设置3个生物学重复。根据定量引物设计的原则,在NCBI-blast 的primer 设计页面进行引物设计和特异性检验(https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome)并由上海生工生物公司进行合成,引物详细信息见表1。PCR 反应程序:95 ℃5 min;95 ℃10 s,57 ℃10 s,72 ℃10 s,45 个循环;95 ℃5 s,65 ℃1 min,97 ℃continuous;40 ℃30 s。
表1 qRT-PCR 验证基因及引物信息
Table 1 Validation gene and primer information for qRT-PCR

基因符号Gene symbol 100260212 100256849 100241136 100249652 100233053 100243167 100247990 109124267 100248210基因ID Gene ID VIT_05s0077g00730 VIT_15s0046g02190 VIT_09s0002g05080 VIT_14s0006g02290 VIT_05s0094g01570 VIT_12s0134g00020 VIT_03s0088g00710 N/A N/A引物序列Primer sequence(5’-3’)F:GATCAGACTAAAGCGGCGGA F:CGACATGTGGGCTTGGAGAA F:GACTCCAGATGGCAGACAGT F:GGGAAGGAGGTCAAGGATGG F:TGGAGCATGTAACCACCCAG F:AGCAATTGCCGGAAAGGACTA F:GTGGGTGGGGAATGCCGA F:TGAATCGGGATCAACCGACA F:GCACCAGTTGTCAGCTCCTT R:CGCAAATCTCGGCTCTCTCT R:TCGGATCTGTTTCGCTCCAC R:GCTTCTGCACCCCATCACTA R:CTCACGAATCTCCGACACCC R:AGGTCGAACAACCAAGTCCC R:ACAAGTCAGCCCACATATTGA R:CCCGGTGGATCGTAGTTGC R:CCAGCTAGGAGGCCAAGAATA R:CTCTTAACCACCAGGGCCAA产物长度Length of product/bp 108 129 102 105 80 145 122 84 95
2 结果与分析
2.1 转录组测序数据整体情况
将测序所得序列去除接头序列、低质量序列和核糖体rRNA序列后,每个样品平均产出6.65 Gb数据,比对基因组的平均比对率为86.08%,比对基因集的平均比对率为79.39%;共检测到基因数为25 545,其中已知的基因为23 220个,预测的新基因为2403个。
对各组重复之间进行相关性分析(图1),除S1_2 相关系数为0.767,低于0.8 外,其余每组样品之间的相关系数均高于0.8,分析数据时将S1_2舍去,以确保结果的可靠性。
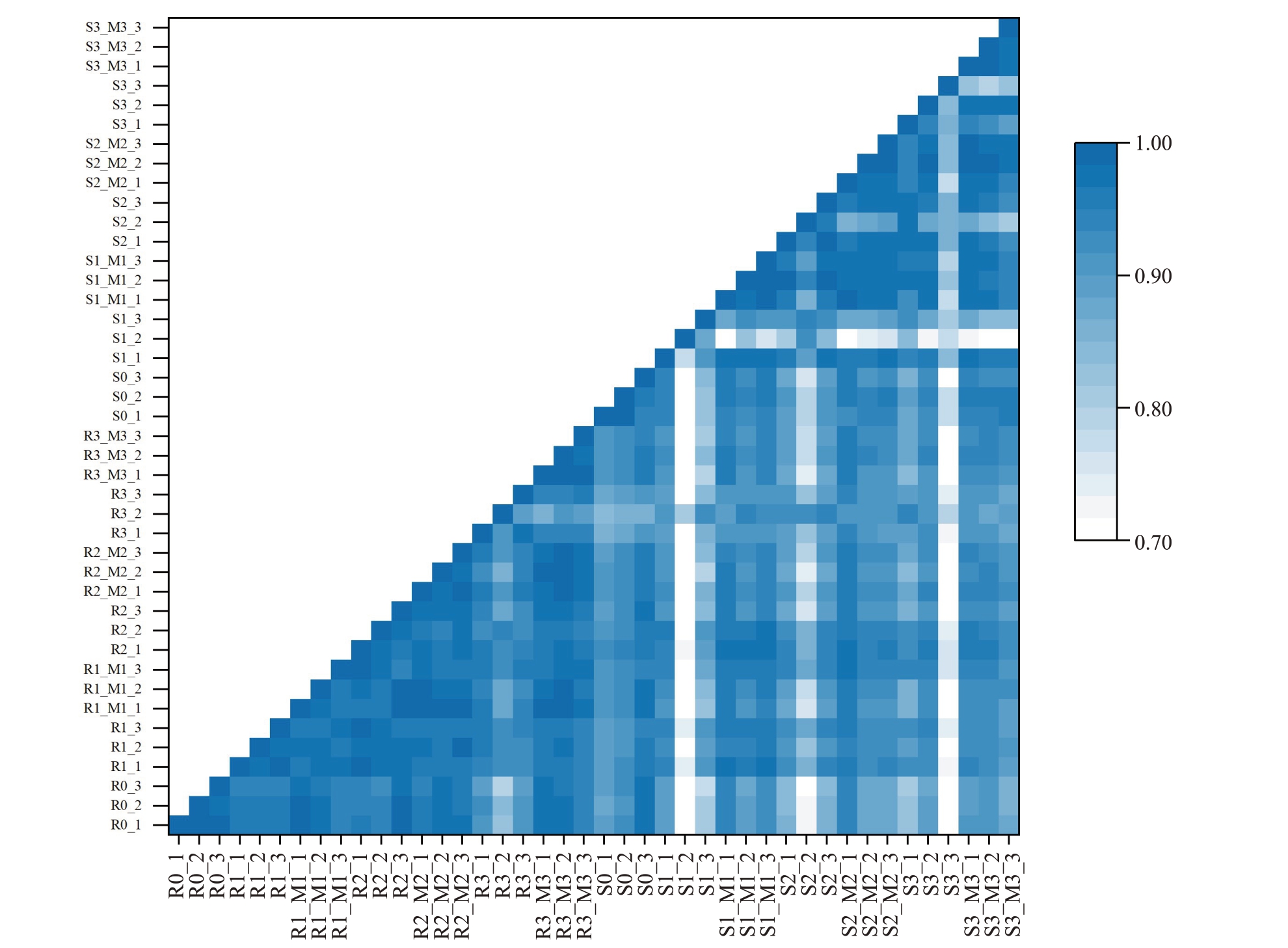
图1 各组重复之间相关性分析
Fig.1 Correlation analysis among groups of repetitions
R0、R1、R2、R3 分别为抗病株系接种胶孢炭疽菌0、24、48、72 h 的样本,R0_M0、R1_M1、R2_M2、R3_M3 为对应时期清水对照。S 代表感病株系,命名方式同上。
R0, R1, R2, and R3 are the samples of the resistant clones inoculated with C. gloeosporioides at 0, 24, 48, and 72 h, respectively, and R0_M0,R1_M1, R2_M2, R3_M3 are the pure water controls in the corresponding period. S represents the susceptible clone, and the nomenclature is the same as above.
