苹果属(Malus Mill.)为蔷薇科(Rosaceae)、苹果亚科(Maloideae),是蔷薇科中经济价值较高的植物。自1754年以来,苹果属一直是人们研究的重要类群之一。苹果属植物用途广泛,可被用于鲜食、加工、砧木、观赏和园林绿化等,至今许多地区仍有苹果属植物野生群落分布,如新疆野苹果和山荆子等[1]。中国是世界苹果属植物的起源中心之一和最大的基因多样性中心,拥有丰富的原生种和类型。中国原产苹果属植物野生种有新疆密集分布区和主要包括四川、云南西部、贵州及西藏东南部地区的横断山脉分布区[2],除此之外,中国的17个省或自治区均有野生种分布[3-4]。物种遗传多样性是其生存适应的基础,可有效评估其现有的生存方式和状态[5],更可以揭示物种的进化历史[6-7]、分析其进化潜力和未来命运[8]、探讨物种稀有或濒危原因[9-10]。中国苹果属植物的遗传多样性和起源演化研究对于世界苹果属植物研究意义重大。
植物叶绿体基因组(cpDNA)具有分子质量小、单亲遗传、多拷贝及碱基序列重组少等特点[11],非编码区进化速度较快,变异主要以点突变和插入/缺失为主[12],适用于系统研究较低的分类阶元(如科、属)[13],可直接进行其遗传多样性检测[14],其母系遗传的特点在植物系统发育研究中具有独特优势[15],因此在揭示植物群体遗传、亲缘关系和系统发育方面都具有较高的可信度,在植物系统发育、遗传多样性和亲缘关系演化等研究中已被广泛应用[16-18]。果树基因组高度杂合,虽然已经发表了3 套苹果基因组[19-21],但是仍然难以组装完整的叶绿体全基因组序列,对苹果cpDNA的研究还主要是对其区段进行研究。国内外学者较早地利用1~2 个叶绿体DNA 区域开展苹果属植物叶绿体DNA 变异和亲缘关系等的研究,Savolainen 等[22]对40 个苹果品种和15 个苹果属野生种的叶绿体DNA的atp-rbcL间隔区和rbcL编码序列的第一个53 密码子长904 bp 的序列进行了测序,研究苹果叶绿体DNA间区的变异和亲缘关系。Robinson等[23]基于matK和ITS序列分析苹果属29个种和苹果品种间的亲缘关系,支持将苹果属分为3 大类。Coart 等[24]通过对苹果属cpDNA 多样性研究,为欧洲栽培苹果的起源和进化提供新的证据。丁芳兵[25]则是利用matK 和ITS 序列证明了湖北海棠与丽江山荆子、三叶海棠有较近的亲缘关系。朱元娣等[26]利用相同的基因区域研究了新疆野苹果与中国苹果的系统发育,并得出叶绿体基因matK 并不适用于栽培苹果种内的系统发育分析的结论;徐榕雪[27]也利用该区域的内含子序列信息,构建了苹果属23个种27份材料系统发育树,但仅获得遗传变异信息位点23 个,推断matK 不太适合近缘种间及种下类群材料的遗传关系分析。李慧峰[28]利用trnL-trnF 序列构建的泰沂山区苹果属植物系统树在一定程度上可以支持表型分类结果,但无法有效解决其复杂遗传背景下的系统发育关系。叶绿体单个基因或单个基因间隔区难以满足具有复杂遗传背景的果树种质资源的遗传分析要求,叶绿体基因组用于系统发育的研究已经发展到多个基因及基因间隔区的多态性位点组合分析的阶段[29-31]。利用叶绿体DNA 对具有复杂遗传背景的苹果属植物的遗传变异分析,需要用大群体、进化速率快的多个区域方可实现。利用多个区段对中国原产多种苹果属植物的研究尚未见报道。
中国苹果起源于新疆野苹果[1],“西洋苹果”可能起源于与新疆野苹果有共同祖先关系的塞威士苹果[19],山荆子可能参与了其驯化过程[32],尚需更多的证据解析苹果属各种的起源演化过程。全世界有22 个国家进行苹果种质资源的收集和保存[33],野外考察获得的原始材料的不断积累,以及新技术的发展,为不断探索“苹果是如何而来”提供了更多方法。但在苹果叶绿体全基因组尚未获得以及测序成本依然较高的情况下,对母系遗传的cpDNA非编码区的研究,仍然为解析苹果属植物系统发育的重要方法。笔者以近十年间考察收集到的苹果属植物为研究试材,采用cpDNA标记方法,从cpDNA水平检测中国原产苹果属植物中15 个种的遗传分化和遗传结构,解析不同种苹果属植物的遗传关系,明确15种苹果属植物的遗传多样性水平,为苹果属植物的起源演化研究和保护利用提供依据。
1 材料和方法
1.1 材料
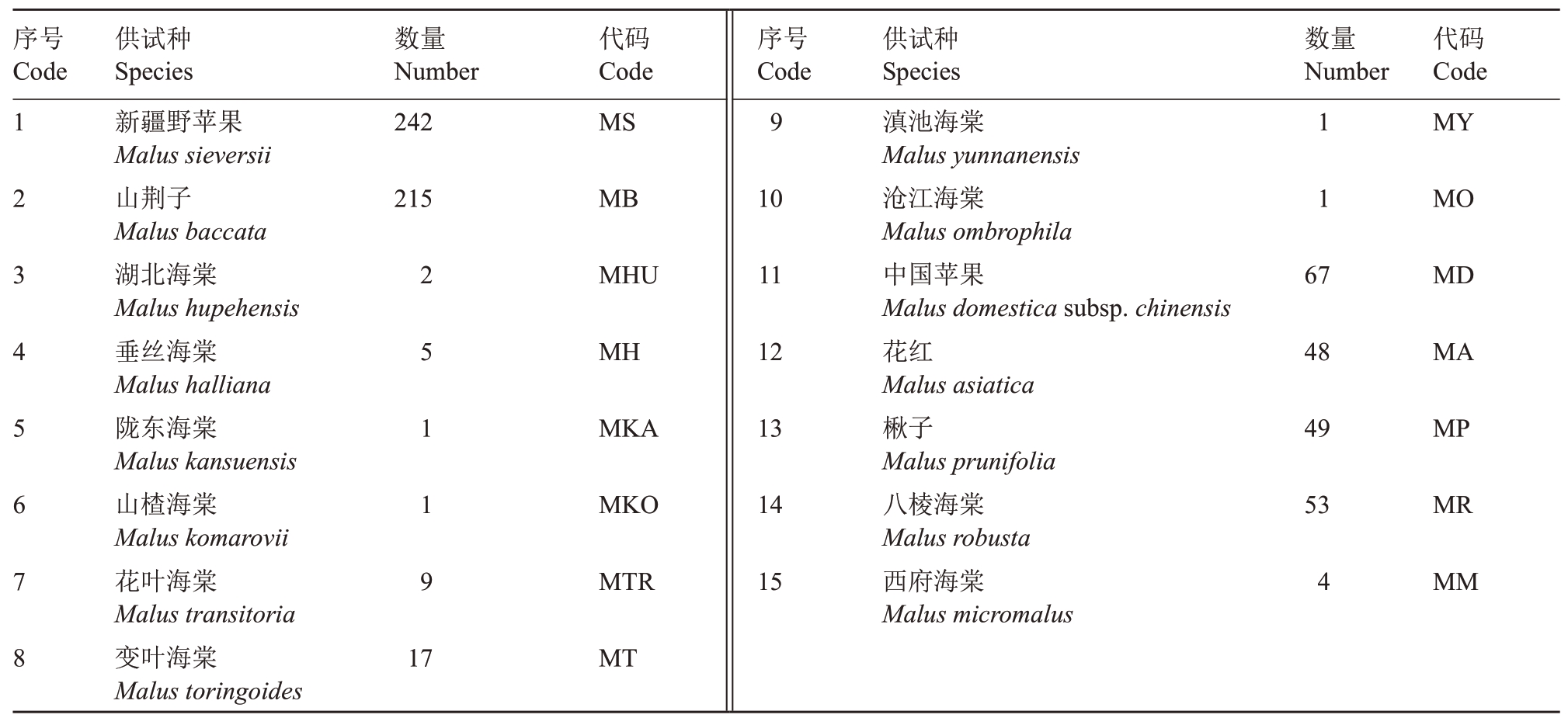
供试722 份苹果属植物分属于15 个种(表1),其中669份采自国家果树种质兴城梨、苹果圃(辽宁兴城),为近十年间野外考察收集并引入圃内无性繁殖后保存的种质,初步通过表型鉴定确定其分类归属。48份采自国家果树种质公主岭寒地果树圃(吉林省公主岭),5份采自国家果树种质云南特有果树及砧木圃(云南省昆明)。
表1 用于叶绿体DNA 分析的苹果属植物15 个种的种质资源
Table 1 15 species of Malus Mill.germplasm resources for cpDNA analysis
?