在0、24、48、72 h时抗病株系和感病株系之间的差异表达基因分别有5527、5924、5451 和7977 个。其中上调基因数分别为1468、1801、1528和1893个,下调基因数为4059、4123、3923 和6084 个(表2)。抗病和感病株系之间下调的差异表达基因数目远大于上调的基因数目。在24 h和72 h上调的差异表达基因数目显著高于0 h;72 h下调的差异表达基因数目是其他时期1.5倍。
表2 抗病株系和感病株系间之间差异表达基因数目统计
Table 2 Statistics of DEGs between resistant and susceptible strain
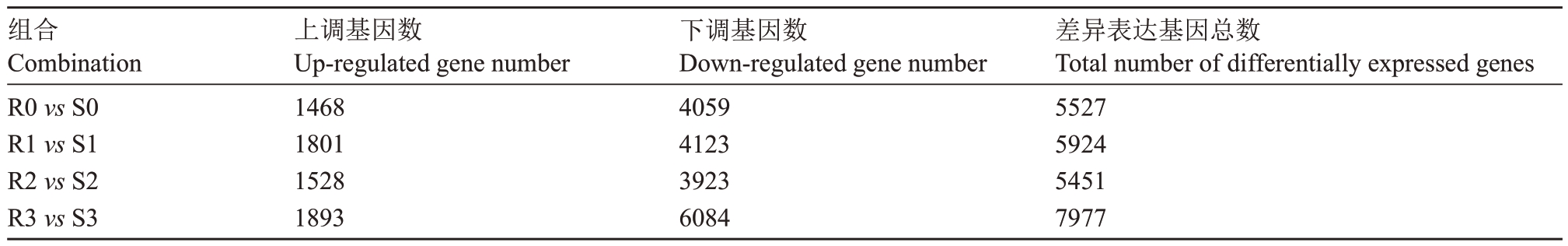
组合Combination R0 vs S0 R1 vs S1 R2 vs S2 R3 vs S3上调基因数Up-regulated gene number 1468 1801 1528 1893下调基因数Down-regulated gene number 4059 4123 3923 6084差异表达基因总数Total number of differentially expressed genes 5527 5924 5451 7977
2.2 Pathway富集分析
基于上述获得的差异表达基因,对抗感株系之间的差异表达基因进行KEGG 代谢通路富集分析,分别取每个时期富集最显著的前20 个通路进行分析,结果如图2。其中4个时期富集最为显著的通路均为植物与病原体相互作用,其次是丝裂原活化蛋白激酶(MAPK)信号途径和类黄酮生物合成。除此之外,在染病后3 个时期富集较为突出的通路主要有ABC转运蛋白、内质网加工蛋白、其他聚糖降解、花青素生物合成、内吞作用、次生代谢产物的生物合成等。
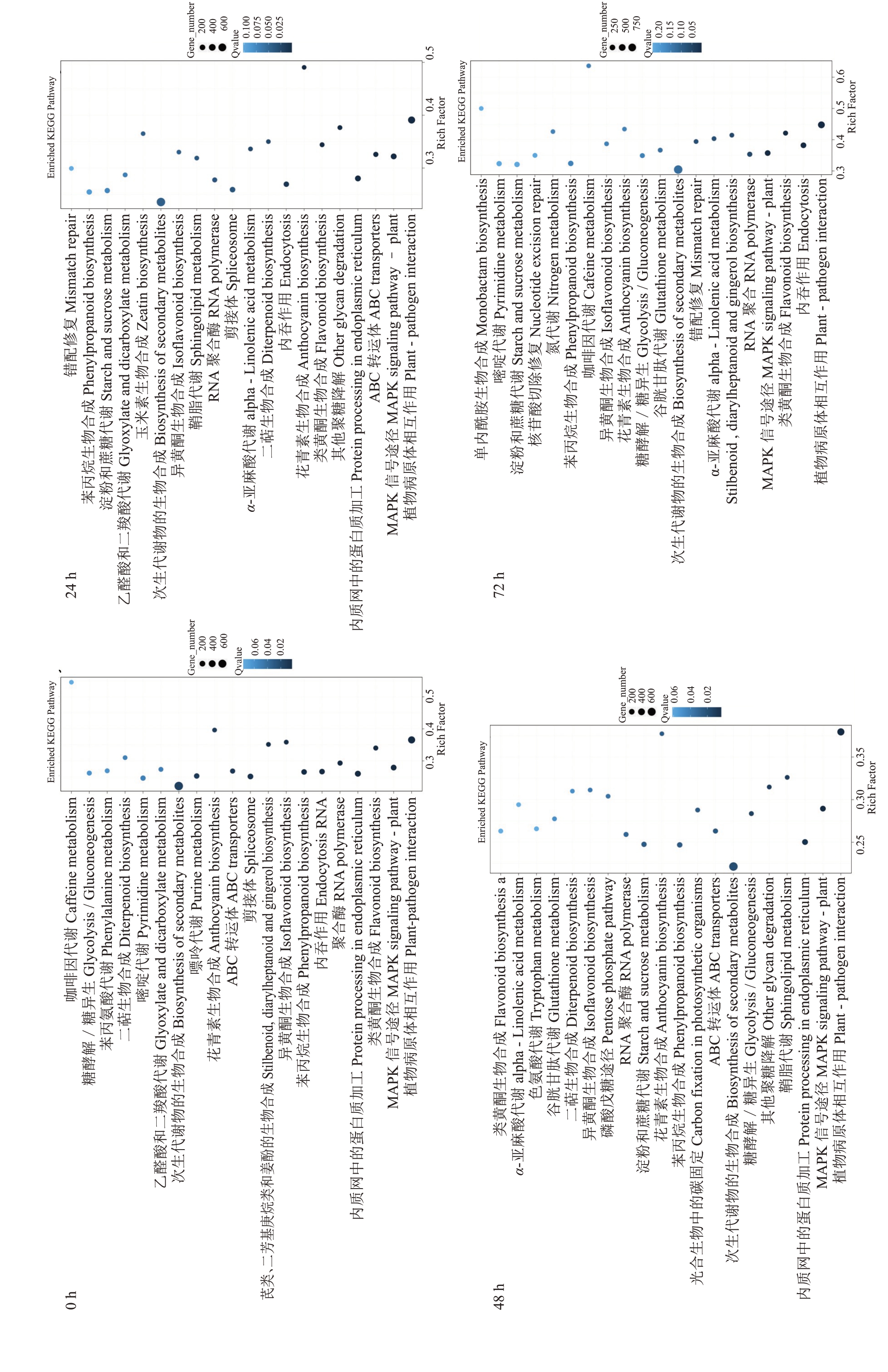
图2 抗感株系差异表达基因0、24、48、72 h pathway 富集分析
Fig. 2 Pathway enrichment analysis of DEGs in resistant and susceptible strains
从上述富集的通路中,选择抗感株系表达差异较大,或受胶孢炭疽菌诱导后表达变化明显的基因,为重点关注的差异表达基因。取其中部分差异基因做热图,从图3可见,差异表达基因主要来自植物与病原体相互作用、类黄酮生物合成和MAPK信号途径3 个通路。其中类黄酮合成中催化酶反-肉桂酸4-单加氧酶(Trans-cinnamate 4-monooxygenase)、查耳酮合酶(Chalcone synthase)、查尔酮异构酶(Chalcone isomerase)、黄烷酮3-羟化酶(Flavanone 3-hydroxylase)、类黄酮3'羟化酶(Flavonoid 3' hydroxylase)、二氢黄酮醇4-还原酶(Dihydroflavonol 4-reductase)在抗病株系染病后期的表达量远高于感病株系;芪合酶(Stilbene synthase)的表达量在感、抗株系染病前期均有升高,且感病株系高于抗病株系。此外还有一些病程相关蛋白、LRR类受体蛋白激酶以及WRKY转录因子、乙烯响应转录因子。
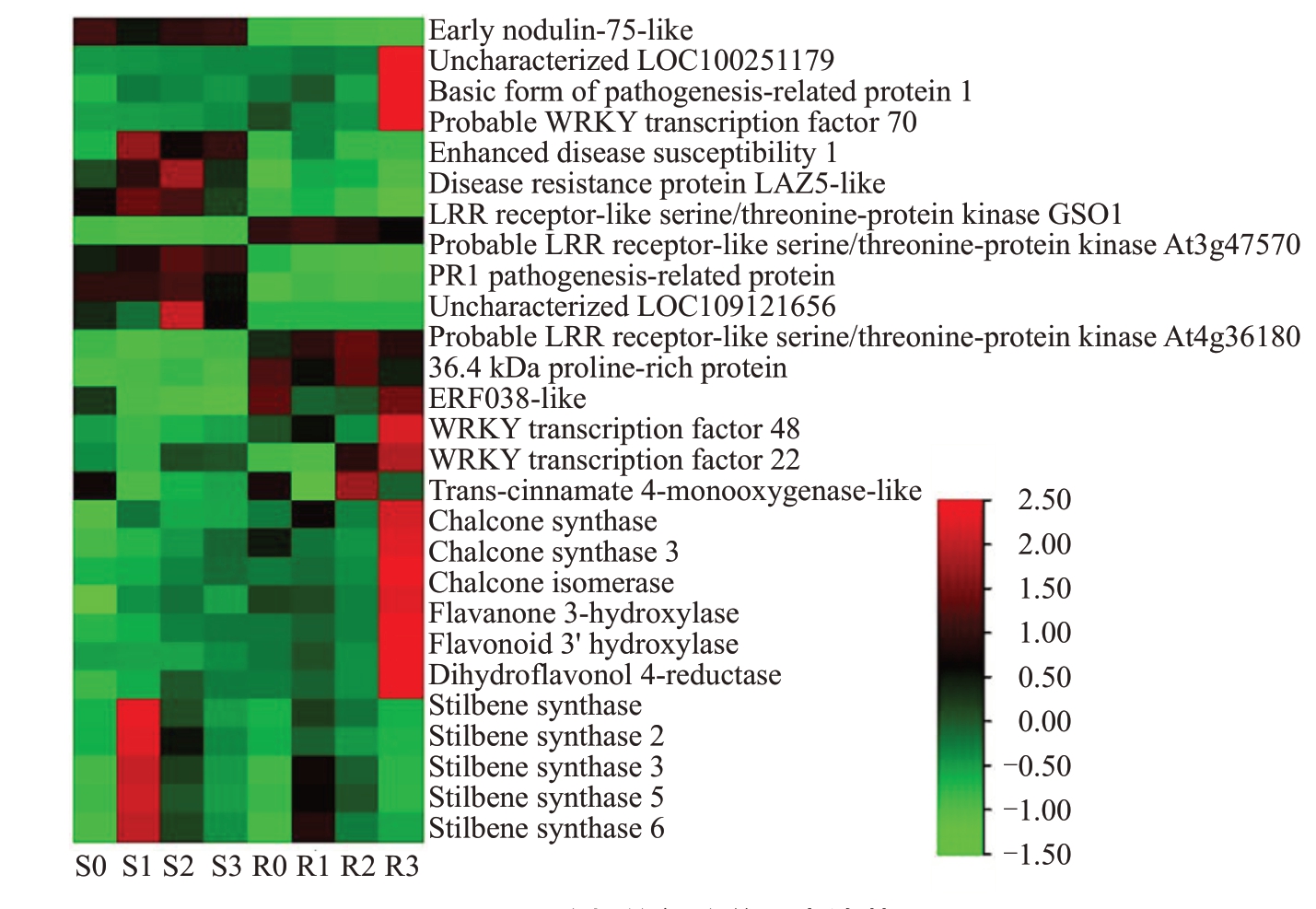
图3 Pathway 分析的部分差异表达基因
Fig.3 Partial DEGs analyzed by pathway analysis
2.3 基于WGCNA的模块筛选
为更好地分析获得感、抗株系之间的差异表达基因,对2.1 获得的差异表达基因进行WGCNA 分析,获得了18 个共表达模块(图4)。根据基因表达模块的趋势与抗病性状的相关性分析,MEdarkgreen、MEsaddlebrown、MElightyellow、MEpink、MEred 模块与抗病性状呈显著正相关。其中MEdarkgreen在R0、R1、R2时期与抗病性状呈显著正相关;MEpink、Mesaddlebrown、MElightyellow 在R0 时期与抗病性状呈显著正相关;MEred模块在R3时期与抗病性状呈显著正相关。对这些抗病相关模块中的差异表达基因进行KEGG 富集分析,其富集的通路主要集中在苯丙烷生物合成、植物病原体相互作用、MAPK信号途径、类黄酮生物合成等。
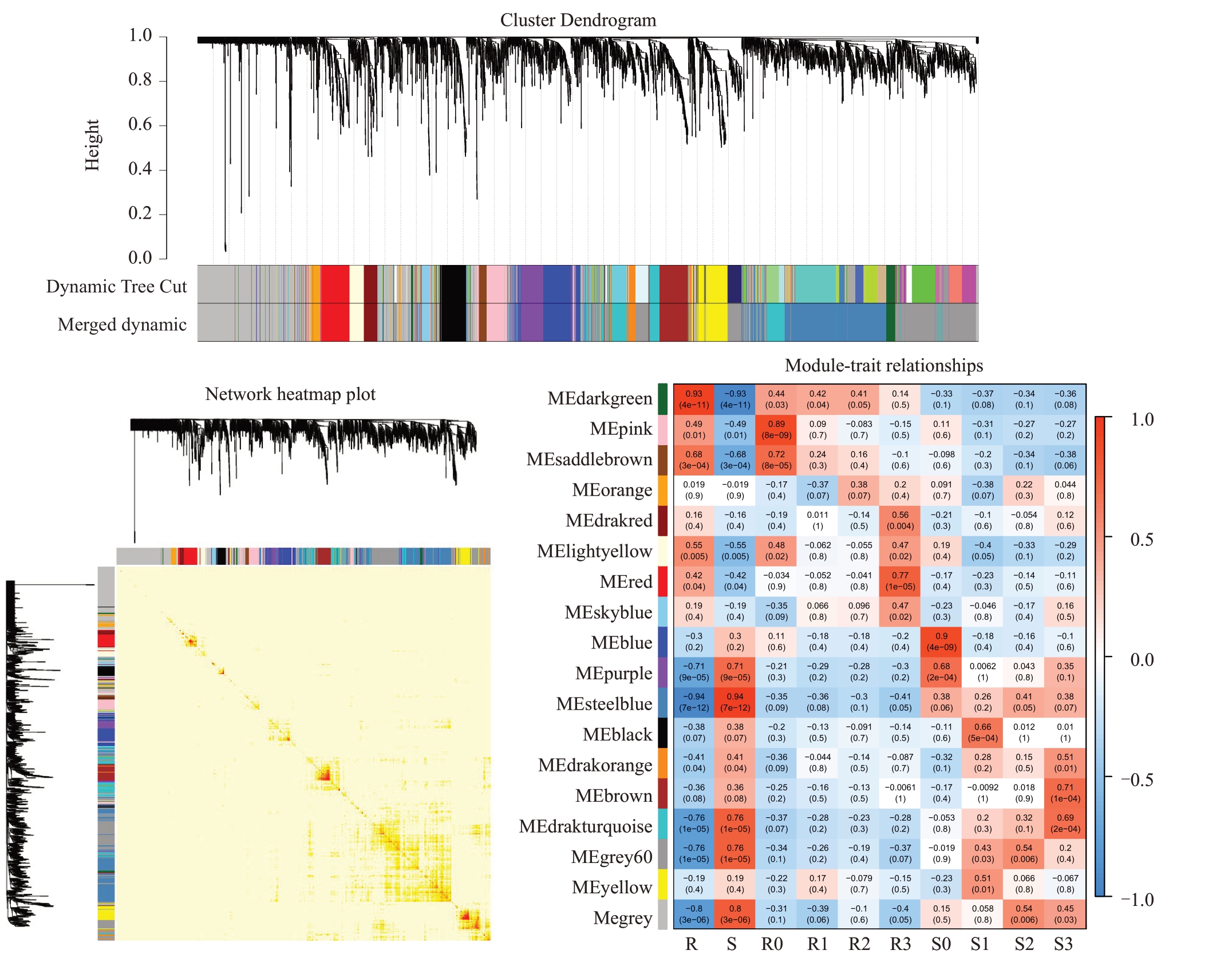
图4 加权共表达网络分析
Fig.4 The Weighted gene co-expression network analysis
2.4 转录因子家族分析
鉴于WGCNA 抗病相关模块和KEGG 富集后的通路中包含大量转录因子,因此对抗感株系之间转录因子差异表达基因进行分析。共获得532个转录因子,分为42个家族。其中含差异表达基因最多的是MYB 家族(19.36%),其次分别为AP2-EREBP(14.85%)、bHLH(9.77%)、NAC(7.33%)、WRKY(6.95%)、FAR1(6.20%)、C2C2(5.26%)、mTERF(4.32%)、C2H2(4.14%)和MASD(3.57%)。
所有转录因子家族差异表达基因的表达模式分析发现(图5),感病株系4个时期的差异表达基因表达量普遍高于抗病株系,这种现象在FAR1 家族中表现得尤为突出,FAR1家族中所有差异表达基因在感病株系中的表达均高于抗病株系。
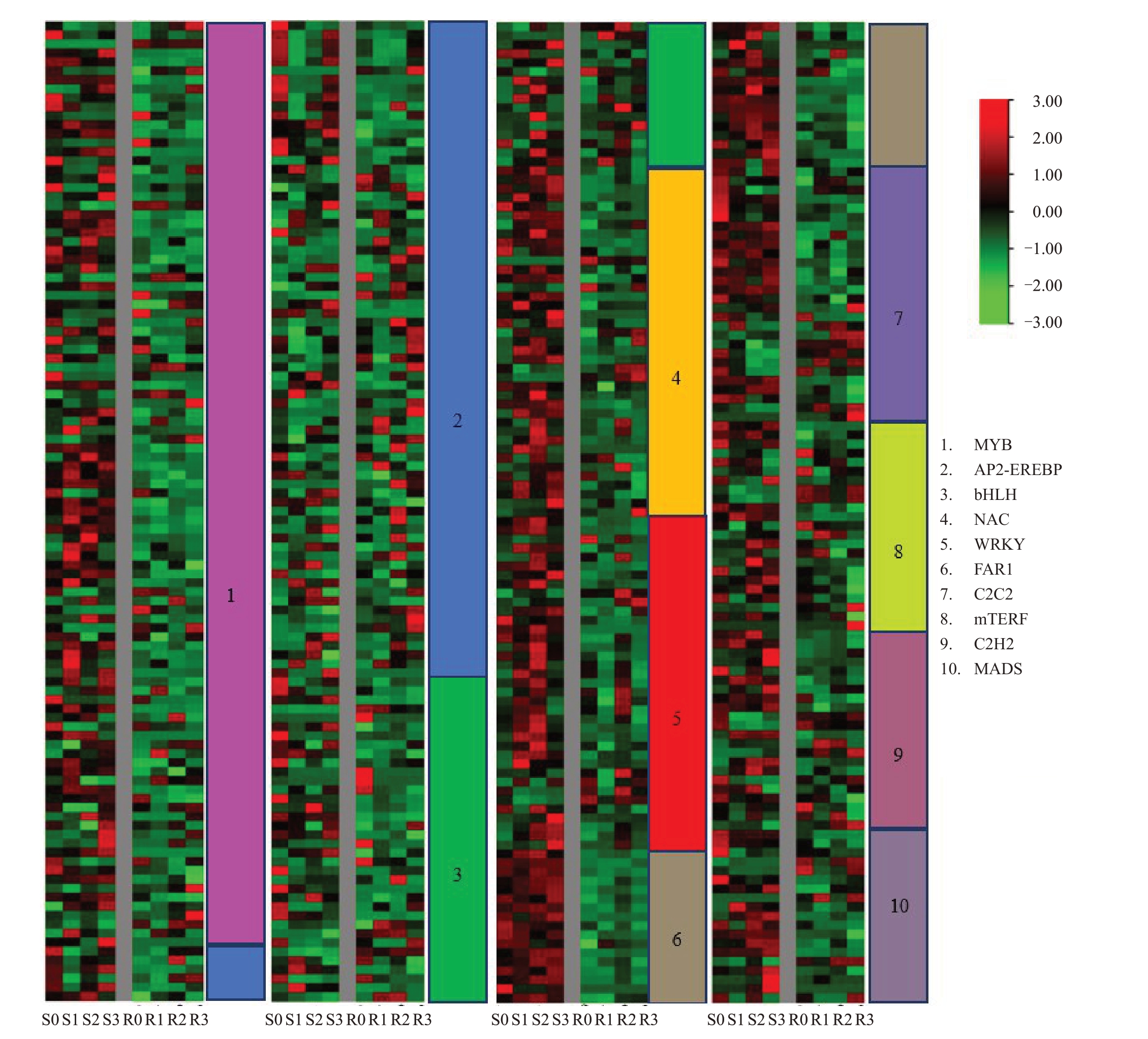
图5 转录因子家族差异表达基因分析
Fig.5 Analysis of DEGs in transcription factor family
将上述转录因子与Pathway 富集结果取交集,并参考表达量和相关基因文献报道,从前5 个转录因子家族中挑选出12 个重点关注的差异表达基因(图6),包含5个乙烯响应转录因子,1个MYB家族转录因子、2 个bHLH 家族转录因子、2 个NAC 家族转录因子和2个WRKY家族转录因子。