1.2 DNA提取及PCR扩增
供试材料春季嫩叶基因组DNA 的提取采用德国QIAGEN的DNeasy Plant Mini Kit。
用于扩增4 个叶绿体DNA 基因间区(trnH-psbA、trnS-trnG spacer+intron、trnT-5’trnL 和5’trnLtrnF)的4 对叶绿体通用引物来源于Shaw 等[34]的序列,优化Volk 等[35]的PCR 反应条件。引物序列、具体反应体系和程序详见高源等[36]的方法,利用美国ABI 3730XL对扩增产物进行正反向测序。
1.3 数据分析
使用MEGA 7.0[37]软件对15 种苹果属植物的4个叶绿体DNA基因间区正反向测序序列进行拼接、比对和合并,构建其基于Da遗传距离(Nei’s Da genetic distance)[38-39]的Neighbour-Joining(NJ)[40]系统发育树。使用DnaSP ver5.10.01[41]软件计算15 种苹果属植物叶绿体DNA的遗传多样性参数,计算种内不同居群间遗传分化系数,包括基因流(Nm)、群体固定系数(Fst)、基因差异分化系数(Gst)和核苷酸分化系数(Nst),评价种内不同居群间的基因交流。使用Arlequin v3.5[42]软件分析苹果属植物种群内和种群间的分子变异(analysis of molecular variation,AMOVA),使用DnaSP ver5.10.01软件计算其基因流和基因分化系数。运用NetWork ver4.6.1.2 分析构建种群间的叶绿体DNA单倍型的邻接网络关系图,具体参见高源等[36]的方法。
2 结果与分析
2.1 叶绿体基因多态性
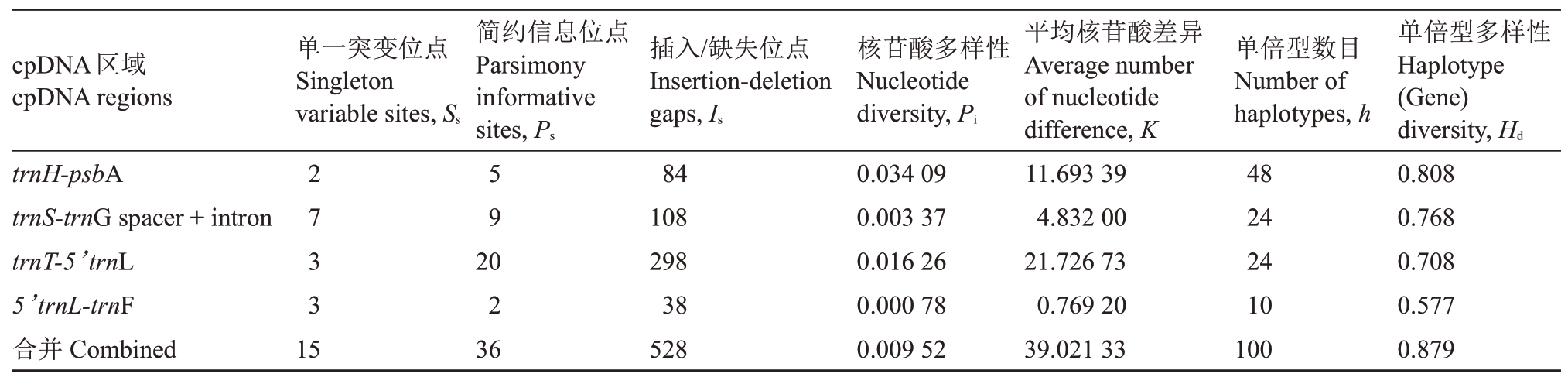
应用4对叶绿体通用引物对15种苹果属植物种质的4个cpDNA 基因间区进行扩增,4个区域trnHpsbA、trnS-trnG spacer + intron、trnT-5’trnL 和5’trnL-trnF 在所有样本中的序列长度分别为343、1 457、1 336 和984 bp。核苷酸多态性最高的区域为trnT-5’trnL,包含3个单一突变位点、20个简约信息位点和298 个插入/缺失位点;核苷酸多态性最低的区域为5’trnL-trnF,仅有3 个单一突变位点、2 个简约信息性位点和8个插入/缺失位点。722份中国原产苹果属植物的4 个cpDNA 基因间区共有变异位点579 个,其中区域trnT-5’trnL 就有321 个,是变异位点最多的区域。trnH-psbA 的核苷酸多样性(Pi)最大,为0.034 09;5’trnL-trnF 的核苷酸多样性(Pi)最小,为0.000 78。平均核苷酸差异(K)最大和最小的区域分别为trnT-5’trnL(K=21.726 73)和5’trnL-trnF(K=0.769 20)。trnH-psbA 区域单倍型数目(h)最多,为48 个,5’trnL-trnF 区域单倍型数目(h)最少,为10个。trnH-psbA的单倍型(基因)多样性(Hd)最高,为0.808;5’trnL-trnF 的单倍型(基因)多样性(Hd)最低,为0.577。722 份苹果属植物种质的4 个cpDNA 区域合并后共有100 个单倍型,核苷酸多样性(Pi)、平均核苷酸差异(K)和单倍型多样性(Hd)分别为0.009 52、39.021 33和0.879(表2)。
表2 15 种苹果属植物的4 个cpDNA 基因间区的多态性
Table 2 The polymorphism of four cpDNA intergenic regions of 15 species of Malus Mill.
?