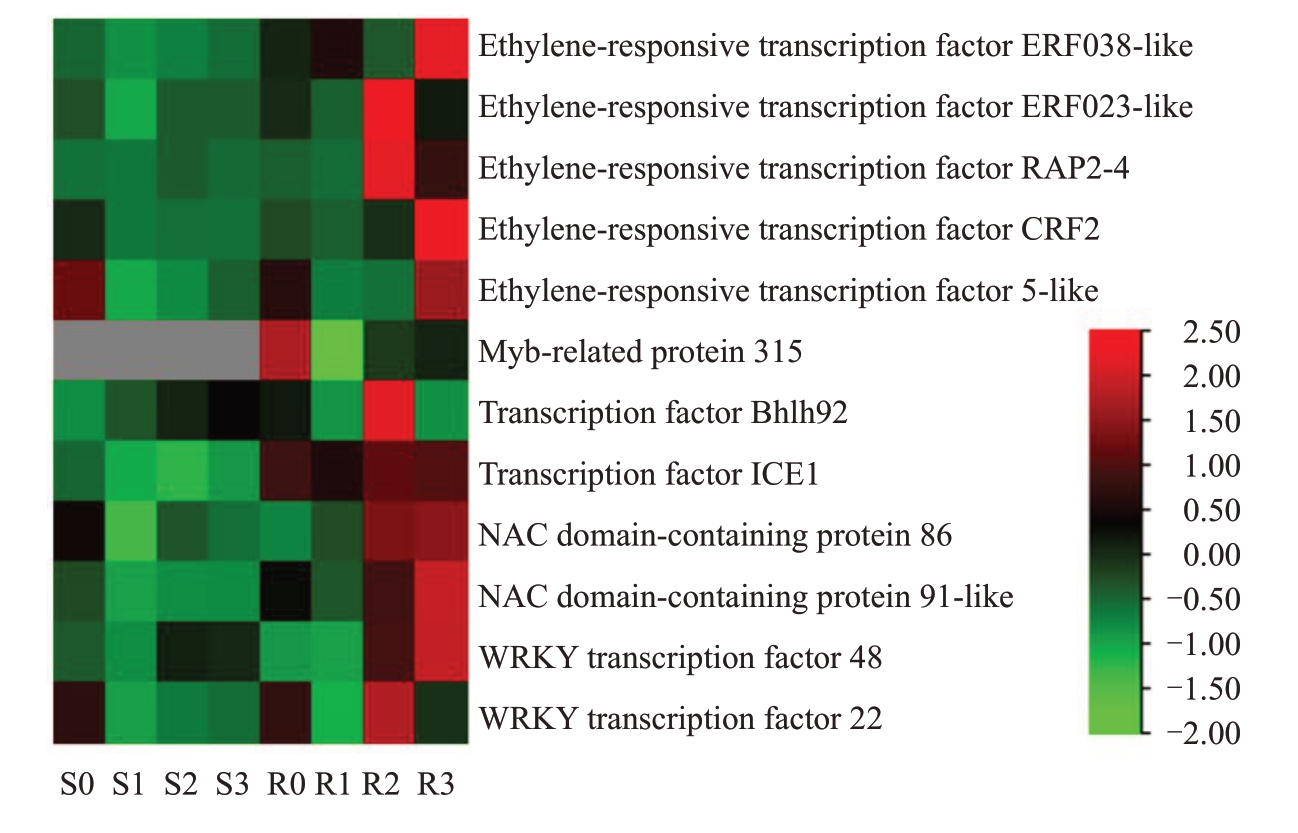
图6 转录因子家族分析的部分差异表达基因
Fig.6 Partial DEGs of transcription factor family analysis
2.5 差异表达基因分析及候选基因的筛选
对抗病和感病株系之间差异表达基因进行维恩分析(图7),通过不同抗性不同接种时间差异表达基因之间的交集,可以获得其连续性差异表达基因。接种胶孢炭疽菌后24、48和72 h,3个时期连续性表达的差异表达基因有2592个,这些差异表达基因的KEGG富集结果中有323个属于次生代谢物的生物合成、307 个属于植物病原体相互作用、143 个属于MAPK信号途径,其他的分别属于苯丙烷生物合成、淀粉和蔗糖代谢、RNA 聚合酶、ABC 转运体、类黄酮生物合成、核苷酸切除修复、异黄酮生物合成、苯丙氨酸代谢等。
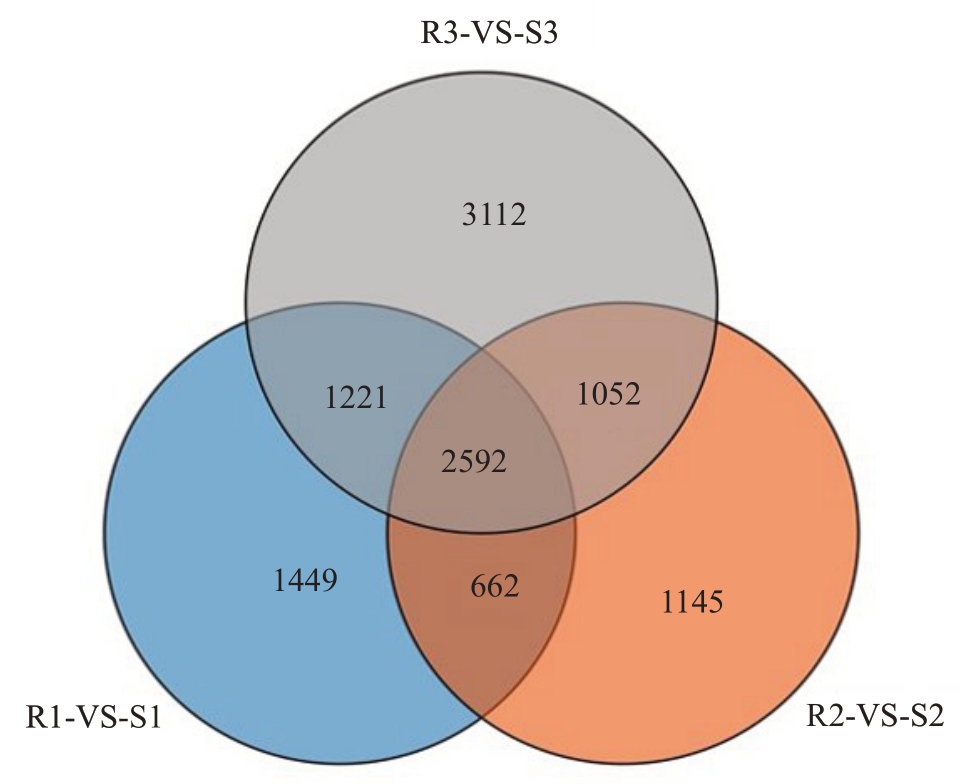
图7 抗病株系与感病株系同时期的连续性差异表达基因
Fig.7 Continuous DEGs between resistant and susceptible strain at the same time
如表3 所示,取Pathway 富集分析、转录因子家族分析筛选到的重点关注基因同连续性共表达基因、染病后关键时期差异表达基因的交集,结合WGCNA中与抗病性相关联的模块,挑选出50个差异表达基因,并参考相关同源基因报道和基因注释,最终筛选出9个抗病候选基因。其中与MAPK信号途径相关的基因最多,包括VIT_05s0077g00730(WRKY48)、VIT_15s0046g02190(WRKY22)、VIT_03s0088g00710(Basic form of pathogenesis-related protein 1)。
表3 9 个抗病候选基因
Table 3 Nine candidate genes for disease resistance
基因符号Gene symbol 100260212 100256849 100241136 100249652 100233053 100243167 100247990 109124267 100248210基因ID Gene ID VIT_05s0077g00730 VIT_15s0046g02190 VIT_09s0002g05080 VIT_14s0006g02290 VIT_05s0094g01570 VIT_12s0134g00020 VIT_03s0088g00710 N/A N/A基因描述Gene description WRKY48 WRKY22 F-box/kelch-repeat protein At1g80440 ERF038-like MLO15(E)-beta-ocimene synthase Basic form of pathogenesis-related protein 1 Probable LRR receptor-like serine/threonine-protein kinase At4g36180 Disease resistance protein LAZ5-like
2.6 实时荧光定量验证
为验证转录组测序和挑选出基因的可靠性,对9个候选基因进行实时荧光定量检测(图8),病原菌侵染后,除probable LRR receptor-like serine/threonine-protein kinase At4g361804在抗病株系48 h后表达量下降趋势快于转录组测序数据外,其他8 个基因实时荧光定量检测结果和转录组测序结果趋势一致,说明转录组数据筛选出差异表达基因具有可靠性。其中WRKY48、WRKY22、(E)-beta-ocimene synthase、F-box/kelch-repeat protein At1g80440 的表达量,在48 h 的抗病株系中较其他时期和抗性株系均有显著提升;ERF038-like、disease resistance protein LAZ5-like的表达量,在所有时期中抗性株系均明显高于感病株系。
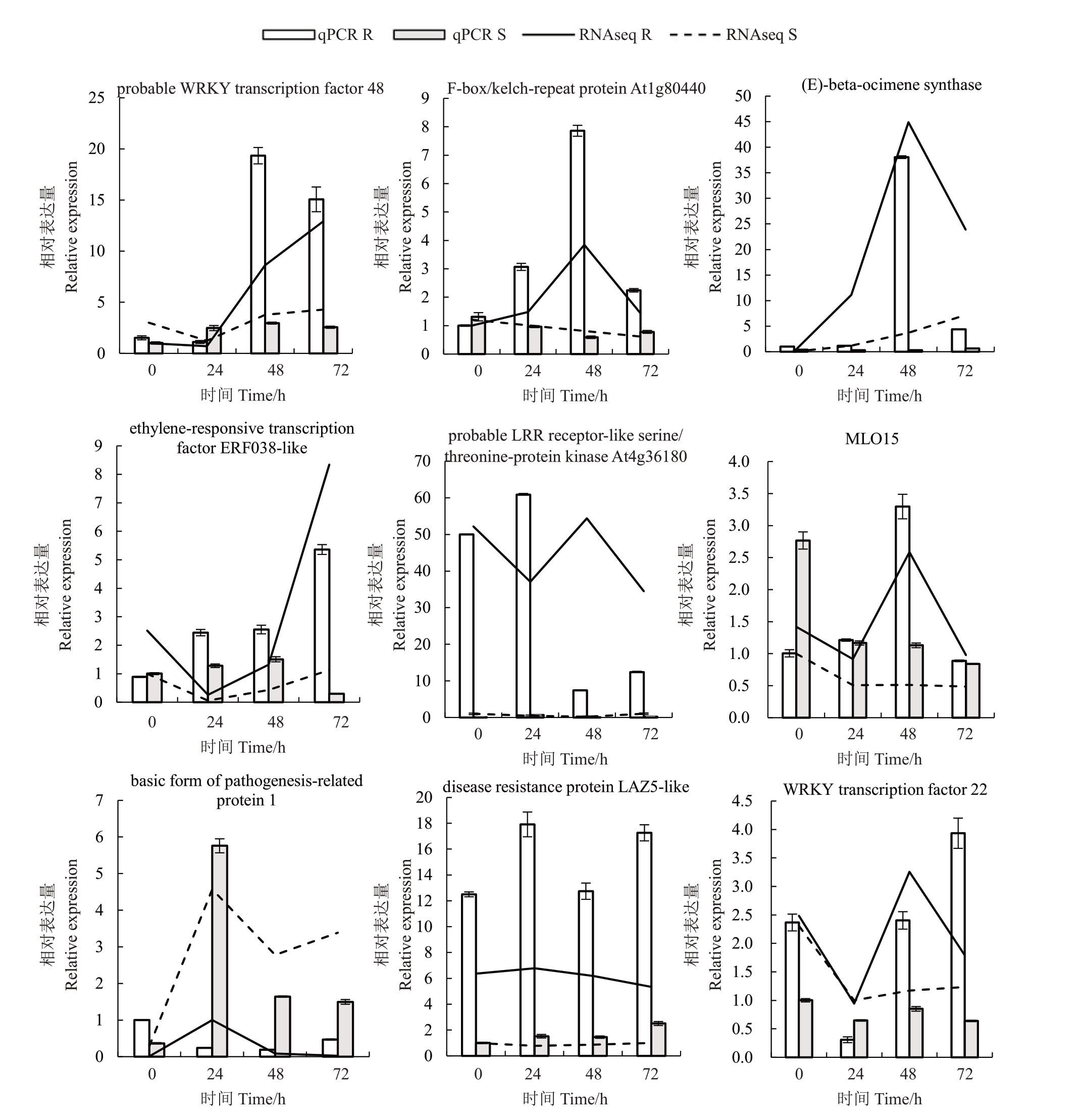
图8 差异表达基因qRT-PCR 鉴定
Fig.8 DEGs were identified by qRT-PCR
3 讨 论
RNA-Seq 技术已经作为一种常见技术,应用于植物抗病响应中相关基因的筛选及抗病机制的解析。如红橘响应褐斑病菌侵染的转录组学分析[21]、杧果响应胶孢炭疽菌的转录组学分析[16,22]等。笔者在本研究中利用RNA-Seq技术,对接种胶孢炭疽菌后不同时间点的抗病和感病葡萄叶片进行转录组测序,分别获得4 个时期两者间差异表达基因5527、5924、5451和7977个。
当胶孢炭疽菌侵染葡萄叶片后,多个代谢通路的基因表达发生改变,其中植物与病原体相互作用、类黄酮生物合成和MAPK信号途径3个通路差异最为显著。前期研究已证实类黄酮参与植物抵抗病原菌过程并作为抗真菌化合物发挥重要作用[23]。在类黄酮生物合成中,底物苯丙氨酸可在苯丙氨酸裂解酶(PAL)、肉桂酸-4-羟基化酶(C4H)、香豆酸-CoA连接酶(4CL)、芪合酶(STS)的作用下形成芪氏化合物[24],而芪氏化合物是一类以二苯乙烯微结构母核的化合物总称,是植物在抵抗胁迫时产生的一种植保素,研究发现其在葡萄抗白粉病中发挥作用[25-26];苯丙氨酸也可作为木质素合成的前体物质或者经查尔酮合酶的催化进入类黄酮的合成[23,27]。关于类黄酮合成通路在响应炭疽病抗性反应中的显著富集,在杧果和草莓的研究中也曾报道[15-17]。在本研究中,受炭疽病病菌的诱导,类黄酮生物合成的关键酶在抗感材料中的表达量表现出显著差异,可见类黄酮生物合成通路对病害的响应在不同植物之间存在相似性。