2.2 中国原产苹果属植物的遗传分化
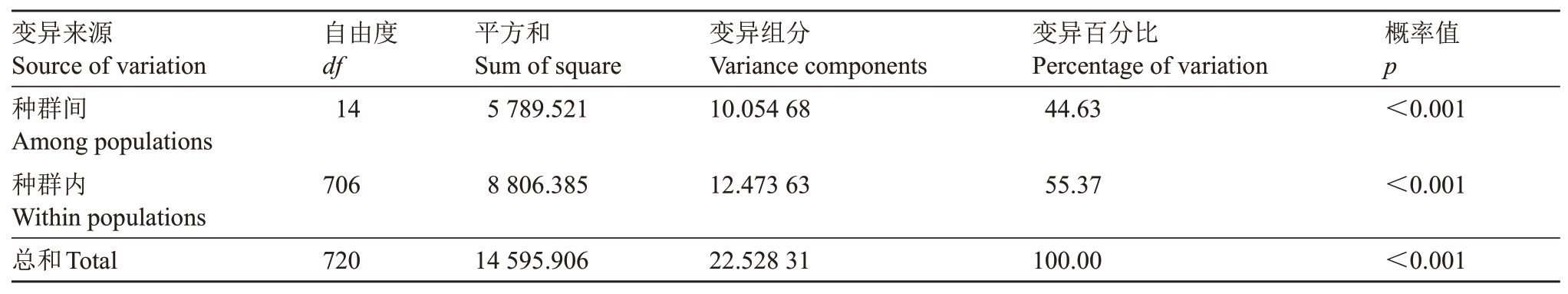
供试的中国原产苹果属植物15个种,将每个种作为一个种群。除去只有一份材料的陇东海棠、滇池海棠、沧江海棠和山楂海棠种群,其余11 个种群分子变异分析的结果表明,种群遗传变异来自于种群间的占44.63%、种群内的占55.37%(表3)。11个种群间的3组遗传分化系数分别为Fst=0.305 57,Nm=0.57;Gst=0.146 70,Nm=1.45;Nst=0.305 74,Nm=0.57,三组遗传分化系数体现出来的种群间的遗传变异分别为30.557%、14.670%和30.574%。虽然种群遗传变异仍然主要来自于种群内,但种群间遗传变异也占有较大的比重。
表3 苹果属11 个种群的分子变异
Table 3 Molecular variance analysis of 11 populations of Malus Mill.
?
以供试苹果属每个种作为种群计算群体间的遗传分化系数,除均只有1份材料的陇东海棠、滇池海棠、沧江海棠和山楂海棠不能参与计算,其余11 个种群两两间的遗传分化系数为0.014~0.840(表4)。新疆野苹果与变叶海棠间的遗传分化系数最高为0.84,楸子与花红间的遗传分化系数最低为0.014。遗传分化系数为0~0.05 的两两种群组合有山荆子和楸子、山荆子和垂丝海棠、花红和中国苹果、花红和楸子、花红和八棱海棠、垂丝海棠和楸子、垂丝海棠和八棱海棠、八棱海棠和楸子;遗传分化系数为0.05~0.15的种群组合有山荆子和花红、山荆子和八棱海棠、中国苹果和楸子、中国苹果和八棱海棠、花红和西府海棠、花红和垂丝海棠;遗传分化系数为0.15~0.25的种群组合有中国苹果和新疆野苹果、中国苹果和山荆子、中国苹果和西府海棠、中国苹果与垂丝海棠、花红和花叶海棠、楸子和西府海棠、楸子和花叶海棠、八棱海棠和西府海棠、西府海棠和湖北海棠、变叶海棠和花叶海棠;遗传分化系数高于0.25的种群组合中,新疆野苹果和变叶海棠遗传分化系数最高,为0.840,其次是新疆野苹果和山荆子的遗传分化系数,为0.646,中国苹果和花叶海棠的遗传分化系数最低,为0.251。
表4 苹果属植物11 个种群间遗传距离和遗传分化系数
Table 4 The genetic distance and differentiation coefficient among 11 populations of Malus Mill.
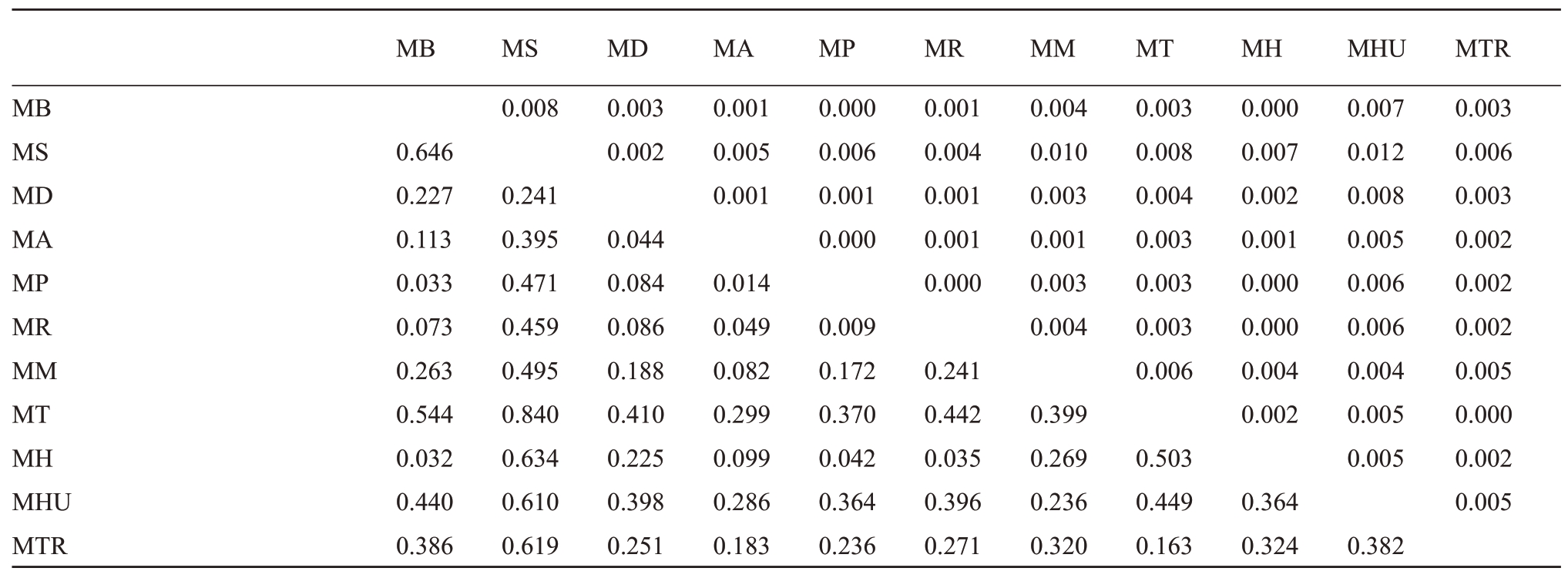
注:对角线下方的值为群体间遗传分化系数(Fst),对角线上方的值为群体间Da 遗传距离。
Note:The data below the diagonal is Fst,and above the diagonal is Da genetic diatance.
?