丝裂原活化蛋白激酶(MAPK)信号途径是MAPK 级联反应中下游过程,MAPK 级联途径通过逐级磷酸化传递胁迫信号,并与乙烯、茉莉酸、脱落酸信号途径交互作用或直接参与植物逆境的应答过程[28],如通过调控乙烯的关键合成酶ACC 合酶,促进乙烯合成[29];此外MAPK 级联途径基因启动子中预测存在WRKY特异性结合原件W-Box以及MYB结合原件MBS,可能被上游的WRKY 或MYB 转录因子激活[30-31]。本研究中差异表达的ERF和WRKY转录因子,作为MAPK 级联反应下游基因,很可能正是受到MAPK信号途径相关胁迫信号的诱导。
FAR1是一类起源于转座子酶的转录因子,在植物光响应、生长发育中发挥重要作用。同时已有报道证实这类转录因子在多种非生物胁迫中也存在积极响应,如FAR1-5可以通过增强抗氧化酶活性和信号激素来增加花生的抗旱能力[32];拟南芥中FHY3/FAR1可通过调控细胞死亡,提高植物抗氧化胁迫能力等[33]。本研究中FAR1 转录因子在感病材料中高表达,可能是因为受病原菌侵染后,引起的细胞死亡和相关激素信号通路参与调控的结果。WRKY 转录因子是一类在植物中常见的与胁迫相关的转录因子,根据序列中WRKY结构的数量和氨锌指基序列类型,可分为I、II、III类,其中III类中部分WRKY转录因子和W-box结合,发挥调控功能。WRKY转录因子对病原菌响应的报道在很多植物中已经有一定研究基础,在拟南芥中,AtWRKY18、AtWRKY33和AtWRKY70 在抗病原菌侵染中具有重要作用[34-36];在葡萄中VlWRKY3、VqWRKY53、VqWRKY56、VdWRKY70 等在抵抗钩丝壳菌或白腐垫壳孢侵染中发挥作用[37-41]。本研究中通过KEGG分析发现,因胶孢炭疽菌侵染产生的差异表达基因WRKY22、WRKY48转录因子,同时参与了植物与病原体相互作用和MAPK 信号途径2 个通路。其中WRKY22受到上游MAPK级联途径逐级磷酸化传递的信号,调控下游病原早期防御响应、相关防御基因的诱导、细胞程序性死亡以及活性氧和PR1蛋白的产生。
4 结 论
胶孢炭疽菌侵染造成葡萄叶片上的炭疽病症状,引起不同抗性株系中基因差异表达。转录组测序分析发现差异表达基因主要集中在病原体相互作用、类黄酮生物合成和MAPK 信号途径等通路,以及AP2-EREBP、MYB、bHLH、NAC 和WRKY 等转录因子家族中,筛选出8个葡萄抗炭疽病候选基因。
[1] 刘晓云,景耀,杨俊秀.植物炭疽菌研究文献综述[J].西北林学院学报,1995,10(4):105-111.LIU Xiaoyun,JING Yao,YANG Junxiu.Literature review of research on plant Colletotrichum[J]. Journal of Northwest Forestry University,1995,10(4):105-111.
[2] 贺普超,王跃进,王国英,任治邦,和纯成.中国葡萄属野生种抗病性的研究[J].中国农业科学,1991,24(3):84-89.HE Puchao,WANG Yuejin,WANG Guoying,REN Zhibang,HE Chuncheng. Research on disease resistance of wild species ofVitisinChina[J].ScientiaAgriculturaSinica,1991,24(3):84-89.
[3] 王跃进,徐炎,张剑侠,周鹏.中国野生葡萄果实抗炭疽病基因的RAPD 标记[J].中国农业科学,2002,35(5):536-40.WANG Yuejin,XU Yan,ZHANG Jianxia,ZHOU Peng. RAPD markers of resistance to anthracnose in Chinese wild grape fruits[J].Scientia Agricultura Sinica,2002,35(5):536-40.
[4] SUZAKI K.Improved method to induce sporulation of Colletotrichum gloeosporioides,causal fungus of grape ripe rot[J].Journal of General Plant Pathology,2011,77(2):81-84.
[5] KUMMUANG N,DIEHL S V,SMITH B J,GRAVES C H.Muscadine grape berry rot diseases in Mississippi-Disease epidemiology and crop reduction[J]. Plant Disease,1996,80(3):244-247.
[6] HONG S K,KIM W G,YUN H K,CHOI K J. Morphological variations,genetic diversity and pathogenicity of Colletotrichum species causing grape ripe rot in Korea[J]. The Plant Pathology Journal,2008,24(3):269-278.
[7] YAN J Y,JAYAWARDENA M,GOONASEKARA I D,WANG Y,ZHANG W,LIU M,HUANG J B,WANG Z Y,SHANG J J,PENG Y L. Diverse species of Colletotrichum associated with grapevine anthracnose in China[J]. Fungal Diversity,2014,71(1):233-246.
[8] 雷龑,林雄杰,陈婷,刘鑫铭,蔡盛华,范国成.福建葡萄炭疽病病原鉴定及致病性分析[J]. 果树学报,2014,31(6):1123-1127.LEI Yan,LIN Xiongjie,CHEN Ting,LIU Xinming,CAI Shenghua,FAN Guocheng. The pathogen identification and pathogenicity analysis of grape anthracnose in Fujian[J]. Journal of Fruit Science,2014,31(6):1123-1127.
[9] SHIRAISHI M,KOIDE M,ITAMURA H,YAMADA M,NAKANO M.Screening for resistance to ripe rot caused by Colletotrichum acutatum in grape germplasm[J]. Vitis,2007,46(4):239-242.
[10] CANNON P F,DAMM U,JOHNSTON P R,WEIR B S.Colletotrichum-current status and future directions[J].Studies in Mycology,2012,73(1):181-213.
[11] WHITELAW W M A,CURTIS S J,HUANG R,STEEL C C,BLANDCHARD C L,ROFFEY P E. Phylogenetic relationships and pathogenicity of Colletotrichum acutatum isolates from grape in subtropical Australia[J]. Plant Pathology,2007,56(3):448-463.
[12] GREER L A,HARPER J D I,SAVOCCHIA S,SAMUELIAN S K,STEEL C C. Ripe rot of south-eastern Australian wine grapes is caused by two species of Colletotrichum:C. acutatum and C. gloeosporioides with differences in infection and fungicide sensitivity[J]. Australian Journal of Grape & Wine Research,2011,17(2):123-128.
[13] PENG L J,SUN T,YANG Y L,CAI L,HYDE K D,BAHKALI A H,LIU Z Y. Colletotrichum species on grape in Guizhou and Yunnan provinces,China[J].Mycoscience,2013,54(1):29-41.
[14] HYDE K D,CAI L,EHC M K,YANG Y L,PRIHASTUTI H.Colletotrichum:A catalogue of confusion[J]. Fungal Diversity,2009,39(1):1-17.
[15] 陈哲,黄静,赵佳,梁宏. 草莓应答炭疽菌侵染的转录组分析[J].植物保护,2020,46(3):138-146.CHEN Zhe,HUANG Jing,ZHAO Jia,LIANG Hong.Transcriptome analysis of strawberry response to anthracnose infection[J].Plant Protection,2020,46(3):138-146.
[16] SUDHEERAN P K,SELA N,CARMELI W M,OVADIA R,PANDA S,FEYGENBERG O,MAURER D,OREN S M,AHARONI A,ALKAN N. Induced defense response in red mango fruit against Colletotrichum gloeosporioides[J]. Horticulture Research,2021,8(1):1-11.
[17] HONG K Q,GONG D Q,ZHANG L B,HU H G,JIA Z W,GU H,SONG K H. Transcriptome characterization and expression profiles of the related defense genes in postharvest mango fruit against Colletotrichum gloeosporioides[J]. Gene,2016,57(1):275-283.
[18] 施云龙.茶树抗炭疽病和抗冻机制及评价研究[D].杭州:浙江大学,2020.SHI Yunlong. Research on the anti-anthracnose and antifreeze mechanism and evaluation of tea trees[D]. Hangzhou:Zhejiang University,2020.
[19] 李静.草莓抗炭疽病遗传图谱构建和抗性相关基因的克隆与表达分析[D].南京:南京农业大学,2012.LI Jing. Construction of genetic map of strawberry resistance to anthracnose and cloning and expression analysis of resistance related genes[D].Nanjing:Nanjing Agricultural University,2012
[20] 傅敏.中国梨炭疽病病原种类多样性及果生刺盘孢与梨寄主的互作研究[D].武汉:华中农业大学,2019.FU Min. Study on the diversity of pathogens of pear Anthracnose in China and the interaction between fruit conidia and pear host[D].Wuhan:Huazhong Agricultural University,2019
[21] 唐科志,周常勇.红橘响应褐斑病菌侵染的转录组学分析[J].中国农业科学,2020,53(22):4584-4600.TANG Kezhi,ZHOU Changyong.Transcriptomic analysis of response of tangerine to brown spot disease infection[J]. Scientia Agricultura Sinica,2020,53(22):4584-4600.