计算试材量多于1 份的11 个种群间的Da遗传距离,11个群体间的Da遗传距离为0~0.012,山荆子和楸子、山荆子和垂丝海棠、变叶海棠和花叶海棠、垂丝海棠和楸子、垂丝海棠和八棱海棠、八棱海棠和楸子、花红和楸子间的遗传距离均为0,湖北海棠和新疆野苹果间的遗传距离最大(0.012),其次是西府海棠和新疆野苹果间的遗传距离(0.010)。
2.3 中国原产苹果属植物遗传结构
计算供试苹果属植物15 个种间的遗传距离并用Neighbour-Joining的方法构建聚类树。通过种群遗传距离的比较,在Neighbor-Joining聚类树中可以识别5个类群,变叶海棠、花叶海棠、湖北海棠、垂丝海棠被归在类群Ⅰ中,楸子、八棱海棠、西府海棠、山荆子和花红归在类群Ⅱ中,中国苹果和新疆野苹果归在类群Ⅲ中,滇池海棠、沧江海棠和陇东海棠归类群Ⅳ中,山楂海棠单独成为类群Ⅴ(图1)。
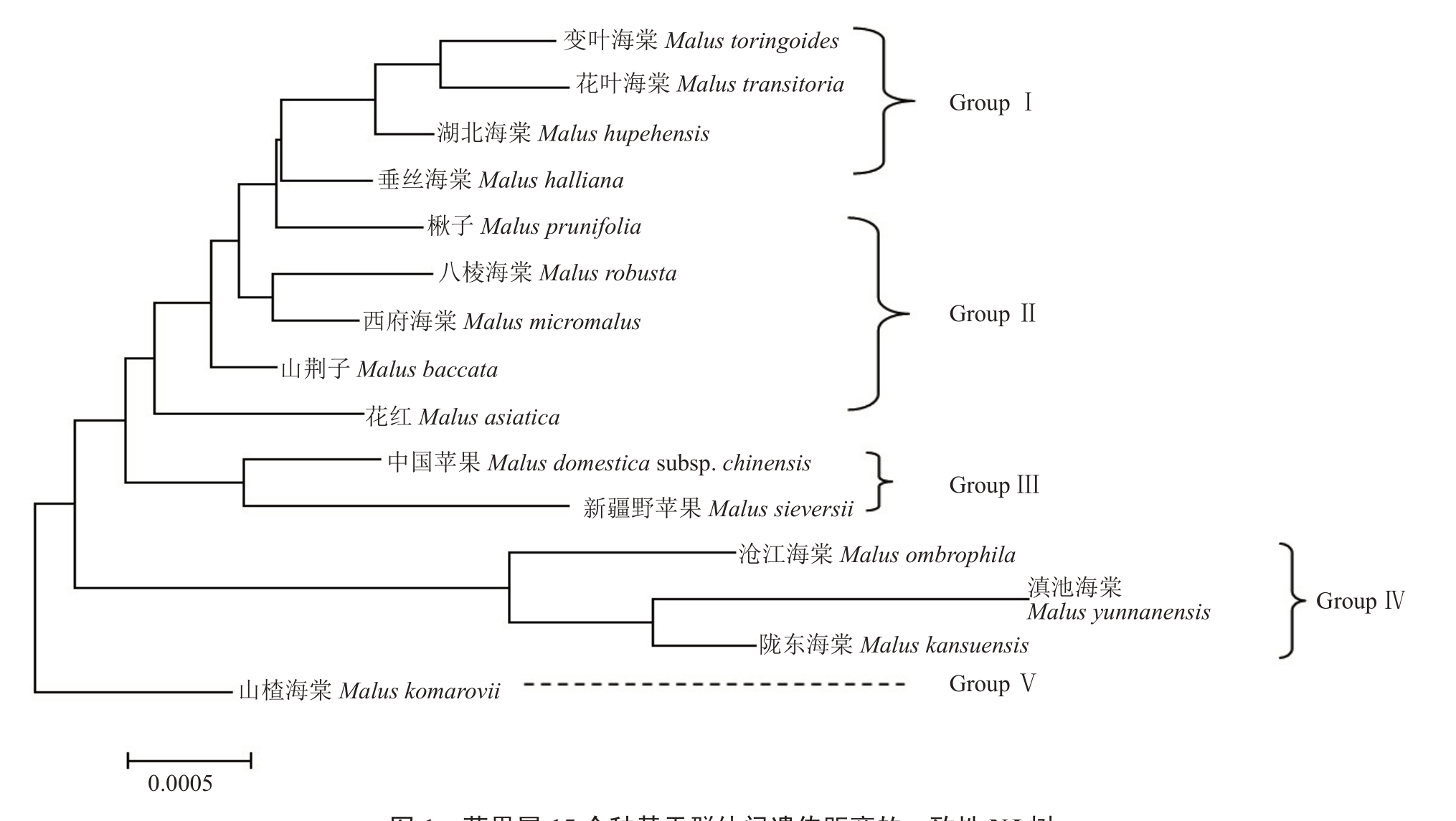
图1 苹果属15 个种基于群体间遗传距离的一致性NJ 树
Fig.1 Consensus neighbor-joining tree of 15 species of Malus Mill.based on the genetic distance of populations
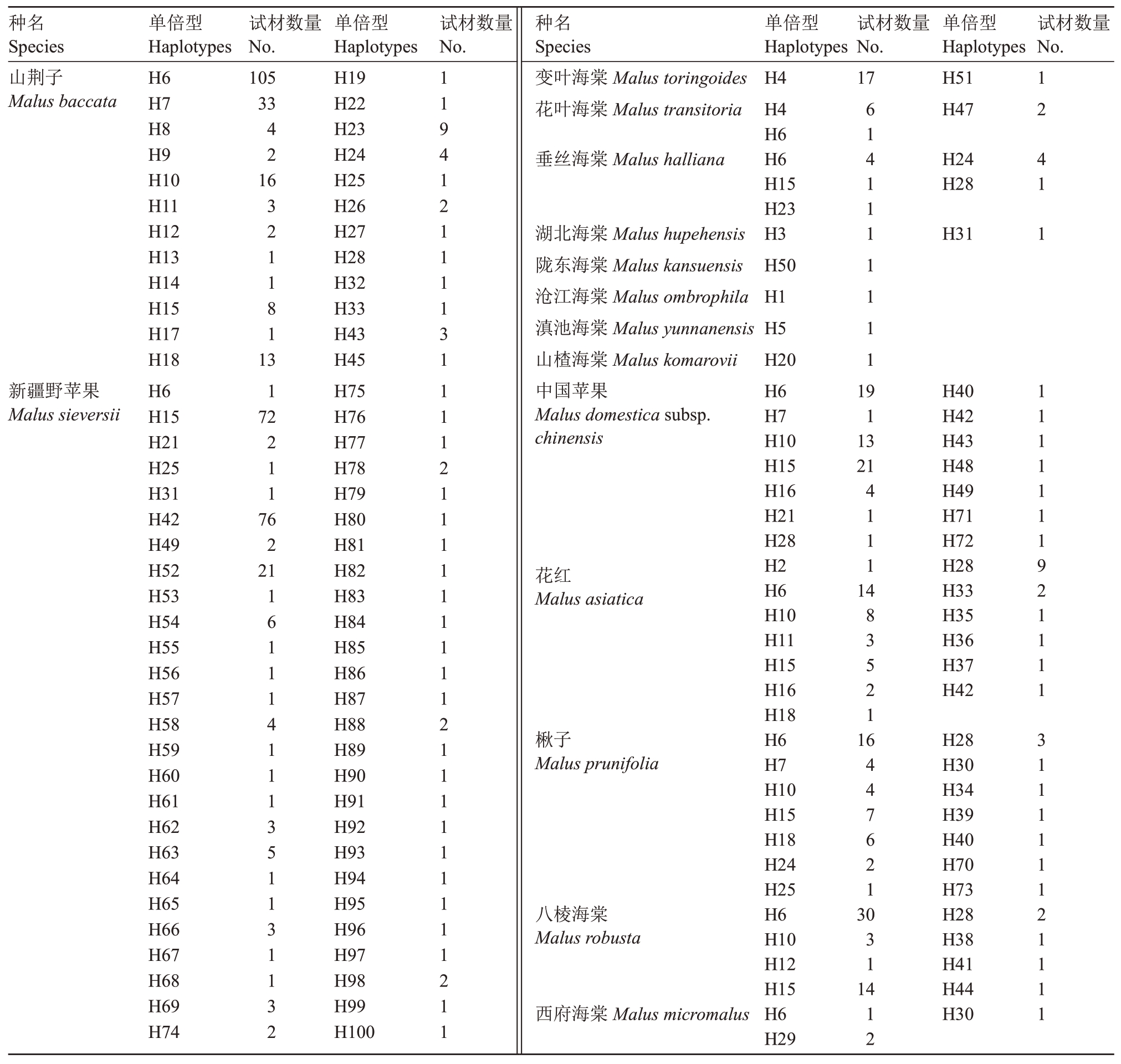
4 个区域合并之后的cpDNA 片段共有单倍型100 个(表5)。新疆野苹果的单倍型最多,242 份新疆野苹果种质有52个单倍型,其中有72份种质集中在单倍型H15,有76份种质集中在单倍型H42,其余新疆野苹果分散在其余的50 个单倍型中。山荆子有24 个单倍型,其中有105 份山荆子集中在单倍型H6中,有10份种质落在单倍型H10中,还有13份种质在单倍型H18 中。变叶海棠主要集中在单倍型H4中,花叶海棠也主要集中在H4中,垂丝海棠主要集中在H6 和H24,湖北海棠的2 份种质中有1 份独享单倍型H3,另外一份属单倍型H31,中国苹果主要集中在单倍型H6、H10和H15,花红以H6中最多,其余分散在12个单倍型中;楸子以H6中最多,其余种质分散在13 个单倍型中;八棱海棠共有8 个单倍型,以H6中种质最多;西府海棠4份种质分成了3个单倍型。陇东海棠、滇池海棠、沧江海棠和山楂海棠均分别独享一个单倍型。
表5 苹果属植物15 个种的单倍型
Table 5 The haplotypes of 15 spcecies of Malus Mill.in China
?
基于苹果属15 个种4 个叶绿体DNA 基因间区合并区域的单倍型,构建其单倍型的关联图(图2)。H和mv分别表示单倍型和中介矢量位点,单倍型网络图的主干(Tarso)由23个中介矢量位点和20个单倍型组成,23个中介矢量位点仍然为缺失单倍型。位于网络图中心区域的单倍型为mv8、mv9、mv16、mv17 和mv18,均为缺失单倍型,其向三个方向扩散,其一是由mv17 向外扩散至H6,并形成由H6、H43、H45和mv11组成的闭合环;其二是由mv8逐步经过单倍型H21、H89、mv6,并形成由mv1、mv4、mv6 和H87 组成的闭合环;其三是由mv16 分化为mv18 和H70,mv18 和H70 的亲缘关系较远,mv18形成2个分支,H70也有2个分支,并在其中一个分支的末端呈星状辐射。
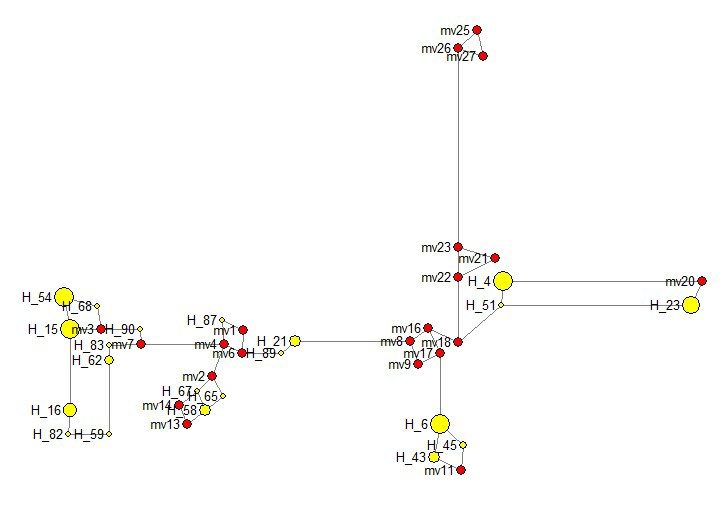
图2 苹果属15 个种基于4 个叶绿体DNA 基因间区合并区域的单倍型中介邻接网络主干图
Fig.2 Tarso of Median-Joining network(MJ)for cpDNA haplotypes of 15 spcecies of Malus Mill.based on four combined cpDNA intergenic regions
3 讨 论
3.1 15种苹果属植物叶绿体基因的遗传多样性
15 种苹果属植物4 个cpDNA 基因间区的核苷酸多样性和单倍型多样性具有种间差异。其中trnH-psbA(Pi=0.034 09,Hd=0.808)的核苷酸多样性和单倍型多样性均为最高,两者最低的区域均为5’trnL-trnF(Pi=0.000 78,Hd=0.577)。15种722份苹果属植物共有579 个突变位点,突变类型为单碱基突变和插入/缺失突变,其中插入/缺失突变位点为528个,占到全部位点突变的91.2%。插入/缺失的发生频率是单碱基突变的10倍以上,插入/缺失是苹果属叶绿体基因组进化的主要动力,这与Clegg等[11]的理论相一致。