[22] 叶子,孙宇,刘志鑫,刘晓妹,蒲金基,张贺,黄海,宋锦佳,安仕芹.杧果叶片响应胶孢炭疽病菌侵染的转录组分析[J/OL].分子植物育种,1-22[2021-08-13]. http://kns. cnki. net/kcms/detail/46.1068.S.20210513.1051.006.html.YE Zi,SUN Yu,LIU Zhixin,LIU Xiaomei,PU Jinji,ZHANG He,HUANG Hai,SONG Jinjia,AN Shiqin. Mangifera indica rubber blade response spore anthrax infection of the transcriptome analysis[EB/OL]. Molecular Plant Breeding,1-22 [2021-08-13]. HTTP:// http://kns. cnki. net/kcms/detail/46. 1068. S.20210513.1051.006.html.
[23] SUDHEERAN P K,OVADIA R,GALSARKER O,MAOZ I,SELA N,MAURER D,FEYGENBERG O,SHAMIR M O,ALKAN A.Glycosylated flavonoids:fruit's concealed antifungal arsenal[J].New Phytologist,2020,225(4):1788-1798.
[24] 孟旭辉,张评浒,张朝凤. 芪类化合物及其合成酶的研究进展[J].中国野生植物资源,2010,29(3):15-20.MENG Xuhui,ZHANG Pinghu,ZHANG Chaofeng. Research progress of astragalus and its synthase[J]. Chinese Wild Plant Resources,2010,29(3):15-20.
[25] 赵凯茜,丁茜,王跃进.中国野生毛葡萄芪合酶基因VqSTS11和VqSTS23 调控抗白粉病的研究[J].园艺学报,2020,47(7):1264-1276.ZHAO Kaiqian,DING Qian,WANG Yuejin. Study on the regulation of Chinese wild Astragalus synthase genes VqSTS11 and VqSTS23 against powdery mildew[J]. Acta Horticulturae Sinica,2020,47(7):1264-1276.
[26] 丁茜,赵凯茜,王跃进.中国野生毛葡萄芪合酶基因表达及对葡萄抗白粉病的影响[J].中国农业科学,2021,54(2):310-323.DING Qian,ZHAO Kaiqian,WANG Yuejin. Expression of Astragalus synthase gene in Chinese wild grapevine and its effect on resistance to powdery mildew[J].Scientia Agricultura Sinica,2021,54(2):310-323.
[27] 李慎昌,孙磊,樊秀彩,张颖,姜建福,刘崇怀.刺葡萄R2R3-MYB 转录因子VdMYB14 调控类黄酮合成功能解析[J].果树学报,2020,37(6):783-792.LI Shenchang,SUN Lei,FAN Xiucai,ZHANG Ying,JIANG Jianfu,LIU Chonghuai. Regulation of flavonoid synthesis by r2R3-MYB transcription factor VdMYB14 in Thorn grape[J].Journal of Fruit Science,2020,37(6):783-792.
[28] 尹兵兵,潘凌云,付畅.逆境下植物MAPK 级联途径基因的表达调控及与其他信号途径的交互作用[J/OL].分子植物育种,1-10[2021-08-16]. http://kns. cnki. net/kcms/detail/46. 1068. S.20210104.1341.007.html.YIN Bingbing,PAN Lingyun,FU Chang. Gene expression and regulation of MAPK cascade pathway and its interaction with other signaling pathways in plants under stress[J/OL]. Molecular Plant Breeding,1-10 [2021-08-16]. HTTP:// http://kns.cnki.net/kcms/detail/46.1068.S.20210104.1341.007.html.
[29] JOO S,LIU Y D,LUETH A,ZHANG S Q.MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the noncatalytic Cterminal domain,which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway[J].The Plant Journal,2008,54(1):129-140.
[30] CUI L C,YANG G,YAN J L,PAN Y,NIE X J. Genome-wide identification,expression profiles and regulatory network of MAPK cascade gene family in barley[J].BMC Genomics,2019,20(1):750-770.
[31] CHEN L,SUN H,WANG F J,YUE D D,SHEN X K,SUN W N,ZHANG X L,YANG X Y. Genome-wide identification of MAPK cascade genes reveals the GhMAP3K14-GhMKK11-GhMPK31 pathway is involved in the drought response in cotton[J]. Plant Molecular Biology:An International Journal on Molecular Biology,Molecular Genetics and Biochemistry,2020,103(10):211-223.
[32] 闫彩霞,李春娟,孙全喜,张浩,王娟,苑翠玲,单世华,赵小波. 花生中FAR1-5 转录因子的克隆和功能分析[J]. 花生学报,2020,49(2):16-20.YAN Caixia,LI Chunjuan,SUN Quanxi,ZHANG Hao,WANG Juan,YUAN Cuiling,SHAN Shihua,ZHAO Xiaobo. Cloning and functional analysis of far1-5 transcription factor in peanut[J]. Journal of Peanut Science,2020,49(2):16-20.
[33] 马琳.拟南芥中光信号蛋白FHY3/FAR1 调控细胞死亡的分子机理研究[D].泰安:山东农业大学,2015.MA Lin. Molecular mechanism of cell death regulated by light signal protein FHY3/FAR1 in Arabidopsis[D]. Tai’an:Shandong Agricultural University,2015.
[34] XU X P,CHEN C H,FAN B F,CHEN Z X. Physical and functional interactions between pathogen- induced Arabidopsis WRKY18,WRKY40,and WRKY60 transcription factors[J].The Plant cell,2006,18(5):1310-1326.
[35] RAMOS R N,MARTIN G B,POMBO M A,ROSLI H G.WRKY22 and WRKY25 transcription factors are positive regulators of defense responses in Nicotiana benthamiana[J]. Plant Molecular Biology,2021,105(1-2):65-82.
[36] BIRKENBIHL R P,DIEZEL C,SOMSSICH I E. Arabidopsis WRKY33 Is a Key Transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection[J]. Plant Physiology,2012,159(1):266-285.
[37] GUO R R,QIAO H B,ZHAO J,WANG X H,TU M X,GUO C L,WAN R,LI Z,WANG X P. The grape VlWRKY3 gene promotes abiotic and biotic stress tolerance in transgenic Arabidopsis thaliana[J].Frontiers in Plant Science,2018,9:545-60.
[38] 王丹.中国野生毛葡萄转录因子WRKY 及bZIP 表达与调控抗病研究[D].杨凌:西北农林科技大学,2019.WANG Dan.Expression and regulation of WRKY and bZIP transcription factors in Chinese wild hairy grape and their resistance to disease[D].Yangling:Northwest A&F University,2019.
[39] 王雅.中国野生毛葡萄VqWRKY56 基因的克隆及抗白粉病功能研究[D].杨凌:西北农林科技大学,2020.WANG Ya. Cloning of VqWRKY56 gene and its resistance to powdery mildew in wild grape of China [D]. Yangling:Northwest A&F University,2020.
[40] 冯虎,张颖,樊秀彩,姜建福,孙海生,刘崇怀.刺葡萄0943 抗白腐病转录因子VdWRKY49 的克隆及序列分析[J].果树学报,2015,32(5):777-786.FENG Hu,ZHANG Ying,FAN Xiucai,JIANG Jianfu,SUN Haisheng,LIU Chonghuai. Cloning and sequence analysis of VdWRKY49,a transcription factor against white rot in grapevine 0943[J].Journal of Fruit Science,2015,32(5):777-786.
[41] 张颖,樊秀彩,孙海生,姜建福,刘崇怀.葡萄种质资源对叶片白腐病的抗性鉴定及评价[J]. 果树学报,2017,34(9):1095-1105.ZHANG Ying,FAN Xiucai,SUN Haisheng,JIANG Jianfu,LIU Chonghuai. Identification and evaluation of resistance of grape germplasm resources to leaf white rot[J]. Journal of Fruit Science,2017,34(9):1095-1105.