中国原产苹果属植物cpDNA 具有高水平的遗传多样性。供试722份种质的4个cpDNA基因间区的遗传多样性(Hd=0.879,Pi=0.008 52),明显高于170 个物种的平均数值(0.670~0.826)[43]。新收集中国原产苹果属植物的遗传多样性水平较高,对于提高国家果树种质兴城梨、苹果资源圃的苹果属植物的遗传多样性水平非常有效。根据各种的居群遗传多样性水平和遗传结构,可以指导进一步的苹果属植物考察和收集工作。
3.2 15种苹果属植物的遗传分化
15种苹果属植物在4个cpDNA区域的Tajima’s D 检验全部为负值,组合后的Tajima’s D 值为-1.503 16,在p >0.10 检测水平上不显著。苹果属植物4个cpDNA区域整体符合中性进化模型,在进化过程中有明显的种群扩张,其遗传进化以种群内突变或者随机漂变为主。
按照种划分的15 个种群以种群内遗传变异为主,种群间与种群内的遗传变异值比较接近,这与高源等[44]利用SSR分子标记方法对苹果属植物研究取得的结果相一致。新疆野苹果与变叶海棠间的遗传分化系数最高(0.84),远远高于高度遗传分化的界定值,基因交流甚少;而楸子与花红间的遗传分化系数最低(0.014),无遗传分化。无遗传分化的类群中以栽培种间为主。究其原因,以代表种花红和楸子为例,其在中国栽培历史久远,并在栽培过程中衍生诸多类型。花红适应范围广,除珠江流域和黑龙江流域诸省栽培较少外,其他如新疆、内蒙古、山西、陕西和河北等15省、自治区的果树志或文献上都有相关描述;而楸子人为分布极广,在中国的华北、西北和华中等地均有栽培[1]。通过野外考察发现花红与楸子的收集区域一致[4],无地理环境隔离。苹果属植物种间基本不存在生殖隔离,依靠不同起源地及不同分布地区之间的环境隔离保持种性,由于某些因素的改变,一旦环境隔离被打破,极易发生种间杂交,产生新的物种或新的类型,沙果和海棠果等即是由此产生的[2]。因此,多数栽培种的人工栽培和无地理隔离可能是遗传分化不明显的重要原因。野生种群间的遗传分化多为高度遗传分化,野生种起源地或分布区域之间的环境隔离难以轻易打破,各种之间种性保持明显,其遗传多样性较高,与野外收集野生种群的表型鉴定结果相符[45-46]。在无遗传分化和中度遗传分化的各种群组合中,除山荆子和垂丝海棠外,其余全部为栽培种,相比于野生种、栽培种之间基因交流更为频繁,山荆子和垂丝海棠在演化过程中起到了一定的作用。这与Duan 等[32]得出的山荆子可能参与了栽培苹果驯化过程的结论相一致。
NJ 聚类是基于最小进化的原则进行的聚类[47],相较于主要基于最小距离原则进行的聚类,当群体突变率变化时,NJ 法构建系统树的准确性高于UPGMA 法[48]。根据NJ 聚类结果,主要分布于云南和四川横断山区的变叶海棠、湖北海棠、花叶海棠和垂丝海棠归为同一组,山荆子与楸子、八棱海棠、西府海棠和花红归为一组,中国苹果与新疆野苹果归为一组,滇池海棠、沧江海棠和陇东海棠归为一组,而山楂海棠独成一组,与其他种的遗传距离均较远。苹果属各种的分类归属与种属关系和地理分布明显相关,这与张春雨等[49-50]对苹果属植物遗传关系的研究结论相同。
3.3 15种苹果属植物的遗传结构
供试种质4个cpDNA 区域合并后,以新疆野苹果单倍型最多,占所有单倍型的一半以上,其中以单倍型H15 和H42 拥有的新疆野苹果数量最多;其次是山荆子,有一半种质集中在H6中。山荆子是苹果属植物中分布最广、变异更为多样的类群[51];而新疆野苹果属第三纪孑遗植物[52],种质类型极为丰富[53]。两者为苹果属植物的发展演化贡献了最多的单倍型。含中国苹果种质数量最多的单倍型是H6、H10 和H15,与含有山荆子和新疆野苹果数量最多的单倍型相一致;而含有花红、楸子、八棱海棠种质数量最多的单倍型均为H6,与含有山荆子种质数量最多的单倍型相一致。栽培种与新疆野苹果和山荆子密切相关。中国苹果、花红、楸子和八棱海棠的部分单倍型晚于部分新疆野苹果的单倍型,由新疆野苹果发展演化而来。中国本土的栽培苹果起源于新疆野苹果[1],并通过现代的测序技术手段证明与新疆野苹果有共祖关系的塞威士苹果是现代栽培苹果(“西洋苹果”)的祖先[19,32],本研究证明了苹果属栽培种部分类型的起源演化与新疆野苹果有着一定的关系。
苹果属的多个野生种独占单倍型,将是野外考察收集的重点。2份湖北海棠在网络图中各独享一个单倍型。陇东海棠、滇池海棠、沧江海棠和山楂海棠各独享一个单倍型。湖北海棠、陇东海棠、滇池海棠和沧江海棠主要在川滇黔藏分布区[2],因此该区域将是野生种的重点考察收集区域。山楂海棠仅在中国长白山自然保护区海拔1 100~1 300 m 的山地有少量分布,为国家二级重点保护濒危种[54],具有苹果属重要的单倍型,应加强保护。变叶海棠和花叶海棠主要存在于同一个单倍型H4 中,两者表型相似[1],变叶海棠种群是变叶海棠与陇东海棠和花叶海棠3个种的杂交复合体[2],因此两者出现在同一个单倍型中并非偶然。变叶海棠在川滇黔藏分布区和青海山区均有分布,而花叶海棠主要在青海山区,因此青海山区将是另一个需要集中考察收集的区域。
4 结 论
苹果属植物不同种具有不同的演化路线,各个种间以及种内起源演化关系错综复杂。山荆子和新疆野苹果的部分种质占据较为古老的单倍型,并在发展演化过程中经历过种群扩张。新疆野苹果与栽培种以及山荆子之间有基因交流。新疆野苹果和山荆子仍是未来苹果属植物种质资源考察收集的重点,中国苹果、花红、楸子和八棱海棠的收集力度需要进一步加大。独占单倍型的野生种主要分布区域将是野外考察的重点区域。
[1] 李育农.苹果属植物种质资源研究[M].北京:中国农业出版社,2001:65-67.LI Yunong. Researches of germplasm resources of Malus Mill.[M].Beijing:China Agricultural Press,2001:65-67.
[2] 张新忠,王忆,韩振海.我国苹果属(Malus Mill.)野生资源研究利用的现状分析[J].中国农业科技导报,2010,12(3):8-15.ZHANG Xinzhong,WANG Yi,HAN Zhenhai.Analysis on present research and utilization status of wild apple germplasm resources (Malus Mill.) in China[J]. Journal of Agricultural Science and Technology,2010,12(3):8-15.
[3] 王昆,刘凤之,高源,王大江,龚欣,刘立军.中国苹果野生种自然地理分布、多型性及利用价值[J]. 植物遗传资源学报,2013,14(6):1013-1019.WANG Kun,LIU Fengzhi,GAO Yuan,WANG Dajiang,GONG Xin,LIU Lijun. The natural distribution,diversity,and utilization of wild apple species in China[J]. Journal of Plant Genetic Resources,2013,14(6):1013-1019.
[4] 王大江,王昆,高源,赵继荣,刘立军,龚欣,李连文.我国苹果属资源现代分布调查初报[J]. 植物遗传资源学报,2017,18(6):1116-1124.WANG Dajiang,WANG Kun,GAO Yuan,ZHAO Jirong,LIU Lijun,GONG Xin,LI Lianwen. Preliminary investigation of modern distribution of Malus resources in China[J]. Journal of Plant Genetic Resources,2017,18(6):1116-1124.
[5] 尚占环,姚爱兴.生物遗传多样性研究方法及其保护措施[J].宁夏农学院学报,2002,23(1):66-69.SHANG Zhanhuan,YAO Aixing.Biological genetic diversity research methods and their protective measures[J]. Journal of Ningxia Agricultural College,2002,23(1):66-69.
[6] BYRNE M,MACDONALD B. Phylogeography and conservation of three oil mallee taxa,Eucalyptus kochii ssp. kochii,ssp.plenissima and E. horistes[J]. Australian Journal of Botany,2000,48(3):305-312.
[7] 陈国庆,黄红文,葛学军.濒危植物矮沙冬青的等位酶多样性及居群分化[J].武汉植物学研究,2005,23(2):131-137.CHEN Guoqing,HUANG Hongwen,GE Xuejun. Allozyme diversity and population differentiation in an endangered plant,Ammopiptanthus nanus(Leguminosae)[J].Journal of Wuhan Botanical Research,2005,23(2):131-137.
[8] 潘丽芹,季华,陈龙清.荷叶铁线蕨自然居群的遗传多样性研究[J].生物多样性,2005,13(2):122-129.PAN Liqin,JI Hua,CHEN Longqing. Genetic diversity of the natural populations of Adiantum reniforme var.sinense[J].Biodiversity Science,2005,13(2):122-129.
[9] BELLUSCI F,PALERMO A M,PELLEGRINO G,MUSACCHIO A. Genetic diversity and spatial structure in the rare,endemic orophyte Campanula pseudostenocodon Lac.(Apennines,Italy),as inferred from nuclear and plastid variation[J]. Plant Biosystems,2008,142(1):24-29.
[10] SHAH A,LI D Z,GAO L M,LI H T,MOLLER M. Genetic diversity within and among populations of the endangered species Taxus fauna (Taxaceae) from Pakistan and implications for its conservation[J].Biochemical Systematics and Ecology,2008,36(3):183-193.
[11] CLEGG M T,BRANDON S G,LEARN G H,MORTON B R.Rates and patterns of chloroplast DNA evolution[J]. Proceedings of the National Academy of Sciences of the United States of America,1994,91:6795-6801.
[12] PALMER J D,JANSEN R K,MICHAELS H J,CHASE M W,MANHART J R. Chloroplast DNA variation and plant phylogeny[J]. Annals of the Missouri Botanical Garden,1988,75(4):1180-1206.
[13] 倪梁红,赵志礼,米玛.药用植物叶绿体基因组研究进展[J].中药材,2015,38(9):1990-1994.NI Lianghong,ZHAO Zhili,MI Ma. Advances in research on chloroplast genome of medicinal plants[J]. Journal of Chinese Medicinal Materials,2015,38(9):1990-1994.
[14] 李宏韬,赵淑青,赵彦修,张慧. 叶绿体基因工程简介[J]. 遗传,2003,25(4):495-498.LI Hongtao,ZHAO Shuqing,ZHAO Yanxiu,ZHANG Hui. The introduction of chloroplast gene engineering[J].Hereditas,2003,25(4):495-498.
[15] 张韵洁,李德铢.叶绿体系统发育基因组学的研究进展[J].植物分类与资源学报,2011,33(4):365-375.ZHANG Yunjie,LI Dezhu. Advances in phylogenomics based on complete chloroplast genomes[J]. Plant Diversity and Resources,2011,33(4):365-375.
[16] BADENES M L,PARFITT D E. Phylogenetic relationship of cultivated Prunus species from an analysis of chloroplast DNA variation[J]. Theoretical & Applied Genetics,1995,90(7/8):1035-1041.
[17] WEN J,BERGGERN S T,LEE C H,ICKERT-BOND S M,YI T S,YOO K,XIE L,SHAW J,POTTER D. Phylogenetic inferences in Prunus (Rosaceae) using chloroplast ndh F and nuclear ribosomal ITS sequences[J]. Journal of Systematics & Evolution,2008,46(3):322-332.
[18] 付涛,王志龙,钱萍仙,李文,袁冬明,严春风.高等植物DNA条形码最新研究进展及其应用[J].核农学报,2016,30(5),887-896.FU Tao,WANG Zhilong,QIAN Pingxian,LI Wen,YUAN Dongming,YAN Chunfeng.The latest research progress and application of the DNA barcode in higher plants[J].Journal of Nuclear Agricultural Sciences,2016,30(5):887-896.
[19] VELASO R,ZHARKIKH A,AFFOURTIT J,……,GOREMYKIN V.The genome of the domesticated apple (Malus × domestica Borkh.)[J].Nature Genetics,2010,4:833-839.
[20] DACCORD N,CELTON J M,LINSMITH G,……,BUCHER E. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development[J]. Nature Genetics,2017,49(7):1099-1108.
[21] ZHANG L Y,JIANG H,HAN X L,LI J J,GAO Y,RICHARDS C M,ZHANG C X,TIAN Y,LIU G M,GUL H,WANG D J,TIAN Y,YANG C X,MENG M H,YUAN G P,KANG G D,WU Y L,WANG K,ZHANG H T,WANG D P,CONG P H.A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour[J]. Nature Communication,2019,10(1):1494.
[22] SAVOLAINEN V,CORBAZ R,MONCOUSIN C,SPICHIGER R,MANEN J F.Chloroplast DNA variation and parentage analysis in 55 apples [J]. Theoretical & Applied Genetics,1995,90(7/8):1138-1141.
[23] ROBINSON J,HARRIS S A,JUNIPER B E. Taxonomy of the genus Malus Mill. (Rosaceae) with emphasis on the cultivated apple,Malus domestica Borkh.[J]. Plant Systematics & Evolution,2001,226(1/2):35-58.
[24] COART E,GLABEKE S,LOOSE M,LARSEN A S,ROLDANRUIZ I. Chloroplast diversity in the genus Malus: New insights into the relationship between the European wild apple [Malus sylvestris(L.)Mill.]and the domesticated apple(Malus domestica Borkh.)[J].Molecular Ecology,2006,15:2171-2182.
[25] 丁芳兵.湖北海棠(Malus hupehensis)及近缘种的matK 和ITS序列分析[D].南京:南京林业大学,2012.DING Fangbing. Sequence analysis of matK and ITS between Malus hupehensis and its relative species[D]. Nanjing: Nanjing Forestry University,2012.
[26] 朱元娣,曹敏格,许正,王昆,张文.基于ITS 和matK 序列探讨新疆野苹果与中国苹果的系统演化关系[J]. 园艺学报,2014,41(2):227-239.ZHU Yuandi,CAO Minge,XU Zheng,WANG Kun,ZHANG Wen. Phylogenetic relationship between Xinjiang wild apple(Malus sieversii Roem.) and Chinese apple (Malus × domestica subsp. chinesnsis) based on ITS and matK sequences[J]. Acta Horticulturae Sinica,2014,41(2):227-239.
[27] 徐榕雪.基于分子标记技术的部分苹果属植物分类地位及亲缘关系研究[D].南京:南京林业大学,2018.XU Rongxue. Taxonomic status and phylogenetic relationships of some Malus plants based on molecular marker technology[D].Nanjing:Nanjing Forestry University,2018.
[28] 李慧峰.泰沂山区苹果属植物系统学研究[D].沈阳:沈阳农业大学,2012.LI Huifeng. Studies on the taxonomy of the genus Malus Mill.(Rosaceae) of Taiyi-Mountains[D]. Shenyang: Shenyang Agricultural University,2012.
[29] BARAKET G,OLFA S,KHALED C,MESSAOUD M,MOHAMED M,MOKHTAR T,AMEL S. Chloroplast DNA analysis in Tunisian fig cultivars (Ficus carica L.): Sequence variations of the trnl-trnf intergenic spacer[J].Biochemical Systematics and Ecology,2009,36(11):828-835.
[30] POTTER D,LUBY J J,HARRISON R E.Phylogenetic relationships among species of Fragaria (Rosaceae) inferred from noncoding nuclear and chloroplast DNA sequences[J]. Systematic Botany,2009,25(2):337-348.
[31] GHADA B,AHMED B A,KHALED C,OLFA S,MESSAOUD M,MOKHTAR T,AMEL S. Molecular evolution of chloroplast DNA in fig (Ficus carica L.): Footprints of sweep selection and recent expansion[J]. Biochemical Systematics and Ecology,2010,38(4):563-575.
[32] DUAN N B,BAI Y,SUN H H,……,CHEN X S. Genome resequencing reveals the history of apple and supports a two-stage for fruit enlargement[J].Nature Communication,2017,8:249.
[33] NYBOM H,GARKAVA-GUSTAVSSON L. Apple gene banksfor breeding,research or public entertainment[J].Acta Horticulturae,2009,814:71-76.
[34] SHAW J,LICKEY E B,BECK J T,FARMER S B,LIU W S,MILLER J,SIRIPUN K C,WINDER C T,SCHILLING E E,SMALL R L.The tortoise and the hare Ⅱ:Relative utility of 21 noncoding chloroplast and sequences for phylogenetic analysis[J].American Journal of Botany,2005,921(1):142-166.
[35] VOLK G M,HENK A D,BALDO A,FAZIO G,CHAO C T,RICHARDS C M. Chloroplast heterogeneity and historical admixture within the genus Malus[J].American Journal of Botany,2015,102(7):1198-1208.
[36] 高源,王大江,王昆,丛佩华,张彩霞,李连文,朴继成.新疆野苹果叶绿体DNA 变异与遗传进化分析[J].植物遗传资源学报,2020,21(3):79-87.GAO Yuan,WANG Dajiang,WANG Kun,CONG Peihua,ZHANG Caixia,LI Lianwen,PIAO Jicheng. Chloroplast DNA variation and genetic evolution of Malus sieversii M. Roem[J].Journal of Plant Genetic Resources,2020,21(3):79-87.
[37] KUMAR S,STECHER G,TAMURA K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets[J].Molecular Biology Evolution,2016,33(7):1870-1874.
[38] NEI M.Genetic distance between populations[J].American Naturalist,1972,106(949):283-292.
[39] NEI M,TAJIMA F,TATENO Y.Accuracy of estimated phylogenetic tree from molecular data[J]. Journal of Molecular Evolution,1983,19:153-170.
[40] SAITO N,NEI M.The Neighbor-joining method:A new method for reconstructing phylogenetic trees[J]. Molecular Biology and Evolution,1987,4(4):406-425.
[41] LIBRADO P,ROZAS J. DnaSP v5:A software for comprehensive analysis of DNA polymorphism data[J]. Bioinformatics,2009,25:1451-1452.
[42] EXCOFFIER L,LISCHER H E.Arlequin Suite ver 3.5:A new series of programs to perform population genetics analyses under linux and windows[J]. Molecular Ecology Resources,2010,10:564-567.
[43] PETIT R,AGUINAGALDE I,DE BEAULIEU J L,……,VENDRAMN G G. Glacial refugia: Hotspots but not melting pots of genetic diversity[J].Science,2003,300:1563-1565.
[44] 高源,王昆,王大江,赵继荣,张彩霞,丛佩华,刘立军,李连文,朴继成.7 个来源地区山荆子的遗传多样性与群体结构分析[J].中国农业科学,2018,51(19):3766-3777.GAO Yuan,WANG Kun,WANG Dajiang,ZHAO Jirong,ZHANG Caixia,CONG Peihua,LIU Lijun,LI Lianwen,PIAO Jicheng. The genetic diversity and population structure analysis on Malus baccata (L.) Borkh. from 7 sources[J]. Scientia Agricultura Sinica,2018,51(19):3766-3777.
[45] 杨锋,刘志,伊凯,刘延杰,王强,孙建设.东北山定子(Malus baccata(L.)Borkh.)野生居群表型遗传多样性分析及生态地理分布研究[J].植物遗传资源学报,2015,16(3):490-496.YANG Feng,LIU Zhi,YI Kai,LIU Yanjie,WANG Qiang,SUN Jianshe.Studies on geographical regions and analysis on genetic diversity of phenotypic of natural population of Malus baccata(L.) Borkh. in Northeast of China[J]. Journal of Plant Genetic Resources,2015,16(3):490-496.
[46] 刁永强,许正,闫鹏,陈淑英,张学超,刘君.新疆野苹果资源类型表型性状鉴定及优异资源的初步筛选[J].经济林研究,2019,37(1):17-24.DIAO Yongqiang,XU Zheng,YAN Peng,CHEN Shuying,ZHANG Xuechao,LIU Jun.Phenotypic characteristic identification and preliminary selection of excellent resources in Malus sieversii resource types[J].Non-wood Forest Research,2019,37(1):17-24.
[47] NEI M,KUMAR S. Molecula revolution and phylogenetics[M].New York:Oxford University Press,2000:269.
[48] TAKEZAKI N,NEI M. Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA[J]. Genetics,1996,144(1):389-399.
[49] ZHANG C Y,CHEN X S,HE T M,LIU X L,FENG T,YUAN Z H. Genetic structure of Malus sieversii population from Xinjiang,China,revealed by SSR markers[J]. Journal of Genetics and Genomics,2007,34(10):947-955.
[50] 张春雨,陈学森,林群,苑兆和,张红,张小燕,刘崇祺,吴传金. 新疆野苹果群体遗传结构和遗传多样性的SRAP 分析[J].园艺学报,2009,36(1):7-14.ZHANG Chunyu,CHEN Xuesen,LIN Qun,YUAN Zhaohe,ZHANG Hong,ZHANG Xiaoyan,LIU Chongqi,WU Chuanjin.SRAP markers for population genetic structure and genetic diversity in Malus sieversii from Xinjiang,China[J]. Acta Horticulturae Sinica,2009,36(1):7-14.
[51] 杜学梅,杨廷桢,高敬东,王骞,蔡华成,李春燕,弓桂花.中国野生山定子的自然分布及利用研究现状[J].中国农学通报,2017,33(5):24-28.DU Xuemei,YANG Tingzhen,GAO Jingdong,WANG Qian,CAI Huacheng,LI Chunyan,GONG Guihua.Wild Malus baccata (L.) Borkh. in China: Natural distribution,utilization and study status[J]. Chinese Agricultural Science Bulletin,2017,33(5):24-28.
[52] 张新时.伊犁野果林的生态地理特征和群落学问题[J].植物学报,1973,15(2):239-253.ZHANG Xinshi. On the eco-geographical characters and the problems of classification of the wild fruit-tree forest in the ili valley of Sinkiang[J]. Acta Botanica Sinica,1973,15(2): 239-253.
[53] 林培钧,崔乃然.天山野果林资源:伊犁野果林综合研究[M].北京:中国林业出版社,2000.LIN Peijun,CUI Nairan. Wild fruit forest resources in Tianshan Mountains: Comprehensive study on Yili wild fruit forest[M].Beijing:China Forestry Press,2000.
[54] 汪松,解焱.中国物种红色名录(第1 卷)[M].北京:高等教育出版社,2004:304-464.WANG Song,XIE Yan. Red list of Chinese species (Volume 1)[M].Beijing:Higher Education Press,2004:304-464.